Abstract
Background
Nephronophthisis-associated ciliopathies (NPHP-AC) comprise a group of autosomal recessive cystic kidney diseases that includes nephronophthisis (NPHP), Senior-Loken syndrome (SLS), Joubert syndrome (JBTS), and Meckel-Gruber syndrome (MKS). To date, causative mutations in NPHP-AC have been described for 18 different genes, rendering mutation analysis tedious and expensive. To overcome the broad genetic locus heterogeneity we devised a strategy of DNA pooling with consecutive massively parallel resequencing (MPR).
Methods
In 120 patients with severe NPHP-AC phenotypes we prepared 5 pools of genomic DNA with 24 patients each which were used as templates in order to PCR-amplify all 376 exons of 18 NPHP-AC genes (NPHP1, INVS, NPHP3, NPHP4, IQCB1, CEP290, GLIS2, RPGRIP1L, NEK8, TMEM67, INPP5E, TMEM216, AHI1, ARL13B, CC2D2A, TTC21B, MKS1, and XPNPEP3). PCR products were then subjected to MPR on a Illumina Genome-Analyzer and mutations were subsequently assigned to their respective mutation carrier via CEL I endonuclease-based heteroduplex screening and confirmed by Sanger sequencing.
Results
For proof of principle we used DNA from patients with known mutations and demonstrated the detection of 22 out of 24 different alleles (92% sensitivity). MPR led to the molecular diagnosis in 30/120 patients (25%) and we identified 54 pathogenic mutations (27 novel) in 7 different NPHP-AC genes. Additionally, in 24 patients we only found single heterozygous variants of unknown significance.
Conclusions
The combined approach of DNA pooling followed by MPR strongly facilitates mutation analysis in broadly heterogeneous single-gene disorders. The lack of mutations in 75% of patients in our cohort indicates further extensive heterogeneity in NPHP-AC.
Keywords: Next-generation sequencing, Ciliopathy, Nephronophthisis
INTRODUCTION
Dysfunction of the primary cilium / basal body complex has been implicated in the pathogenesis of NPHP-associated ciliopathies (NPHP-AC) including nephronophthisis (NPHP), Senior Loken syndrome (SLSN), Joubert syndrome (JBTS), and Meckel-Gruber syndrome (MKS).[1] NPHP-AC are rare, genetically heterogeneous, autosomal recessive inherited disorders, which share a broad phenotypic spectrum as a result of ciliary/centrosomal defects in various cell types, e.g. retinal photoreceptors or renal tubular epithelial cells.[2] In NPHP, kidney tubular basement membrane disintegration, tubular atrophy, interstitial fibrosis, and cyst formation results in progressive renal failure during childhood or adolescence. About 15% of patients develop extrarenal manifestations, in particular, progressive retinal dystrophy, referred to as SLSN. In patients with JBTS, midbrain-hindbrain malformations and cerebellar vermis hypoplasia /aplasia results in numerous neurological features including developmental delay, intellectual disability, muscle hypotonia, ataxia, oculomotor apraxia, nystagmus, and irregular breathing patterns in neonates.[3] The most severe manifestation of the NPHP-AC clinical spectrum is seen in fetuses with Meckel-Gruber syndrome, a perinatally lethal ciliopathy, characterized by central nervous system malformations (typically occipital encephalocele), bilateral postaxial hexadactyly, hepatobiliary ductal plate malformation, and multicystic dysplastic kidneys. Mutations in 18 different recessive genes have been identified as the molecular cause in NPHP-AC (Table 1). Twelve genes have been implicated in NPHP and/or SLSN (NPHP1, INVS, NPHP3, NPHP4, IQCB1, CEP290, GLIS2, RPGRIP1L, NEK8, TMEM67, TTC21B, and XPNPEP3).[4–15] Ten are known to cause Joubert syndrome (AHI1, TMEM216, INPP5E, NPHP1, CEP290, RPGRIP1L, TMEM67, ARL13B, CC2D2A, TTC21B).[16–25] Mutations in 5 genes (MKS1, TMEM67, CEP290, RPGRIP1L, CC2D2A, TMEM216) have been shown to cause MKS [12, 17, 26–29]. Multiple allelism within the NPHP-AC phenotypic spectrum has been recurrently reported for many of these genes, especially CEP290, RPGRIP1L, TMEM67, CC2D2A, TTC21B, and TMEM216. For example, hypomorphic missense mutations in the gene TMEM67 (MKS3/NPHP11) are implicated in NPHP with liver fibrosis and JBTS type 6, whereas truncating mutations in TMEM67/MKS3 have been reported in MKS cases with severe developmental and dysplastic phenotypes.[14, 22, 27] The presence of multiple allelism and broad heterogeneity together with extensive phenotypic clinical overlap in patients with NPHP-AC requires extensive mutational analysis efforts in order to identify the underlying molecular etiology. The challenge of analyzing increasing numbers of candidate genes associated with disease in large cohorts of patients can now be met by applying next-generation sequencing technologies. We chose to perform single PCR reactions with well established primer pairs, rather than trying implementing multiplexing, in order to amplify all coding exons (376) of 18 known NPHP-AC disease genes. In order to reduce the calculated number of 45,120 PCR reactions, required to amplify all 376 exons in 120 individuals, we devised a pooling strategy by generating 5 DNA pools derived from 24 individuals each. Identification of the mutation carrying individuals was done by CEL1 endonuclease-based heteroduplex analysis and Sanger sequencing.[30] We demonstrate that the approach of pooling DNA samples in combination with MPR is a robust, economic, and highly effective method for examining larger patient cohorts for mutations, especially in diseases with broad genetic locus heterogeneity.
Table 1.
Genes investigated by massively parallel resequencing in 120 patients with severe NPHP-AC
| Gene | Locus / Protein | Chromosome | Accession # | Exon Count |
Coding Exon Count |
Open reading frame size [bp] |
|---|---|---|---|---|---|---|
| NPHP1 | NPHP1 / Nephrocystin 1 | 2 | NM_000272 | 20 | 20 | 2,202 |
| INVS | NPHP2 / Inversin | 9 | NM_014425 | 17 | 16 | 3,198 |
| NPHP3 | NPHP3 / Nephrocystin 3 | 3 | NM_153240 | 27 | 27 | 3,993 |
| NPHP4 | NPHP4 / Nephroretinin | 1 | NM_015102 | 30 | 29 | 4,281 |
| IQCB1 | NPHP5 / IQ motif containing B1 | 3 | NM_001023570 | 15 | 13 | 1,797 |
| CEP290 | NPHP6 / Centrosomal protein 290kDa | 12 | NM_025114 | 54 | 53 | 7,440 |
| GLIS2 | NPHP7 / GLIS family zinc finger 2 | 16 | NM_032575 | 6 | 6 | 1,575 |
| RPGRIP1L | NPHP8 / RPGRIP1-like | 16 | NM_015272 | 27 | 26 | 3,948 |
| NEK8 | NPHP9 / NIMA related kinase 8 | 17 | NM_178170 | 15 | 15 | 2,079 |
| TMEM67 | NPHP11 / Meckelin | 8 | NM_153704 | 28 | 28 | 2,988 |
| INPP5E | JBTS1 / Inositol polyphosphate-5-phosphatase | 9 | NM_019892 | 10 | 10 | 1,935 |
| TMEM216 | JBTS2 / Transmembrane protein 216 | 11 | AK303687 | 5 | 5 | 438 |
| AHI1 | JBTS3 / Jouberin | 6 | NM_001134831 | 28 | 26 | 3,591 |
| ARL13B | JBTS8 / ADP-ribosylation factor-like 2-like 1 | 3 | NM_182896 | 11 | 10 | 1,287 |
| CC2D2A | JBTS9 / Coiled-coil and C2 domain containing 2A | 4 | NM_001080522 | 37 | 35 | 4,863 |
| TTC21B | Tetratricopeptide repeat domain 21B | 2 | NM_024753 | 29 | 29 | 3,951 |
| MKS1 | MKS1 / Meckel syndrome type 1 | 17 | NM_017777 | 18 | 18 | 1,680 |
| XPNPEP3 | NPHPL1 / X-prolyl aminopeptidase 3 | 22 | NM_022098 | 10 | 10 | 1,524 |
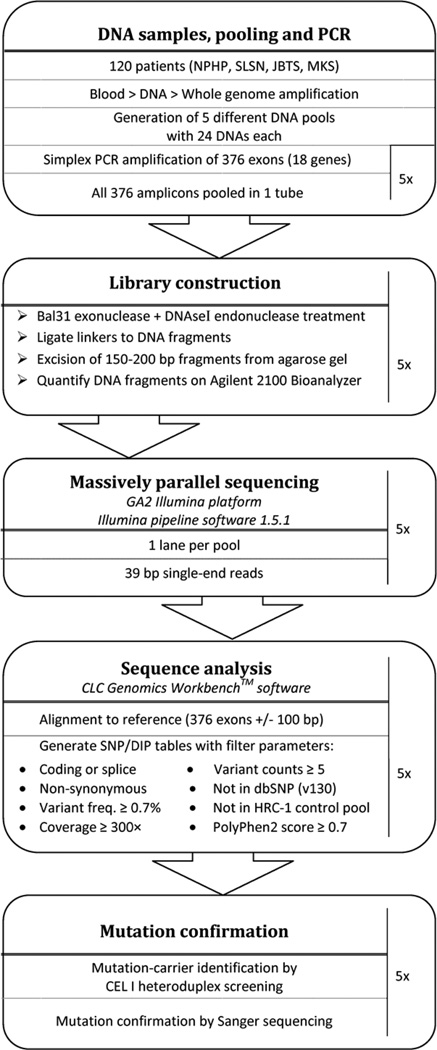
An overview of the various steps undertaken, including DNA pooling, PCR, MPR, and mutation carrier identification is depicted in a flowchart (Figure 1).
Figure 1.
Flowchart illustrating the various steps of the DNA pooling and massive parallel resequencing approach, which was applied in order to perform mutation analysis for 18 genes in 120 patients with nephronophthisis associated ciliopathies.
MATERIALS AND METHODS
Human subjects
We obtained blood samples, pedigrees, and clinical information after receiving informed consent (www.renalgenes.org) from all patients. Approval for experiments on humans was obtained from the University of Michigan Institutional Review Board. The cohort of 120 patients with severe NPHP-AC included 54 (45%) patients diagnosed as JBTS with kidney involvement, 14 (12%) patients with SLSN, 6 (5%) patients with MKS, and 46 (38%) patients with NPHP and early onset of end-stage renal disease before age 7 years. The cohort consisted of 56 (47%) familial cases vs. 64 (53%) sporadic cases. Consanguinity was known to be present in 30 (25%) families. As a first diagnostic step, homozygous deletions of NPHP1 were excluded in all patients by multiplex PCR.[30] Total genome homozygosity mapping was performed in 114 of 120 patients. In patients exhibiting long runs of homozygosity (most likely by descent) at known NPHP-AC loci, exon sequencing for respective genes was carried out and was negative. Ninety-six healthy control DNA samples (Human Random Control DNA panel-1, HRC-1) were obtained from the European Collection of Cell Cultures (ECACC, Salisbury, United Kingdom).
