A number of single nucleotide polymorphisms (SNPs) in the genes IKZF1, ARID5B, CEBPE and CDKN2A have been implicated in the propensity to develop acute lymphoblastic leukemia (ALL) in childhood (< 15 years of age)1–4 but since the genetic background of adult and childhood ALL is different, it is not clear if these are also risk factors for adult-onset ALL. We analyzed seven SNPs (two in IKZF1: rs11978267, rs4132601; three in ARID5B: rs10821936, rs10994982, rs7089424; and one each in CEBPE: rs2239633 and CDKN2A: rs3731217) which were reported to confer an increased risk for childhood ALL in 322 immunologically and genetically well-characterized adult or adolescent ALL patient samples. All samples were obtained from BCR-ABL-negative patients at time points when they were in molecular remission, thus excluding mutations acquired during onset of disease and randomly selected from a large collection of archived samples obtained during diagnostic procedures in the framework of the German Multicenter ALL Study Group (GMALL) trial 07/2003 (Clinicaltrials.gov identifier:00198991). This trial has been approved by various local and central ethics committees and our study complied with the principles set forth in the Declaration of Helsinki. The median age of the patient cohort was 38 years (range 16–76 years), and 58.4% of the patients were male. Flow cytometry immunophenotyping and molecular genetic analyses for BCR-ABL, MLL-AF4, MLL-ENL, TCF3-PBX1, and ETV6-RUNX1 were performed at primary diagnosis as described previously.5,6 Assessment of minimal residual disease was made as described.7,8 BCR-ABL-positive samples and mature B-ALL (sIg+, or MYC-IGH+) samples were excluded. Twenty-six patients were MLL-AF4-positive, 5 MLL-ENL-positive, 2 TCF3-PBX1-positive, and 2 ETV6-RUNX1-positive. Patient samples were selected in such a way as to match the distribution of immunophenotypes observed in the last ten years in the GMALL study group, thereby taking into consideration that BCR-ABL-positive patients had been excluded (Table 1).
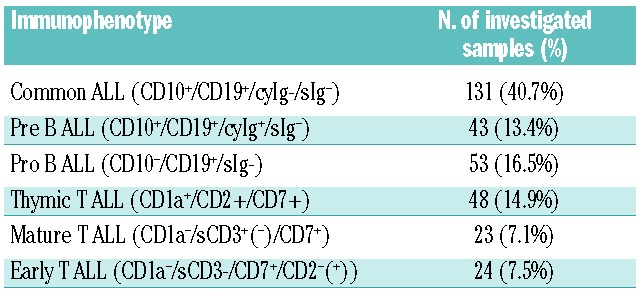
Table 1.
Immunophenotypes of the investigated patients. BCR-ABL-positive patients (25% of the total) were excluded.
The control group comprised a total of 1516 healthy individuals of German origin collected through 2004 at the Institute of Transfusion Medicine in Mannheim, Germany (genotyped for rs4132601, rs7089424, rs2239633, rs3731217).4,9 Since no German control data were available for ARID5B rs10994982, this SNP was compared to a cohort comprising 14,311 individuals of European descent without ALL, collected by the Wellcome Trust Case Control Consortium (WTCCC); these results are only reported for completeness and are not listed in Table 2.10 Details of the control groups have been described previously.2,4 The SNP Genotyping Assays (Applied Biosystems, Darmstadt/Germany, Cat # 4351379) were used with the TaqMan Genotyping Master Mix (Applied Biosystems, Cat # 4371355) on a RotorGene cycler (QIAGEN, Hilden/Germany) using basically the conditions recommended by the supplier.
Table 2.
SNP genotypes in patients and controls. Absolute and relative frequencies of different SNP genotypes in patients (entire cohort, B-lineage patients, T-lineage patients) and controls, with corresponding odds ratios and P values. The following genotypes were risk factors for B-lineage ALL: rs4132601 ‘G/T’ and ‘G/G’, rs7089424 ‘G/G’, rs2239633 ‘C/C’. None of the genotypes was a risk factor for T-lineage ALL.