Whole genome amplification and DNA pooling
In order to normalize various DNA samples of dissimilar quality, whole genome amplification (WGA) was performed by individually amplifying 10 ng genomic DNA of 120 different individuals and 96 healthy control samples. Using Phi29 based DNA polymerase strand displacement amplification according to the manufacturer’s instructions (GenomiPhi DNA amplification kit, GE Healthcare). Subsequently, DNA was purified using 96 well spin columns (Rapid 96TM PCR purification system, Marligen-Biosciences). Genomic DNA of 24 individuals was pooled at 2 µg each and diluted to 60 ng/µl. Five pools were generated to represent 120 individuals. As a control, another equimolar DNA pool was generated, by pooling 96 DNA samples derived from healthy individuals of Caucasian origin [Human Random Control DNA Panel-1 (HRC-1)].
PCR amplification
All 376 exons of the genes NPHP1 (20 exons), INVS (16), NPHP3 (28), NPHP4 (30), IQCB1 (15), CEP290 (54), GLIS2 (6), RPGRIP1L (27), NEK8 (15), TMEM67 (28), INPP5E (10), TMEM216 (5), AHI1 (27), ARL13B (10), CC2D2A (37), TTC21B (29), MKS1 (18), and XPNPEP3 (10) were individually amplified using pairs of exon flanking primers (Supplementary Table 1) in each of the DNA pools as the PCR-template. A 12 µl total volume single PCR reaction was set up with 120 ng (2 µl) of pooled DNA derived from 24 different patients (5 ng of each DNA), 2.5 pmol of each forward and reverse primer, and 6 µl HotStar-Taq Polymerase mixture (Qiagen). DNA amplification was performed on a thermal cycler (Mastercycler, Eppendorf) using Thermo-Fast® 96-well plates (ABgene), applying the following touchdown PCR protocol for all PCR reactions: Initial denaturation at 94°C for 15 min, followed by 24 cycles with an annealing temperature decreasing 0.7°C per cycle, starting at 72°C for 30 sec; denaturation at 94°C for 30 sec, and extension at 72°C for 1 min. An additional 20 cycles with fixed annealing temperature were added: 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min with a final extension of 72°C for 10 min. Two µl of each reaction was analyzed by agarose gel-electrophoresis (1.2% agarose, 120V, 1 hour).
Enzymatic modification of PCR products prior to MPR
For each pool, we combined 5 µl of each of the 376 exonic PCR products ranging from 139 bp to 1,236 bp and purified the DNA fragments on 3 separate columns of a QIAquick PCR Purification Kit (Qiagen). In order to generate random start positions for next-generation sequencing, to reduce the presence of primer sequences and adjacent intronic sequences, and to reduce fragment sizes, we digested the mixture of PCR products with BAL-31 exonuclease and DNase I endonuclease. Ten µg of the purified PCR fragment mixture was digested with 5 units of BAL-31 exonuclease (New England Biolabs) for 5 min at 30°C in a 200 µl reaction. The reaction was stopped by adding EGTA to a final concentration of 20 mM and immediately heat inactivated at 65°C for 10 min, followed by Qiaquick PCR column purification (Qiagen). Remaining DNA (about 2 µg) was further digested by incubating with 1 unit DNAse I (Roche) and freshly prepared reaction buffer [2x reaction buffer contains 20 mM Tris-Cl (pH 7.5), 2 mM CaCl2, and 20 mM MnCl2] for 3 min at 16°C. In the presence of Mn2+ ions, the DNase I enzyme cleaves both DNA strands at approximately the same site. [31] The reaction was stopped by adding 2 µl EDTA (500 mM) and immediately heat inactivated at 65°C for 10 min. Afterwards, DNA was purified using Qiaquick columns (Qiagen) and eluted with 30 µl EB buffer.
Next-generation sequencing on a Solexa/Illumina GA2 platform
Library construction of the modified PCR fragment mixture was performed using “Genomic DNA Sample Prep Kit” according to the manufacturer’s instructions (Illumina, San Diego, CA). Fragments were separated on a 1.5% agarose gel and excised in the 150–200 bp range. Fragments were then purified and subjected to 10 rounds of PCR amplification using complementary linker specific primers. The amount and size distribution of each sample was analyzed on a Bioanalyzer 2100 (Agilent Technologies, Inc). For next-generation sequencing, single-stranded DNA fragments were annealed to a flow cell surface in a cluster station (Illumina) and subjected to 46 cycles of bridge amplification. Fragments were run on a single lane of a Solexa/Illumina Genome Analyzer II platform, generating about 15–20 million single-end sequence reads of 39 bases each. Image analysis and base calling was generated by the Genome Analyzer Pipeline 1.5.1 with default parameters. Illumina specific FASTQ files containing sequence information and quality scores for each base call were exported for downstream analyses.
Sequence read mapping and variant calling
Sequence alignment was performed with ‘CLC Genomics Workbench™’ software (CLC-bio, Aarhus, Denmark) using imported and annotated human reference genome assembly hg18 (NCBI build 36) chromosome sequence files as a reference. We annotated all 24 human chromosomal reference data sets with gene transfer files (GTF) after downloading dbSNP(v130) from UCSC and by annotating all obligatory splice sites. After importing the concatenated FASTQ (Illumina) files generated by the GAII pipeline into the CLC-software, sequence reads were mapped to exonic coding regions plus adjacent 100 bp intronic sequence of all 376 initially PCR-amplified targets. Gapped alignments with default parameters applying bioinformatic costs for mismatches of “2” and indel costs of “3” was performed. Variant calls were obtained using the following filter parameters: Coverage ≥ 300X, variant frequency of at least 0.7%, and a minimum variant count of 5 reads. The variant analysis included coordinates of obligatory splice sites, and all variants predicted to change the amino acid sequence (missense, nonsense, and coding indels). When calling variants from normal reference sequence (VRS), we excluded all synonymous variants and all variants localized in non-coding exonic regions or in introns (except obligatory splice sites). Furthermore, we excluded known SNPs (v130) and all variants identified in the healthy control pool MPR experiment processed in parallel using identical filter parameters (coverage, minor allele frequency, and minimal counts). To prioritize for downstream analysis we scored missense variants according to the information of evolutionary conservation and the likelihood of a potential protein damaging effect using PolyPhen2 software predictions (http://genetics.bwh.harvard.edu/pph/).[32] All variants with a predicted “probable” or “possible” damaging effect and a score above 0.7 (PolyPhen2) were further analyzed in the original unpooled DNA samples of 24 pooled individuals by CEL1 heteroduplex analysis and/or direct Sanger sequencing.
Identifying mutation carriers out of 24 pooled individuals
In order to identify the carrier(s) of the selected VRS (114 altogether) from a pool of 24 patients, standard mutation detection techniques were applied. The mutation containing exon was amplified by PCR using genomic DNA of all 24 patients as template in separate reactions. Subsequently, the PCR products were analyzed using CEL I heteroduplex screening or using direct Sanger sequencing.[30] In all cases in which only one mutated allele was discovered, we screened for a potential second mutated allele by Sanger sequencing all exons of the respective gene.
RESULTS
Proof of principle for pooled DNA sample analysis
In a pilot project we tested the DNA pooling and next-generation sequencing strategy by pooling the DNA of 18 patients (36 alleles) with known mutations in 18 different NPHP-AC causing genes (Table 1, Supplementary Table 2). The equimolar DNA pool contained altogether 14 different heterozygous and 10 different homozygous mutations/variants. From a total of 24 different mutations/variants, 9 were missense, 8 were nonsense, 2 were small insertion/deletions, 2 were synonymous rare polymorphisms, and 3 were affecting obligatory splice sites. After MPR of 376 different pooled PCR products derived from the DNA pool of 18 different mutation carriers, the analysis revealed confirmation of 22 out of 24 known mutations/variants (92%) (Supplementary Table 2). For the two mutations (p.S360T in NPHP3, and R85X in TMEM216) that were not detected, there was a lack of coverage of the respective exons with only two or zero variant sequence reads, respectively (Supplementary Table 2). Mutations were detected after filtering for variants affecting amino acid residues, reading frames, or obligatory splice sites by setting parameters for variant calls for minor allele frequency to ≥ 0.7%, minimum coverage to ≥ 300×, and the minimum number of reads (counts) to ≥ 5 as described in methods.
Statistics on Solexa/Illumina GA2 sequence runs of pooled DNA samples
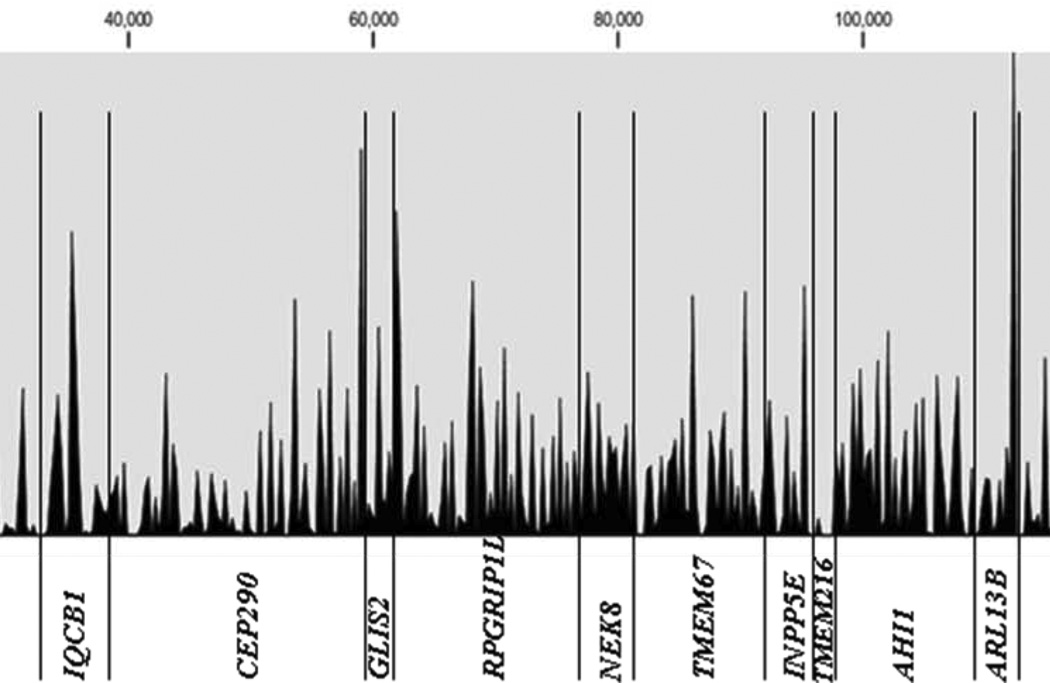
Next-generation sequencing of 376 different PCR products generated by using pooled DNA samples was carried out on a Illumina/Solexa GA2 platform (one pool per lane of a flow cell). In addition to 5 experimental patient’s DNA pools (Patient-pool 1>5) generated by pooling 24 samples each, we sequenced the PCR products derived from a pool of 96 healthy control individuals (HRC1-individuals) and one positive control pool derived from 18 patients with known mutations. Statistical features of Solexa/Illumina sequence runs representing the 5 experimental patient pools are shown in Table 2. One lane of a 8-lane flow cell GA2 run delivered on average about 16.7 million (range: 15.5 M to 19.9 M) short reads of 39 bases. After gapped alignment to the human reference sequence, 85% of all reads mapped back to one of the 376 targeted amplicons (18 genes). The sequence concatenation of all 376 amplicons amounts to a total length of 147.5 kb, 52.8 kb of which were exonic coding regions. The median coverage for coding nucleotide sites was 4,186X (mean: 5,753X), which translates into a median depth of 87X (mean: 120X) per site per single allele, respectively. An example of the overall coverage of all concatenated exons derived from 18 NPHP-AC genes is shown in Figure 3 for patient pool #3. The overall maximum coverage of 54,386X was found in patient pool #5. On average, about 95% of the targeted coding regions were sufficiently covered at least 300-fold in order to reliably call a heterozygous mutation/variant present on only one out of 48 pooled alleles derived from 24 patients.
Table 2.
Statistics on Solexa/Illumina massively parallel resequencing runs of 5 DNA pools derived from 120 patients with NPHP-AC
| Patient Pool |
DNAs pooled |
# of reads (39 bases) |
Mean coverage depth of coding regions |
Median coverage depth of coding regions |
Maxi-mum coverage depth of coding regions |
Coding bases covered ≥300× [%] |
VRS | SNP130 | VRS in HRC1-pool |
VRS minus SNPv130 minus HRC1-pool |
VRS (PolyPhen-2 score ≥0.7) |
Confirmed by Sanger sequencing |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pool #1 | 24 | 15,600,763 | 6,239 | 4,567 | 53,284 | 97.6 | 67 | 26 | 2 | 39 | 32 | 24 (75%) |
| Pool #2 | 24 | 16,580,432 | 5,561 | 4,429 | 34,507 | 95.6 | 85 | 27 | 17 | 41 | 16 | 8 (50%) |
| Pool #3 | 24 | 15,536,857 | 4,690 | 3,809 | 29,767 | 91.4 | 121 | 25 | 14 | 82 | 36 | 18 (50%) |
| Pool #4 | 24 | 15,741,392 | 4,842 | 2,877 | 48,372 | 95.1 | 57 | 19 | 8 | 30 | 19 | 15 (79%) |
| Pool #5 | 24 | 19,882,361 | 7,432 | 5,250 | 54,386 | 97.5 | 49 | 22 | 8 | 19 | 11 | 8 (73%) |
| Sum | 120 | 83,341,805 | NA | NA | NA | NA | 379 | 119 | 49 | 211 | 114 | 74 (65%) |
| Mean | 24 | 16,668,361 | 5,753 | 4,186 | 44,063 | 95.4 | 86 | 25 | 10 | 42 | 23 | 15 |
VRS, variants from reference sequence; SNP, single nucleotide polymorphism, HRC-1, Human random control DNA panel-1.
Figure 3.
Obtained coverage depth along the entire concatenated sequence of all 376 amplicons (145 kb) derived from 18 different NPHP-AC genes. Shown as an example are the results from the MPR mutation analysis performed on 1 lane of a Solexa/Illumina flowcell in patient pool #3 (DNA pool of 24 patients). Note that exonic but also partial intronic regions of all amplicons are shown. The median coverage depth within exonic coding regions (total of 54 kb) is 4,186×. About 95% of all coding bases are covered at least 300-fold.
Mutation carrier identification by CEL I endonuclease heteroduplex screening and Sanger sequencing
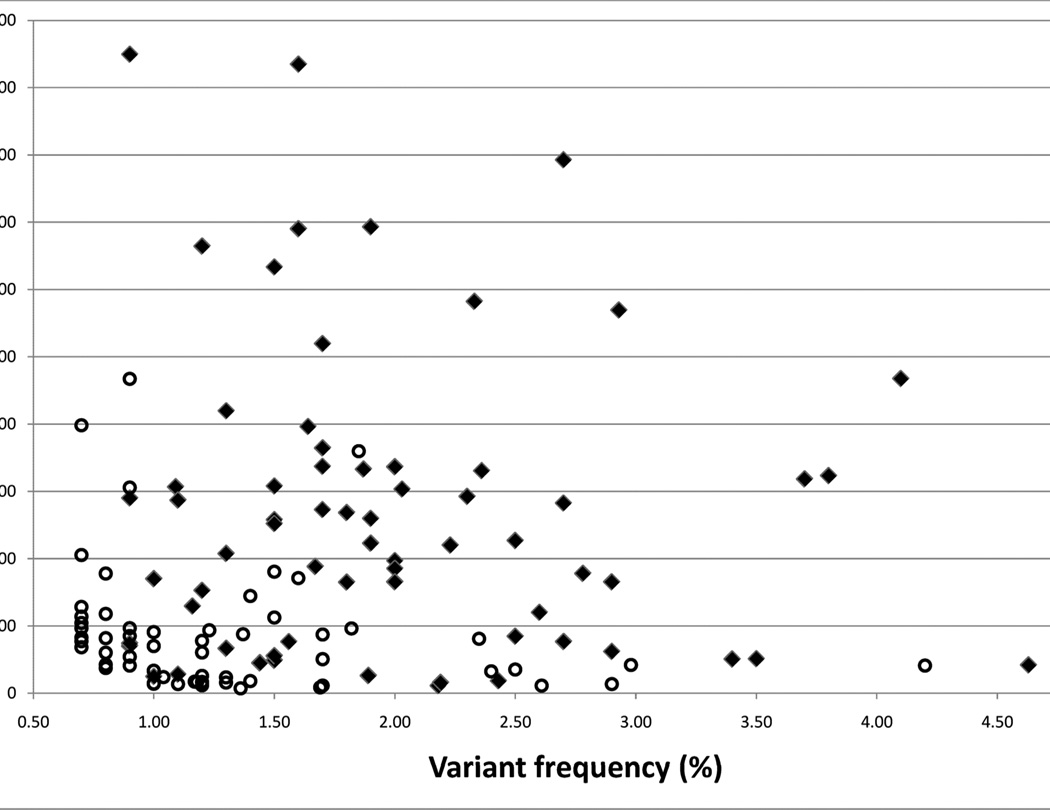
MPR of PCR products of 120 individuals revealed initially a total of 379 variants within coding regions of 18 genes analyzed, 119 of which were known SNPs (Table 2). Performing MPR on a healthy control sample pool in parallel reduced the remaining 260 variants by another 49 variants (Table 2). These variants were either unknown SNPs identified in the human random control pool or false calls due to software base calling or alignment artifacts. Of the remaining 211 variants, 114 truncating mutations (nonsense, frameshift), canonical splice sites and missense changes with a predicted “damaging” impact at the protein level (PolyPhen2 score >0.7) were followed up by identification of the contributing individual. CEL I heteroduplex screening and direct Sanger sequencing were subsequently performed for the 114 variants in order to identify the mutation carrier(s) out of the respective pool of 24 patients. This carrier analysis led to the confirmation of 75 (65%) mutations/variants (“true positives”), whereas 40 variants could not be confirmed (“false positives”) (Table 2). The distribution of` “false” versus “true positive” were plotted in respect to the variant allele frequency (x-axis) and the absolute sequence reads (counts) present for each of the 114 variants analyzed (Figure 4). The likelihood for a variant allele to be “true positives” was found to increase as expected, with higher absolute allele counts and/or higher relative variant allele frequency. In order not to miss mutations we choose relaxed SNP call parameters of 0.7% for the variant allele frequency with at least 5 absolute counts (reads). This has especially increased “false positive” variants with allele frequencies between 0.7% and 1% (Figure 4). CEL I heteroduplex analysis helped to identify 44 out of altogether 74 variants during the initial screen, but failed in 14 cases to identify the change, as subsequently revealed by Sanger sequencing (in Table 4, 5 indicated as “CEL I negative”). All other variants/mutations have been identified solely by Sanger sequencing without CEL I prescreening. For patients in whom initially only one mutation has been identified, we searched for a second mutated allele by direct Sanger sequencing all exons of the respective gene. This approach led to the identification of 10 additional mutations.
Figure 4.
Sanger sequencing confirmation of “true mutations/variants” (filled diamonds) identified by MPR on Solexa/Illumina platform are plotted versus variants not confirmed “false positive” (open circles). The alignment of a total of about 83 million sequence reads (39 bp each) to the human reference sequence of targeted 376 exons using “CLC Genomics Workbench™” software revealed 114 variants/mutations fulfilling the following criteria: i) absent from SNP database SNPv130, ii) absent from a control pool of 96 DNAs of healthy individuals, and iii) damaging impact at protein level predicted by PolyPhen2 with scores above 0.7. The variant frequency (x-axis) is plotted against the variant counts (y-axis) for each of the 114 variants analyzed. Seventy four variants/mutations have been confirmed by Sanger sequencing (filled diamonds) and we identified the respective mutation carrier out of a pool of 24 patients. Note that Sanger sequencing confirmation (filled diamonds) has been successful for only 3 variants with allele frequencies below 1%. The expected frequency of a heterozygous change found in a pool of 24 patients DNA is about 2.1%. Changes with frequencies above 1% and high absolute counts (>50) are almost always confirmed by Sanger sequencing.
Table 4.