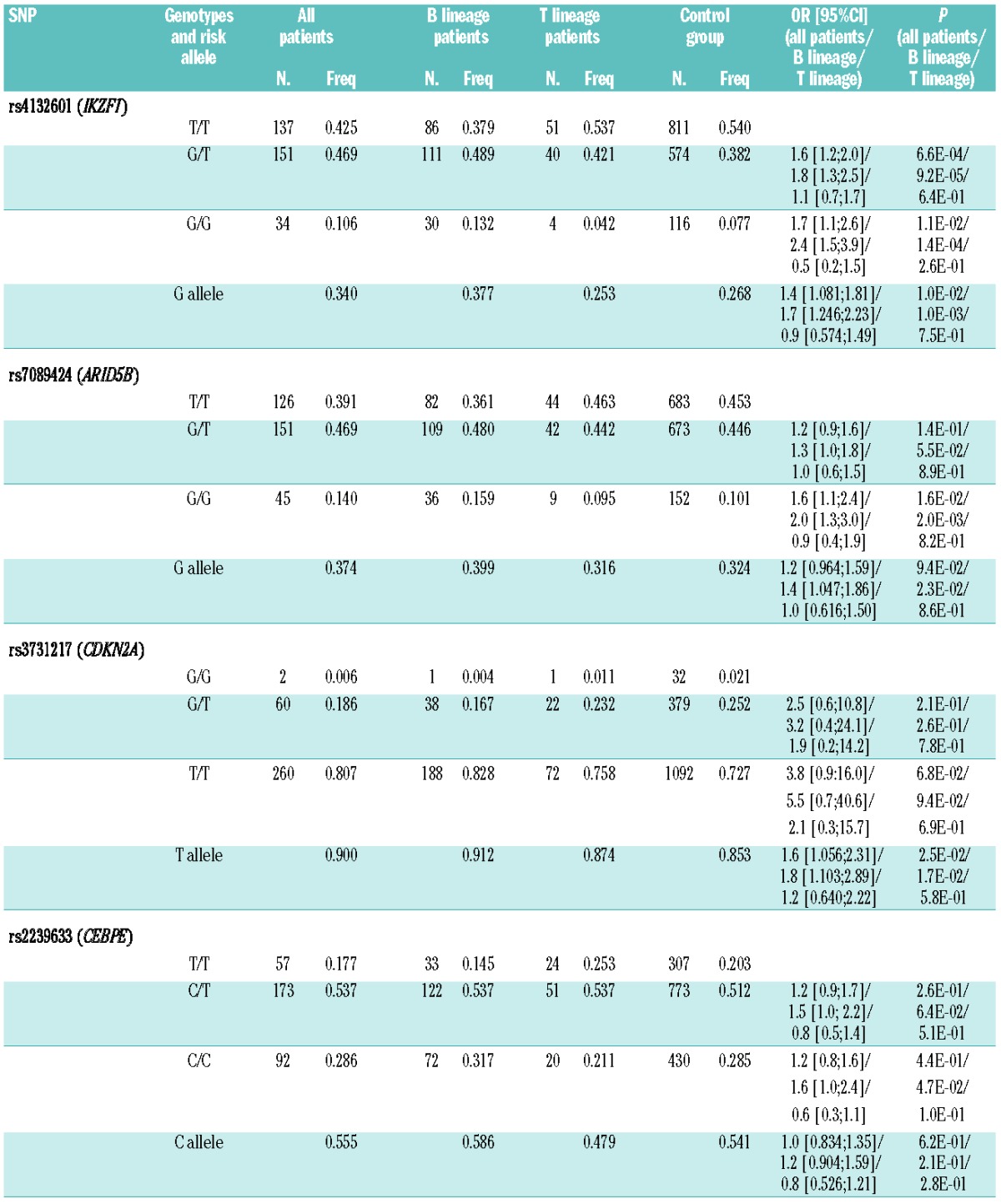
Some SNPs in the same gene were very closely correlated suggesting that they belonged to gene haplotypes. Linkage disequilibrium (LD) analysis revealed the following correlations: IKZF1 rs4132601–rs11978267: D′=0.9931, r2=0.9794; ARID5B rs7089424–rs10821936: D′=0.9731, r2=0.9407, ARID5B rs10821936–rs10994982: D′=0.9305, r2=0.5017; and ARID5B rs7089424–rs10994982: D′=0.9009, r2=0.4735 (see Online Supplementary Appendix). Therefore, rs11978267 and rs10821936 were not considered in further analysis since they added nothing new.
The observed allele frequencies are summarized in Table 2. All presumed “risk alleles” were more frequent in B-lineage ALL patients as compared to non-ALL controls while there was no difference in frequencies in T-lineage ALL. For the following genotypes, the differences were statistically significant in B-lineage ALL patients: IKZF1 rs4132601 (‘G/T’, P=9.2E-05; ‘G/G’, P=1.4E-04), ARID5B rs7089424 (‘G/G’, P=2.0E-03), and CEBPE rs2239633 (‘C/C’, P=4.7E-02). Four genotype differences narrowly missed the significance threshold: ARID5B rs7089424 ‘G/T’ (P=0.055), CDKN2A rs3731217 ‘T/T’ (P=0.094), and CEBPE rs2239633 (‘C/T’, P=0.064). Male patients had lower overall frequencies of all risk genotypes than female patients which was largely but not entirely explained by the higher percentage of T-lineage ALL in males.11 Females with MLL-negative B-lineage ALL appeared at particular high risk with ORs of 4.74 (P=6.75E-07; 2.567;8.768) for rs4132601 ‘G/G’ and 1.87 (P=1.5E-02; 1.130;3.086) for rs4132601 ‘G/T’, but the numbers were small (19 and 37 of a total of 84 MLL-negative female B-lineage ALL patients with ‘G/G’ and ‘G/T’ genotype) and, therefore, this finding needs to be validated in an independent sample set. The previous pediatric studies reported a moderate risk for childhood-onset ALL with ORs of 1.8–2.8 (IKZF1 rs4132601),1,4 1.6–3.2 (ARID5B rs7089424),1,4 1.1–1.6 (CEBPE rs22396339),1,4 and 1.4 (CDKN2A rs3731217).3 In our adult cohort, we observed comparable ORs for rs4132601 (1.5–2.0) and rs22396339 (1.6) but the ORs for rs7089424 were lower and only significant for the bialleleic SNP (Table 2). rs10994982 showed no difference as compared to the WTCCC control group.