Genotypes and phenotypes of 24 patients carry a single heterozygous variant/mutation in at least one of 11 NPHP-AC genes
| Patient | Diagnosis | Origin | Consan-guinity | Gene | Nucleotide Changea (Zygosity) |
Amino Acid Change |
Mutant allele frequency |
Evolutionary conservation | Poly-Phen-2 scoreb |
Mutation Assignmentc |
Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D. rerio | C. intest. | C.elegans. | |||||||||||
| F437-22 | NPHP | Italy | − | NPHP3 | c.3422T>C (h) | p.L1141P | 78/5161 (1.5%) | + | − | + | 0.996 | Dir.seq | [8] |
| F299-22 | NPHP, LF | Germany | − | NPHP4 | c.271T>C (h) | p.F91L | 101/5196 (1.9%) | + | + | + | 0.998 | CEL I neg. | [41] |
| F521-21 | JBTS | Arab | + | NPHP4 | c.944G>A (h) | p.T315M | 244/6468 (3.8%) | − | − | − | 0.980 | CEL I | novel |
| A396-21 | NPHP | Turkey | + | IQCB1 | c.1271A>G (h) | p.Q424R | 227/13809 (1.6%) | + | + | − | 0.980 | CEL I | novel |
| A75-21d | NPHP | Poland | nd | CEP290 | c.254A>C (h) | p.N85T | 13/1487 (0.9%) | + | − | − | 0.995 | CEL I | novel |
| F631-21 | JBTS | Germany | − | RPGRIP1L | c.1177G>A (h) | p.E393K | 115/6747 (1.7%) | + | − | − | 0.980 | CEL I | [42] |
| A75-21d | NPHP | Poland | nd | RPGRIP1L | c.1964A>G (h) | p.Y655C | 191/12672 (1.5%) | + | − | − | 1.000 | CEL I | novel |
| A1042-21 | JBTS | Germany | − | TMEM67 | c.1334T>C (h) | p.V445A | 135/6734 (2.0%) | + | − | − | 0.992 | CEL I | novel |
| A108-22 | JBTS | ND | nd | TMEM67 | c.1911C>T (h) | p.F637L | NA | − | + | − | 0.999 | Dir.seq | novel |
| A380-21d | SLSN | Italy | + | TMEM67 | c.2009C>T (h) | p.T670I | 421/15859 (2.7%) | − | + | + | 0.992 | CEL I neg. | novel |
| F962-23f | NPHP4 | Swiss | nd | TMEM67 | c.2374A>G (h) | p.R792G | 37/3058 (1.2%) | + | + | + | 1.000 | CEL I neg. | novel |
| A386-21 | JBTS | Turkey | − | TMEM67 | c.2461G>A (h) | p.G821S | NA | + | + | + | 0.980 | Dir.seq | [14] |
| A1421-21d | JBTS | Egypt | − | TMEM67 | c.2461G>A (h) | p.G821S | 98/4402 (2.2%) | + | + | + | 0.980 | CEL I | [14] |
| A805-21 | NPHP | Pakistan | + | ARL13B | c.1169G>A (h) | p.R390L | NA | + | − | − | 0.993 | Dir.seq | novel |
| F459-22 | JBTS, TMEM67 |
USA | nd | CC2D2A | c.1519A>G (h) | p.K507E | 154/5654 (2.7%) | + | − | − | 0.780 | CEL I | novel |
| F16-21 | SLSN | France | − | CC2D2A | c.1519A>G (h) | p.K507E | 154/5654 (2.7%) | + | − | − | 0.780 | CEL I | novel |
| A3208-21 | JBTS | UK | − | CC2D2A | c.1519A>G (h) | p.K507E | 61/3307 (1.8%) | + | − | − | 0.780 | CEL I | novel |
| A559-22 | NPHP | Arab | + | CC2D2A | c.2161C>T (h) | p.P721S | 122/7296 (1.7%) | + | + | − | 0.999 | CEL I neg. | [40] |
| A944-21 | NPHP | Pakistani | + | CC2D2A | c.2161C>T (h) | p.P721S | 383/9355 (4.1%) | + | + | − | 0.999 | CEL I | [40] |
| A1347-21 | NPHP | Turkey | + | CC2D2A | c.2161C>T (h) | p.P721S | 383/9355 (4.1%) | + | + | − | 0.999 | CEL I | [40] |
| A1421-21d | JBTS | Egypt | − | CC2D2A | c.3056G>A (h) | p.R1019Q | 272/11649 (2.3%) | + | + | + | 1.000 | CEL I | novel |
| A1152-21 | SLSN | Arab | − | TTC21B | c.2258C>T (h) | p.P753L | 24/1534 (1.6%) | + | + | − | 0.999 | CEL I | novel |
| A2430-21 | MKS | UK | − | TTC21B | c.2588G>A (h) | p.R863Q | 35/1016 (3.4%) | − | + | + | 0.733 | CEL I | novel |
| A1502-21 | JBTS | USA | − | TTC21B | c.3004C>G (h) | p.L1002V | 125/6667 (1.9%) | + | + | − | 0.922 | CEL I | novel |
| F132-24 | NPHP | USA | − | MKS1 | c.857T>C (h) | p.D286G | 156/6617 (2.4%) | + | − | − | 0.983 | CEL I | novel |
| A1859-21 | JBTS | Austria | − | XPNPEP3 | c.463C>T (h) | p.R155W | 99/5373 (1.8%) | + | − | + | 0.907 | CEL I | novel |
All mutations were absent from 96 healthy control subjects. Direct Sanger sequencing for all exons in the respective gene was performed in order to identify a second mutated allel but was negative. Mutation numbering is based on cDNA position in reference sequences indicated in Table 1 with +1 corresponding to the A of the ATG translation initiation codon.
PolyPhen-2 (Polymorphism Phenotyping v2) is a tool which predicts possible impact of an amino acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations (http://genetics.bwh.harvard.edu/pph2/). Scores were given between 0 (benign) and 1 (possibly damaging). We analyzed only missense changes with PolyPhen scores above 0.7.
The mutation carrier assignment (from a pool of 24) was performed by heteroduplex based CEL I endonuclease screening (CEL I) or by direct Sanger sequencing of all 24 DNA samples (Dir. Seq) for the respective exon. In cases CEL I analysis was inconclusive or negative (CEL I neg.), all 24 DNA samples from the respective DNA pool were directly sequenced.
Patients A75-21 and A1421-21 carry heterozygous variants/mutations in 2 different NPHP-AC genes. (h), heterozygous; inf., infantile; JBTS, Joubert syndrome; NPHP, nephronophthisis; MKS, Meckel-Gruber syndrome; nd, no data; C.intest., Ciona intestinalis; D.rerio, Danio rerio; C.elegans, Caenorhabditis elegans
Detection of disease causing mutations in 30 out of 120 patients
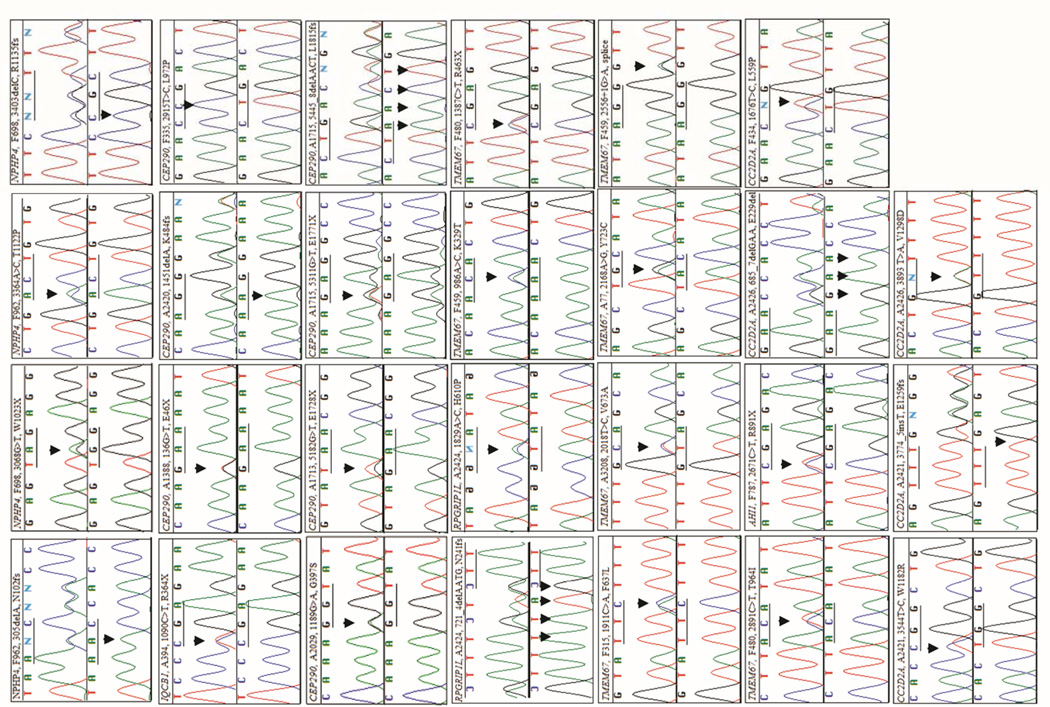
MPR mutation analysis of all coding exons of 18 different NPHP-AC genes (Table 1) in 120 ascertained patients with a severe recessive NPHP-AC disease led to the identification of 57 mutated alleles, comprising 43 different mutations in 30 unrelated patients (Table 3). Twenty four patients carried a compound heterozygous mutation. In 3 patients with a consanguineous background, a homozygous disease allele was identified in CEP290 (p.L972P and p.G1890X twice) (Table 3). In 3 patients, only 1 mutated allele (IQCB1, p.R364X; CEP290, p.K484fsX8; and AHI1, p.R891X) has been identified so far, even though all exons of the respective mutated gene have been sequenced directly. Mutations were discovered in the genes NPHP4 (2 patients), IQCB1/NPHP5 (1 patient), CEP290/NPHP6 (9 patients), RPGRIP1L/NPHP8 (1 patient), TMEM67/NPHP11 (10 patients), AHI1 (1 patient), CC2D2A (3 patients), and TTC21B (3 patients) (Table 3). The category of mutated alleles identified in the present study are as following: 14 nonsense mutations, 11 small insertion/deletions leading to a frameshift, 1 inframe deletion, 3 splice site mutations, and 25 missense mutations (Table 3). Twenty seven of these mutations were novel findings in the genes NPHP4 (p.N102fsX76, p.W1023X, p.T1122P, p.R1135fsX10), IQCB1 (p.R364X), CEP290 (p.E46X, p.G397S, p.K484fsX8, p.L972P, p.E1728X, p.E1771X, p.L1815fsX4), RPGRIP1L (p.N241fsX25, p.H610P), TMEM67 (p.K329T, p.R463X, p.F637L, p.V673A, p.Y723C, p.T964I, c.2556+1G>A splice), AHI1 (p.R891X), and CC2D2A (p.E229del, p.L559P, p.W1182R, p.E1259fsX1, p.V1298D) (Table 3, Figure 2). All mutations were absent from 96 healthy control individuals. In all cases where DNA of relatives was available, recessive mutations segregated with the affected status and segregated from parents as expected. In 17 out of 30 families segregation analysis has been performed and paternal (p) or maternal (m) inheritance has been indicated for respective mutations accordingly in Table 3. The heterozygous TMEM67 mutation p.C615R was found recurrently in 5 different families of German origin. Comparison of SNP genotype linkage data, generated by 250k (StyI) Affymetrix SNP analysis, is compatible with extensive haplotype sharing between respective patients and extends up to 4.2 Mb (204 SNP markers), indicating inheritance from an ancestral founder (data not shown).
Table 3.
Genotypes and phenotypes of 38 patients (30 families) with mutations in NPHP4, IQCB1, CEP290, RPGRIP1L, TMEM67, AHI1, CC2D2A and TTC21B and respective MPR statistics
| Patient | Diagnosis | Kidney ESRD (yrs) |
Eye | Brain | Liver | Other | Origin | Gene | Nucleotide Change(a) (Zygosity State) |
Amino Acid Change (Segregation) |
Mutant Allele Frequency |
Mutation Assignment(b) |
Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F962-23 | NPHP | NPHP— (12) |
- | - | - | - | Germany | NPHP4 | c.3364A>C (h) | p.T1122P (p) | 90/6,163 (1.5%) | CEL I | novel |
| c.305delA (h) | p.N102fsX76 (m) | 14/684 (2.0%) | CEL I | novel | |||||||||
| F698 | −21: NPHP | NPHP (21) | - | - | - | - | Belgium | NPHP4 | c.3068G>A (h) | p.W1023X (m) | 43/1,694 (2.5%) | CEL I | novel |
| −22: NPHP | NPHP (11) | - | - | - | - | c.3403delC (h) | p.R1135fsX10 | 56/7,148 (0.8%) | CEL I | novel | |||
| A394 | −21: SLS | - | RD, Nys | - | - | - | Germany | IQCB1 | c.1090C>T (h) | p.R364X | 77/3,945 (2.0%) | CEL I neg. | novel |
| −22: SLS | NPHP (10) | LCA, Nys | - | ? | ? | ? | Seq 15 exons | ? | |||||
| F15 | −21: SLS | NPHP (11) | LCA, Nys | Atx, MR | - | - | France | CEP290 | c.5649-50insA (h) | p.L1884fsX23 (p) | NA | Seq 53 exons | [33] |
| −24: SLS | NPHP (15) | LCA | Atx, MR | - | - | c.5850delT (h) | p.F1950fsX15 (m) | 34/4,784 (0.7%) | CEL I | [34] | |||
| F335 | −22: SLS | NPHP (13) | LCA | - | - | - | France | CEP290 | c.2915T>C (H) | p.L972P (p, m) | 63/2,404 (2.6%) | CEL I neg. | novel |
| −25: SLS | NPHP (>3) | LCA | - | - | - | ||||||||
| F394-27 | JBTS | NPHP (4) |
RD, Nys, Col |
CVA, Atx, MR |
- | MicCep | Turkey | CEP290 | c.5668G>T (H) | p.G1890X (*, m) | 67/3,316 (2.0%) | CEL I | [10] |
| A1188-21 | JBTS | NPHP (12) |
- | CVA, MR | - | - | Pakistani | CEP290 | c.5668G>T (H) | p.G1890X | 36/1,028 (3.5%) | CEL I | [10] |
| A2029-21 | JBTS | NPHP (2) |
LCA | MTS, MR | HF | - | USA | CEP290 | c.5668G>T (h) | p.G1890X | 92/5460 (1.7%) | Dir. seq. | [10] |
| c.1189G>A (h) | p.G397S | 33/3408 (1.0%) | CEL I | novel | |||||||||
| A1388-21 | JBTS | NPHP (8) |
Nys | CVA, MR | - | deafness | Germany | CEP290 | c.5649-50insA (h) | p.L1884fsX23 | NA | Seq 53 exons | [33] |
| c.136G>T (h) | p.E46X | 99/3,565 (2.8%) | Dir. Seq | novel | |||||||||
| A1713-21 | −21: JBTS | NPHP | LCA | CVH | - | died 3 yrs | Germany | CEP290 | c.6277delG (h) | p.V2093fsX4 | 9/373 (2.4%) | CEL I neg. | [35] |
| −22: JBTS | NPHP | LCA | CVH | - | - | c.5182G>T (h) | p.E1728X | 5/229 (2.2%) | Dir. Seq | novel | |||
| A1715-21 | JBTS | NPHP (10) |
RD | MR, Oph | - | CFA | India | CEP290 | c.5445-8delAACT (h) | p.L1815fsX4 (p) | NA | Seq 53 exons | novel |
| c.5311G>T (h) | p.E1771X (m) | 9/370 (2.4%) | CEL I neg. | novel | |||||||||
| A2420 | −21: MKS | MDK | - | OEC | HDD | - | UK | CEP290 | c.1451delA (h) | p.K484fsX8 | 33/4,111 (0.8%) | CEL I | novel |
| −22: MKS | MDK | - | OEC | HDD | - | ? | ? | ? | Seq 18 exons | ? | |||
| A2424 | −21: MKS | MDK | - | OEC, DWM |
- | PD | UK | RPGRIP1L | c.1829A>C (h) | p.H610P | 83/4,463 (1.9%) | CEL I | novel |
| −22: MKS | MDK | - | OEC, DWM |
- | PD | c.721-4delAATG (h) | p.N241fsX25 | NA | Seq 26 exons | novel | |||
| F90-21 | JBTS | NPHP (4) |
Nys, RD | CVA, MR, Atx, |
HF | PD, MicCep |
Germany | TMEM67 | c.1843T>C (h) | p.C615R | 39/843 (4.6%) | CEL I neg. | [36] |
| c.755T>C (h) | p.M252T (p) | 36/1,243 (2.9%) | CEL I | [37] | |||||||||
| F96-22 | JBTS | NPHP (22) |
Col, RD | CVA, MR, Atx |
- | - | Germany | TMEM67 | c.755T>C (h) | p.M252T (m) | 42/1,538 (2.7%) | CEL I | [37] |
| c.2498T>C (h) | p.I833T (p) | 165/13,292 (1.2%) |
CEL I | [38] | |||||||||
| F278-21 | JBTS | NPHP (14) |
- | Atx, MR | HF, CP |
- | Germany | TMEM67 | c.1843T>C (h) | p.C615R | 234/6,373 (3.7%) | CEL I neg. | [36] |
| c.755T>C (h) | p.M252T | 13/1,409 (0.9%) | CEL I | [37] | |||||||||
| F315-21 | JBTS | NPHP (9) |
Nys | CVH, MR | HF | - | Germany | TMEM67 | c.1843T>C (h) | p.C615R | 234/6,373 (3.7%) | CEL I neg. | [36] |
| c.1911C>A (h) | p.F637L | 17/1,115 (1.5%) | CEL I | novel | |||||||||
| F480-22 | JBTS | NPHP (0.2) |
- | CVH | - | - | Germany | TMEM67 | c.1387C>T (h) | p.R463X (m) | 7/320 (2.2%) | Dir. Seq | novel |
| c.2891C>T (h) | p.T964I (p) | 10/528 (1.9%) | CEL I | novel | |||||||||
| F459-22 | JBTS | NPHP (30) |
Col | CVH, ATX, MR |
HF, BDP |
hearing loss |
USA | TMEM67 | c.986A>C (h) c.2556+1G>A (h) |
p.K329T (p) splice |
95/3,313 (2.9%) 172/10,393 (1.7%) |
CEL I CEL I |
novel novel |
| F631-21 | JBTS | NPHP (21) |
Col | MR, | HF | - | Germany | TMEM67 | c.1045T>C (h) | p.L349S | NA | Seq 28 exons | [37] |
| c.1843T>C (h) | p.C615R (p) | 39/843 4.6%) | CEL I neg. | [36] | |||||||||
| A77-21 | JBTS | NPHP (7) |
OA | CVH, MR |
HF | aortic stenosis |
Germany | TMEM67 | c.622A>T (h) c.2168A>G (h) |
p.R208X p.Y723C (m) |
123/6,069 (2.0%) 13/905 (1.4%) |
CEL I CEL I |
[37] novel |
| A971-21 | JBTS | NPHP (7) |
Nys | CVH, MR, Atx |
HF, CP |
- | Germany | TMEM67 | c.622A>T (h) | p.R208X | NA | Seq 28 exons | [37] |
| c.1843T>C (h) | p.C615R | 234/6,373 (3.7%) | CEL I neg. | [36] | |||||||||
| A3208-21 | JBTS | NPHP (28) |
Col | MTS, MR | - | epilepsy | UK | TMEM67 | c.1351C>T (h) | p.R451X | 75/5,041 (1.5%) | CEL I | [39] |
| c.2018T>C (h) | p.V673A | 270/13,865 (1.9%) |
CEL I | novel | |||||||||
| F787-21 | JBTS | - | - | CVH | - | - | Germany | AHI1 | c.2671C>T (h) | p.R891X | 112/4,540 (2.5%) | CEL I | novel |
| ? | ? | ? | Seq 28 exons | ? | |||||||||
| F434-21 | JBTS | - | OMAC | CVH, HT, Atx, MR |
- | - | Germany | CC2D2A | c.517C>T (h) | p.R173X (p) | 63/3,771 (1.7%) | Dir. Seq | [40]- |
| c.1676T>C (h) | p.L559P (m) | 67/6,139 (1.1%) | CEL I | novel | |||||||||
| A2421-21 | MKS | MDK | - | OEC | HDD | - | UK | CC2D2A | c.3544T>C (h) | p.W1182R | 5/500 (1.0%) | CEL I | novel |
| c.3774_5insT (h) | p.E1259fsX1 | NA | Seq 35 exons | novel | |||||||||
| A2426-21 | MKS | MDK | - | OEC | - | CFA, OHy |
UK | CC2D2A | c.685_7delGAA (h) | p.E229del | NA | Seq 35 exons | novel |
| c.3893T>A (h) | p.V1298D | 295/18,699 (1.6%) |
CEL I | novel | |||||||||
| F244-25 | infantile NPHP |
NPHP (1.5) |
- | MR, Oph | HF | SI, VC, Bro |
Turkey | TTC21B | c.626C>T (h) | p.P209L (m) | 334/11,391 (2.9%) |
Dir. Seq | [25] |
| c.1654-7delTGTC (h) | p.C552fsX1 (p) | NA | Seq 29 exons | [25] | |||||||||
| F514- | −21: NPHP | NPHP (7) | - | - | PSC | - | Swiss | TTC21B | c.448T>C (h) | p.W150R (m) | 53/5,802 (0.9%) | CEL I | [25] |
| −22: NPHP | NPHP(2) | - | - | PSC | SI, VUR | c.3264-3C>G (h) | splice site (p) | NA | Seq 29 exons | [25] | |||
| A34-21 | infantile NPHP |
NPHP (2) |
RD | - | HF | SI, short phalanges |
Portuguese | TTC21B | c.2758-2A>G (h) | splice site (m) | 92/4,905 (1.9%) | CEL I neg. | [25] |
| c.626C>T (h) | p.P209L (p) | 334/11,391 (2.9%) |
Dir. Seq | [25] |
All mutations were absent from at least 96 healthy control subjects. Mutation numbering is based on cDNA position in reference sequences indicated in Table 1 with +1 corresponding to the A of the ATG translation initiation codon.