In summary, the ORs were similar for pediatric and adult ALL, and thus the risk conferred, appears to be lifelong and not limited to childhood. The lower ORs for ARID5B in adult ALL might be explained by the lower frequency of hyperdiploid B-lineage ALL in adults.1 To address the question as to how these intronic SNPs contribute to leukemogenesis, we investigated CD34+ stem cells for cryptic splice variants caused by the IKZF1 and ARID5B SNPs but no splice variants were detected.
Recently, Peyrouze et al. reported their findings when investigating 150 adult ALL patients for almost the same six SNPs in the context of the GRAALL study group.12 No association of any of the investigated SNPs was found but the study had some limitations, most notably a relatively small patient number and an apparently heterogeneous (including BCR-ABL+) patient population and control group. Since these genetic risk factors confer only a moderate risk for adult- or childhood-onset ALL, efforts have been made to set up an international consortium to also identify low-penetrance susceptibility alleles.13 The identification and analysis of such low-penetrance variants may further facilitate the pathogenetic understanding of pediatric and adult ALL. Last, but not least, it has to be kept in mind that ethnic background may influence the risk and thus the hitherto reported risk factors may not be valid for all regions of the world.14
Acknowledgments
The authors thank all involved clinics and patients of the GMALL study group for their support and all involved laboratory technicians for their work. The authors thank Prof. Rajiv Kumar (DKFZ, Heidelberg, Germany) for communicating detailed rs3731217 genotype results of the control group. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk.
Footnotes
Funding: TB was supported by grant 2011-05 from the Alfred & Angelika Gutermuth Foundation, Frankfurt and by grant 12/09f from the German José Carreras Leukemia Foundation. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1006–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Treviño LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1001–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42(6):492–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prasad RB, Hosking FJ, Vijayakrishnan J, Papaemmanuil E, Koehler R, Greaves M, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood. 2010;115(9):1765–7 [DOI] [PubMed] [Google Scholar]
- 5.Burmeister T, Meyer C, Schwartz S, Hofmann J, Molkentin M, Kowarz E, et al. The MLL recombinome of adult CD10-negative B-cell precursor acute lymphoblastic leukemia: results from the GMALL study group. Blood. 2009;113(17):4011–5 [DOI] [PubMed] [Google Scholar]
- 6.Burmeister T, Gökbuget N, Schwartz S, Fischer L, Hubert D, Sindram A, et al. Clinical features and prognostic implications of TCF3-PBX1 and ETV6-RUNX1 in adult acute lymphoblastic leukemia. Haematologica. 2010;95(2):241–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brüggemann M, Raff T, Flohr T, Gökbuget N, Nakao M, Droese J, et al. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood. 2006;107(3):1116–23 [DOI] [PubMed] [Google Scholar]
- 8.Burmeister T, Marschalek R, Schneider B, Meyer C, Gökbuget N, Schwartz S, et al. Monitoring minimal residual disease by quantification of genomic chromosomal breakpoint sequences in acute leukemias with MLL aberrations. Leukemia. 2006;20(3):451–7 [DOI] [PubMed] [Google Scholar]
- 9.Gast A, Bermejo JL, Flohr T, Stanulla M, Burwinkel B, Schrappe M, et al. Folate metabolic gene polymorphisms and childhood acute lymphoblastic leukemia: a case-control study. Leukemia. 2007;21(2):320–5 [DOI] [PubMed] [Google Scholar]
- 10.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Vlierberghe P, Palomero T, Khiabanian H, van der Meulen J, Castillo M, van Roy N, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42(4):338–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peyrouze P, Guihard S, Grardel N, Berthon C, Pottier N, Pigneux A, et al. Genetic polymorphisms in ARID5B, CEBPE, IKZF1 and CDKN2A in relation with risk of acute lymphoblastic leukaemia in adults: a Group for Research on Adult Acute Lymphoblastic Leukaemia (GRAALL) study. [letter]. Br J Haematol 2012;159(5):599–613 [DOI] [PubMed] [Google Scholar]
- 13.Sherborne AL, Hemminki K, Kumar R, Bartram CR, Stanulla M, Schrappe M, et al. Rationale for an international consortium to study inherited genetic susceptibility to childhood acute lymphoblastic leukemia. Haematologica. 2011;96(7):1049–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H, Cheng C, Devidas M, Pei D, Fan Y, Yang W, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J Clin Oncol. 2012;30(7):751–7 [DOI] [PMC free article] [PubMed] [Google Scholar]