The mutation carrier assignment was performed by heteroduplex based CEL I endonuclease screening (CEL I) or by direct Sanger sequencing of the respective 24 DNA samples (Dir. Seq). In cases CEL I analysis was inconclusive or negative (CEL I neg.) all 24 DNA samples from the respective pool were directly sequenced for one exon. In cases where only one mutated allele was found, all exons of the respective gene were sequenced and exon numbers are indicated (Seq # exons).
second mutation not detected;
no maternal or paternal DNA available; Atx, ataxia; BDP, bilary ductal proliferation; Bro, bronchiectasis; CFA, craniofacial abnormalities; CP, cholangio-dysplasia; CVA, cerebellar vermis aplasia; CVH, cerebellar vermis hypoplasia; DWM, Dandy walker malformation; ESRD, end-stage renal disease; (h), heterozygous; (H), homozygous; HDD, hepatic developmental defects; HF, hepatic fibrosis; HT, muscle hypotonia; LCA, Leber congenital amaurosis; (m), heterozygous mutation identified in mother; MDK, multicystic dysplastic kidneys; MicCep, microcephaly; MR, mental retardation; MTS, molar tooth sign; NA, no data available; Nys, nystagmus; OA, opticus atrophy; OEC, occipital encephalocele; OHy, oligohydramnios; OMAC,oculomotor apraxia type Cogan; Oph, ophistotonus; (p), heterozygous mutation identified in father; PD, polydactyly; PSC, primary sclerosing cholangitis; RD, retinal dystrophy; SI, situs inversus; VC, vitium cordis; VUR, vesicoureteral reflux.
Figure 2.
Sequence chromatograms of 28 different novel mutations identified in the genes NPHP4 (4), IQCB1 (1), CEP290 (8), RPGRIP1L (2), TMEM67 (7), AHI1 (1), and CC2D2A (5) in individuals with a NPHP-AC. Gene name, patient identifier, nucleotide change, and inferred amino acid alteration are given above sequence traces. Wild type sequence chromatograms are shown below mutated sequences. All mutations were absent from at least 96 healthy control individuals. Note, that no second mutation has been identified in patients A394, A2420, and F787 in whom we identified a heterozygous truncating mutation in the genes IQCB1 (p.R364X), CEP290 (p.K484fsX8), and AHI1 (p.R891X), respectively. All mutations were found in the heterozygous state with the exception of a homozygous CEP290 missense mutation (p.L972P) in patient F335.
Single heterozygous variants of unknown significance
Mutation analysis by MPR of 18 known disease genes revealed additional 26 single heterozygous missense mutations/variants of unknown significance in 24 patients out of the 120 patients analyzed (Table 4). All changes were absent from at least 96 healthy control individuals. For all 26 missense changes Sanger sequencing was performed for all exons of the respective disease gene, but failed to detect a second mutated allele. Patients of families A75 and A1421 carried heterozygous missense mutations/variants in different disease genes. In a patient from Poland (A75) with nephronophthisis, a heterozygous p.N85T mutation/variant in CEP290 together with a p.Y655C change in the gene RPGRIP1L was detected. The positions of both missense changes were evolutionary conserved in vertebrates including zebrafish. PolyPhen2 software predicted a probable protein damaging effect for both missense changes (Table 4). Individual A1421 from Egypt with Joubert syndrome and liver fibrosis carries the heterozygous missense changes in TMEM67 (p.G821S) and CC2D2A (p.R1019G). Interestingly, the mutation p.G821S has been found homozygously in patients with nephronophthisis with liver fibrosis, published recently (Otto et al., 2009). The amino acids of both missense changes found in family A1421 are highly conserved in evolution including the nematode Caenorhabditis elegans. Altogether, we identified 21 different single heterozygous mutations/variants in the genes NPHP3 (1), NPHP4 (2), IQCB1 (1), CEP290 (1), RPGRIP1L (2), TMEM67 (5), ARL13B (1), CC2D2A (3), TTC21B (3), MKS1 (1), and XPNPEP3 (1). The mutation p.G821S in TMEM67 and heterozygous changes p.K507E and p.P721S in the gene CC2D2A have been found recurrently (Table 4). Five out of the 21 different single heterozygous changes have been published previously as disease causing in a recessive setting and are documented in the Human Gene Mutation Database at the Institute of Medical Genetics in Cardiff (HGMD®, “Biobase” (http://www.biobase-international.com/) (Table 4).
DISCUSSION
Applying next-generation sequencing technology helped us to identify the disease causing mutations in 30 out of 120 patients with severe NPHP-AC. Altogether, we identified 43 different mutations, 28 of which were novel findings within 9 different NPHP-AC genes. Additionally, we found single heterozygous missense changes of unknown significance in 25 patients in 11 out of 18 genes analyzed. We demonstrated that high throughput sequencing using pooled DNA samples is highly efficient in detecting rare mutations in large cohorts of patients with diseases of broad genetic locus heterogeneity. Applying MPR, we screened 376 coding exons derived from 18 different NPHP-AC genes in 120 patients, which comprises more than 45,000 different amplified DNA fragments. Alternative methods, to screen such a large amount of single PCR products, like standard Sanger sequencing, although accurate and reliable, is prohibitively expensive ($180,000) and would require 45,000 single PCR amplifications. CEL1 heteroduplex screening alone without high-throughput sequencing on the other hand is less expensive (estimated costs $12,000), however it would require laborious and time consuming analyses of about 450 different 96-well plates. Another alternative approach is exon capture in combination with next-generation sequencing, which might be considered when a large number of exons (or the total exome) have to be analyzed for a reasonable number of DNA samples (about $4,000/exome/sample). In comparison, the cost of generating sequence data by MPR of 5 pooled samples (376 exons, 120 patients) and subsequent confirmation analyses in the present study was in the range of $6,000. The disadvantage of MPR is the high inherent error rate of about 0.5% per base call. This is problematic especially when samples are pooled and the expected variant frequencies are very low, which certainly will result in high false positive rates. Out of 114 mutations/variants which were predicted after alignment and next-generation sequencing software analysis, only 74 (65%) have been confirmed by Sanger sequencing. Recently, we applied the pooling and MPR strategy also to a cohort of 105 patients with Bardet-Biedl syndrome and screened for mutations in 12 BBS genes. Soft- and hardware improvements of the Illumina sequencers are constantly increasing the number and quality of sequence reads. Interestingly, 100% of the exonic bases in all BBS genes were covered sufficiently (> 300×) with an average coverage of 19,711 after analyzing the most recent sequence run with 28 million short reads. After alignment 11 out of 12 (92%) of the predicted mutations have been directly confirmed by Sanger sequencing (manuscript in preparation). The increased number of sequence reads and the resulting increased base coverage seems to reduce false positive calls dramatically. Better coverage can be also obtained by generating longer (e.g.78 bases) and/or paired-end sequence reads. This will further reduce calling false positives and will strongly reduce the number of confirmation experiments using CEL I endonuclease digestion.
In order to reliably detect a heterozygous variant/mutations in a pool of 48 alleles (expected frequency: 2.1%) after generating a total of up to 800 Mb worth of sequence, we choose cutoffs for minor allele frequency of 0.7% and a minimum count of 5 reads. In a pilot project with 18 pooled DNA samples from patients with already known mutations we were able to identify all but 2 out of 24 known mutated alleles using the outlined cutoff parameters. In both of these missed alleles, the coverage depth was insufficient due to PCR amplification problems.
The question arises, how many mutations might have been missed in the experimental pooling and MPR approach in the 120 patients investigated. We have recognized insufficient coverage below 300× for about 5% of all coding nucleotides out of a total of the 52.8 kb sequenced and expect therefore at least 5% of mutations to be missed. MPR of PCR amplicons generated from pooled DNA samples revealed initially 114 potential mutations using CLC genomics workbench software for alignment and analysis. Seventyfour (65%) of these variants have been finally confirmed by Sanger sequencing, 41 of which represented single heterozygous changes only, without a second mutated allele identified initially. However, Sanger sequencing of all exons of the respective gene in carriers of these single heterozygous changes revealed the second recessive mutation in 10 patients. This indicates that we have missed at least 12% (10/87) of mutations/variants during the initial analysis. The calculated likelihood to miss both mutations of a patient with a compound heterozygous mutation is only in the range of 1–2% (0.12×0.12). Besides the mutations we missed because of low coverage depth, there were 3 different 4 bp deletions with sufficient coverage which were missed using the CLC next-generation sequence software. To address the problem of detection of small indels (4 and more bases) in short reads (39 bp) the use of other software packages have to be considered like “NextGENe®” from the company Softgenetics or the program “Novoalign” from Novocraft. For longer reads of e.g.78 bp, we found that CLC next-generation sequence software reliably calls indels of up to 6 bases. To date, the mutation rate of indels (4 or more bases) in NPHP related genes published in the HGMD® ‘Biobase’ mutation database (release 25th June 2010) is about 6%. Out of 457 different mutations published in the 18 NPHP related genes, 28 (6.1%) fall into this category of 4 or more bases inserted or deleted.
MPR revealed 24 patients with only 1 heterozygous missense change in one gene or 2 missense changes in 2 different genes (3 families, Table 4). We speculate that in most of these cases a second mutated allele has been missed, although all exons have been analyzed by Sanger sequencing. Examples could be, gross rearrangements, copy number variations, deep intronic splice affecting changes, promotor mutations, or polyadenylation signal variants, which were not detectable by solely coding exon sequencing. Some of the changes found, might represent only rare polymorphism without any disease relevance. We have not found any evidence that oligogenicity is involved in NPHP-AC, although we sequenced 120 patients for all known relevant 18 NPHP-AC genes.
The approach of MPR of pooled samples presented here is robust, cost efficient, and best suited for screening large cohorts for mutations in genetically heterogeneous diseases. The lack of sensitivity seen so far for the MPR of pooled sample approach makes a clinical diagnostic application impracticable. In cases where numerous changes are expected or in a clinical mutation diagnostic setting, “barcoding tags” offer an alternative and should be considered. However, that approach requires additional library preparations, large amounts of single PCR reactions or establishment of very complex multiplex PCRs reactions.[43] Anticipated future sequencing technology improvements will allow parallel mutation analysis of higher numbers of genes and DNA samples. This is especially of interest because the lack of mutations in 75% of patients in our cohort indicates further extensive heterogeneity in NPHP-AC.
Supplementary Material
ACKNOWLEDGEMENTS
The authors sincerely thank the affected individuals and their families for participation and we thank the physicians who contributed to this study. We acknowledge R.H. Lyons for excellent next-generation sequencing. F.H. is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and a Frederick G. L. Huetwell Professor. This research was supported by grants from the National Institutes of Health to F.H. (DK1069274, DK1068306, DK064614).
We thank all the physicians and researchers of the “GESELLSCHAFT FÜR
PÄDIATRISCHE NEPHROLOGIE (GPN)” study group for participation
Members of the GPN study group are: C. Bergmann (Aachen, Germany); K. Zerres (Aachen, Germany); J. Gellermann (Berlin, Germany); A. Münch (Berlin, Germany); L. Neumann (Berlin, Germany); M.J. Schürmann (Berlin, Germany); I. Franke (Bonn, Germany); B. Beck (Cologne, Germany); K. Josefiak (Cologne, Germany); D. Michalk (Cologne, Germany); Dr. Stapenhorst (Cologne, Germany); T. Ronda (Cologne, Germany); M. Weber (Cologne, Germany); T. Erler (Cottbus, Germany); B. Weidner (Cottbus, Germany); K.E. Bonzel (Essen, Germany); A-M. Wingen (Essen, Germany); J. Dippell (Frankfurt, Germany); J. Kirschner (Freiburg, Germany); R. Korinthenberg (Freiburg, Germany); M. Mall (Freiburg, Germany); H. Omran (Freiburg, Germany); G. Wolff, (Freiburg, Germany); S. Fuchs (Hamburg, Germany); A. Gal (Hamburg, Germany); M. van Husen (Hamburg, Germany); S. Lüttgen (Hamburg, Germany); D.E. Müller-Wiefel (Hamburg, Germany); J. Drube (Hannover, Germany); J.H.H. Ehrich (Hannover, Germany); S. Fründ (Hannover, Germany); J. Strehlau (Hannover, Germany); G.F. Hoffmann (Heidelberg, Germany); D. Kiepe (Heidelberg, Germany); C. Kneppo (Heidelberg, Germany); S. Rieger (Heidelberg, Germany); B. Tönshoff (Heidelberg, Germany); R. Bambauer (Homburg, Germany); R. Klüte (Ibbenbüren, Germany); M. Heckel (Kronach, Germany); A. Greiner (Leipzig, Germany); N. Jeck (Marburg, Germany); R. Roos (München, Germany); M, Bulla (Münster, Germany); S. Fründ (Münster, Germany), B. Frye (Münster, Germany); E. Harms (Münster, Germany); E. Kuwertz-Broeking (Münster, Germany); B. Wittwer (Münster, Germany); R. Sanwald (Pforzheim, Germany); H-J. Stolpe (Rostock, Germany); J. Höpfner (Schweinfurt, Germany); M. Holder (Stuttgart, Germany); H-E. Leichter (Stuttgart, Germany); G. Baynam (Subiaco, Australia); C. Edwards (Subiaco, Australia); H. Peters (Victoria, Australia); C. Jones (Victoria, Australia); A. Janecke (Innsbruck, Austria); G. Sunder-Plassmann (Vienna, Austria); K. Devriendt, Leuven, Belgium); J. Chow (Vancouver, Canada); P. Trnka (Vancouver, Canada); K. Õunap (Tartu, Estonia); T. Apostolou (Athene, Greece); B. Afroze (Kuala Lumpur, Malaysia); N. Lock Hock (Kuala Lumpur, Malaysia); M. Eccles (Otago, New Zealand); J.W. Dixon (Wellington, New Zealand; S. Hashmi (Karachi, Pakistan); D. Drozdz (Kraków, Poland); A. Pogan (Kraków, Poland); A. Peco-Antic (Belgrade, Serbia); B. Milosevic (Novi Sad, Serbia); V. Stojanovic (Novi Sad, Serbia); E. Holmberg (Umea, Sweden); I. Kern (Geneva, Switzerland); P.H. Axwijk (Amsterdam, The Netherlands); N. Knoers (Nijmegen, The Netherlands); F. Ozaltin (Ankara, Turkey); N. Besbas (Ankara, Turkey); M. Koyun (Antalya, Turkey); A. Nayir (Istanbul, Turkey); H. Kayserili (Istanbul, Turkey); S. Ozturk (Istanbul, Turkey); D. Pehlivan (Istanbul, Turkey); R. Farrington (Cambridge, UK); F.L. Raymond (Cambridge, UK); R. Sandford (Cambridge, UK); J. Whittaker (Cambridge, UK); B. Kerr (Manchester, UK); M. Cadnapaphornchai (Denver, CO, USA); G. Hidalgo (Detroit, MI, USA); S. Andreoli (Indiananapolis, IN, USA); B. Mills (Indiananapolis, IN, USA); M. Bendel-Stenzel (Minneapolis, MN, USA); N. Stover (Portland, OR, USA); R. Weleber (Portland, OR, USA); M. DeBeukelaer (Toledo, OH, USA); C. Kozma (Washington, DC, USA); R. Schonberg (Washington, DC, USA); M. Bitzan (Winston-Salem, NC, USA).
Footnotes
STATEMENT
I [Friedhelm Hildebrandt] the Corresponding Author of this article (“Mutation Analysis of 18 Nephronophthisis-associated Ciliopathy Disease Genes using a DNA Pooling and Next-Generation Sequencing Strategy”) has the right to grant on behalf of all authors and does grant on behalf of all authors, a license to the BMJ Publishing Group Ltd and its licensees, to permit this Contribution (if accepted) to be published in Journal of Medical Genetics (JMG) and any other BMJ Group products and to exploit all subsidiary rights, as set out in our license set out at: (http://jmg.bmj.com/site/about/licence.pdf)
Competing Interest: None to declare.
REFERENCES
- 1.Hildebrandt F, Zhou W. Nephronophthisis-associated ciliopathies. J Am Soc Nephrol. 2007;18:1855–1871. doi: 10.1681/ASN.2006121344. [DOI] [PubMed] [Google Scholar]
- 2.Hildebrandt F, Otto E. Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat Rev Genet. 2005;6:928–940. doi: 10.1038/nrg1727. [DOI] [PubMed] [Google Scholar]
- 3.Parisi MA. Clinical and molecular features of Joubert syndrome and related disorders. Am J Med Genet C Semin Med Genet. 2009;151C:326–340. doi: 10.1002/ajmg.c.30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hildebrandt F, Otto E, Rensing C, Nothwang HG, Vollmer M, Adolphs J, Hanusch H, Brandis M, et al. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet. 1997;17:149–153. doi: 10.1038/ng1097-149. [DOI] [PubMed] [Google Scholar]
- 5.Otto E, Hoefele J, Ruf R, Mueller AM, Hiller KS, Wolf MT, Schuermann MJ, Becker A, Birkenhager R, Sudbrak R, Hennies HC, Nurnberg P, Hildebrandt F. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am J Hum Genet. 2002;71:1161–1167. doi: 10.1086/344395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mollet G, Salomon R, Gribouval O, Silbermann F, Bacq D, Landthaler G, Milford D, Nayir A, Rizzoni G, Antignac C, Saunier S. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat Genet. 2002;32:300–305. doi: 10.1038/ng996. [DOI] [PubMed] [Google Scholar]
- 7.Otto EA, Schermer B, Obara T, O’Toole JF, Hiller KS, Mueller AM, Ruf RG, Hoefele J, Beekmann F, Landau D, Foreman JW, Goodship JA, Strachan T, Kispert A, Wolf MT, Gagnadoux MF, Nivet H, Antignac C, Walz G, Drummond IA, Benzing T, Hildebrandt F. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olbrich H, Fliegauf M, Hoefele J, Kispert A, Otto E, Volz A, Wolf MT, Sasmaz G, Trauer U, Reinhardt R, Sudbrak R, Antignac C, Gretz N, Walz G, Schermer B, Benzing T, Hildebrandt F, Omran H. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- 9.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole JF, Helou J, Attanasio M, Utsch B, Sayer JA, Lillo C, Jimeno D, Coucke P, De Paepe A, Reinhardt R, Klages S, Tsuda M, Kawakami I, Kusakabe T, Omran H, Imm A, Tippens M, Raymond PA, Hill J, Beales P, He S, Kispert A, Margolis B, Williams DS, Swaroop A, Hildebrandt F. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Løken syndrome and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 10.Sayer JA, Otto EA, O’Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG, Allen SJ, Wilkinson CJ, Nigg EA, Shou C, Lillo C, Williams DS, Hoppe B, Kemper MJ, Neuhaus T, Parisi MA, Glass IA, Petry M, Kispert A, Gloy J, Ganner A, Walz G, Zhu X, Goldman D, Nurnberg P, Swaroop A, Leroux MR, Hildebrandt F. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–681. doi: 10.1038/ng1786. Individuals with mutations in XPNPEP3, which encodes a mitochondrial protein, develop a nephronophthisis-like nephropathy. [DOI] [PubMed] [Google Scholar]
- 11.Attanasio M, Uhlenhaut NH, Sousa VH, O’toole JF, Otto E, Anlag K, Klugmann C, Treier AC, Helou J, Sayer JA, Seelow D, Nurnberg G, Becker C, Chudley AE, Nurnberg P, Hildebrandt F, Treier M. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- 12.Delous M, Baala L, Salomon R, Laclef C, Vierkotten J, Tory K, Golzio C, Lacoste T, Besse L, Ozilou C, Moutkine I, Hellman NE, Anselme I, Silbermann F, Vesque C, Gerhardt C, Rattenberry E, Wolf MT, Gubler MC, Martinovic J, Encha-Razavi F, Boddaert N, Gonzales M, Macher MA, Nivet H, Champion G, Bertheleme JP, Niaudet P, McDonald F, Hildebrandt F, Johnson CA, Vekemans M, Antignac C, Ruther U, Schneider-Maunoury S, Attie-Bitach T, Saunier S. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 13.Otto EA, Trapp ML, Schultheiss UT, Helou J, Quarmby LM, Hildebrandt F. Mutations in NIMA-related kinase NEK8 affects ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol. 2008a;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto EA, Tory K, Attanasio M, Zhou W, Chaki M, Paruchuri Y, Wise EL, Wolf MT, Utsch B, Becker C, Nürnberg G, Nürnberg P, Nayir A, Saunier S, Antignac C, Hildebrandt F. Hypomorphic mutations in meckelin (MKS3/TMEM67) cause nephronophthisis with liver fibrosis (NPHP11) J Med Genet. 2009;46:663–670. doi: 10.1136/jmg.2009.066613. [DOI] [PubMed] [Google Scholar]
- 15.O’Toole JF, Liu Y, Davis EE, Westlake CJ, Attanasio M, Otto EA, Seelow D, Nurnberg G, Becker C, Nuutinen M, Kärppä M, Ignatius J, Uusimaa J, Pakanen S, Jaakkola E, van den Heuvel LP, Fehrenbach H, Wiggins R, Goyal M, Zhou W, Wolf MT, Wise E, Helou J, Allen SJ, Murga-Zamalloa CA, Ashraf S, Chaki M, Heeringa S, Chernin G, Hoskins BE, Chaib H, Gleeson J, Kusakabe T, Suzuki T, Isaac RE, Quarmby LM, Tennant B, Fujioka H, Tuominen H, Hassinen I, Lohi H, van Houten JL, Rotig A, Sayer JA, Rolinski B, Freisinger P, Madhavan SM, Herzer M, Madignier F, Prokisch H, Nurnberg P, Jackson PK, Khanna H, Katsanis N, Hildebrandt F. J Clin Invest. 2010;120:791–802. doi: 10.1172/JCI40076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferland RJ, Eyaid W, Collura RV, Tully LD, Hill RS, Al-Nouri D, Al-Rumayyan A, Topcu M, Gascon G, Bodell A, Shugart YY, Ruvolo M, Walsh CA. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 17.Valente EM, Logan CV, Mougou-Zerelli S, Lee JH, Silhavy JL, Brancati F, Iannicelli M, Travaglini L, Romani S, Illi B, Adams M, Szymanska K, Mazzotta A, Lee JE, Tolentino JC, Swistun D, Salpietro CD, Fede C, Gabriel S, Russ C, Cibulskis L, Sougnez C, Hildebrandt F, Otto EA, Held S, Diplas BH, Davis EE, Mikula M, Strom CM, Ben-Ze’ev B, Lev D, Sagie TL, Michelson M, Yaron Y, Krause A, Boltshauser E, Elkhartoufi N, Roume J, Shalev S, Munnich A, Saunier S, Inglehearn C, Saad A, Alkindy A, Thomas S, Vekemans M, Dallapiccola B, Katsanis N, Johnson CA, Attié-Bitach T, Gleeson JG. Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat Genet. 2010 doi: 10.1038/ng.594. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS, Abdel-Aleem A, Rosti RO, Kayserili H, Swistun D, Scott LC, Bertini E, Boltshauser E, Fazzi E, Travaglini L, Field SJ, Gayral S, Jacoby M, Schurmans S, Dallapiccola B, Majerus PW, Valente EM, Gleeson JG. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parisi MA, Bennett CL, Eckert ML, Dobyns WB, Gleeson JG, Shaw DW, McDonald R, Eddy A, Chance PF, Glass IA. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am J Hum Genet. 2004;75:82–91. doi: 10.1086/421846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, Fazzi E, Signorini S, Louie CM, Bellacchio E, Bertini E, Dallapiccola B, Gleeson JG International Joubert Syndrome Related Disorders Study Group. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 21.Arts HH, Doherty D, van Beersum SE, Parisi MA, Letteboer SJ, Gorden NT, Peters TA, Märker T, Voesenek K, Kartono A, Ozyurek H, Farin FM, Kroes HY, Wolfrum U, Brunner HG, Cremers FP, Glass IA, Knoers NV, Roepman R. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 22.Baala L, Romano S, Khaddour R, Saunier S, Smith UM, Audollent S, Ozilou C, Faivre L, Laurent N, Foliguet B, Munnich A, Lyonnet S, Salomon R, Encha-Razavi F, Gubler MC, Boddaert N, de Lonlay P, Johnson CA, Vekemans M, Antignac C, Attie-Bitach T. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am J Hum Genet. 2007a;80:186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attié-Bitach T, Holden KR, Dobyns WB, Traver D, Al-Gazali L, Ali BR, Lindner TH, Caspary T, Otto EA, Hildebrandt F, Glass IA, Logan CV, Johnson CA, Bennett C, Brancati F, Valente EM, Woods CG, Gleeson JG International Joubert Syndrome Related Disorders Study Group. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorden NT, Arts HH, Parisi MA, Coene KL, Letteboer SJ, van Beersum SE, Mans DA, Hikida A, Eckert M, Knutzen D, Alswaid AF, Ozyurek H, Dibooglu S, Otto EA, Liu Y, Davis EE, Hutter CM, Bammler TK, Farin FM, Dorschner M, Topçu M, Zackai EH, Rosenthal P, Owens KN, Katsanis N, Vincent JB, Hildebrandt F, Rubel EW, Raible DW, Knoers NV, Chance PF, Roepman R, Moens CB, Glass IA, Doherty D. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am J Hum Genet. 2008;83:559–571. doi: 10.1016/j.ajhg.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis EE, Zhang Q, Diplas BH, Davey LM, Hartley J, Stoetzel C, Szymanska K, Muzny DM, Young AC, Wheeler DA, Cruz P, Morgan M, Lewis LR, Cherukuri P, Maskeri B, Mullikin JC, Blakesley RW, Blakesley RW, Lewis RA, Bergmann C, Otto EA, Saunier S, Scambler PJ, Beales PL, Gleeson JG, Maher ER, Attié-Bitach T, Dollfus H, Johnson CA, Green ED, Gibbs RA, Hildebrandt F, Pierce EA, Katsanis N. Mutations in IFT139 contribute both causal and modifying alleles across the ciliopathy spectrum. Manuscript submitted for publication. 2010 [Google Scholar]
- 26.Kyttälä M, Tallila J, Salonen R, Kopra O, Kohlschmidt N, Paavola-Sakki P, Peltonen L, Kestilä M. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat Genet. 2006;38:155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- 27.Smith UM, Consugar M, Tee LJ, McKee BM, Maina EN, Whelan S, Morgan NV, Goranson E, Gissen P, Lilliquist S, Aligianis IA, Ward CJ, Pasha S, Punyashthiti R, Malik Sharif S, Batman PA, Bennett CP, Woods CG, McKeown C, Bucourt M, Miller CA, Cox P, Algazali L, Trembath RC, Torres VE, Attie-Bitach T, Kelly DA, Maher ER, Gattone VH2nd, Harris PC, Johnson CA. The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat Genet. 2006;38:191–196. doi: 10.1038/ng1713. [DOI] [PubMed] [Google Scholar]
- 28.Baala L, Audollent S, Martinovic J, Ozilou C, Babron MC, Sivanandamoorthy S, Saunier S, Salomon R, Gonzales M, Rattenberry E, Esculpavit C, Toutain A, Moraine C, Parent P, Marcorelles P, Dauge MC, Roume J, Le Merrer M, Meiner V, Meir K, Menez F, Beaufrère AM, Francannet C, Tantau J, Sinico M, Dumez Y, MacDonald F, Munnich A, Lyonnet S, Gubler MC, Génin E, Johnson CA, Vekemans M, Encha-Razavi F, Attié-Bitach T. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet. 2007b;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tallila J, Jakkula E, Peltonen L, Salonen R, Kestilä M. Identification of CC2D2A as a Meckel syndrome gene adds an important piece to the ciliopathy puzzle. Am J Hum Genet. 2008;82:1361–1367. doi: 10.1016/j.ajhg.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otto EA, Helou J, Allen SJ, O’Toole JF, Wise EL, Ashraf S, Attanasio M, Zhou W, Wolf MT, Hildebrandt F. Mutation analysis in nephronophthisis using a combined approach of homozygosity mapping, CEL I endonuclease cleavage, and direct sequencing. Hum Mutat. 2008b;3:418–426. doi: 10.1002/humu.20669. [DOI] [PubMed] [Google Scholar]
- 31.Anderson S. Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids Res. 1981;9:3015–3027. doi: 10.1093/nar/9.13.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tory K, Lacoste T, Burglen L, Morinière V, Boddaert N, Macher MA, Llanas B, Nivet H, Bensman A, Niaudet P, Antignac C, Salomon R, Saunier S. High NPHP1 and NPHP6 mutation rate in patients with Joubert syndrome and nephronophthisis: potential epistatic effect of NPHP6 and AHI1 mutations in patients with NPHP1 mutations. J Am Soc Nephrol. 2007;18:1566–1575. doi: 10.1681/ASN.2006101164. [DOI] [PubMed] [Google Scholar]
- 34.Perrault I, Delphin N, Hanein S, Gerber S, Dufier JL, Roche O, Defoort-Dhellemmes S, Dollfus H, Fazzi E, Munnich A, Kaplan J, Rozet JM. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum Mutat. 2007;28:416. doi: 10.1002/humu.9485. [DOI] [PubMed] [Google Scholar]
- 35.Brancati F, Barrano G, Silhavy JL, Marsh SE, Travaglini L, Bielas SL, Amorini M, Zablocka D, Kayserili H, Al-Gazali L, Bertini E, Boltshauser E, D’Hooghe M, Fazzi E, Fenerci EY, Hennekam RC, Kiss A, Lees MM, Marco E, Phadke SR, Rigoli L, Romano S, Salpietro CD, Sherr EH, Signorini S, Stromme P, Stuart B, Sztriha L, Viskochil DH, Yuksel A, Dallapiccola B, Valente EM, Gleeson JG International JSRD Study Group. CEP290 mutations are frequently identified in the oculo-renal form of Joubert syndrome-related disorders. Am J Hum Genet. 2007;81:104–113. doi: 10.1086/519026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tallila J, Salonen R, Kohlschmidt N, Peltonen L, Kestilä M. Mutation spectrum of Meckel syndrome genes: one group of syndromes or several distinct groups? Hum Mutat. 2009;30:E813–830. doi: 10.1002/humu.21057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khaddour R, Smith U, Baala L, Martinovic J, Clavering D, Shaffiq R, Ozilou C, Cullinane A, Kyttälä M, Shalev S, Audollent S, d’Humières C, Kadhom N, Esculpavit C, Viot G, Boone C, Oien C, Encha-Razavi F, Batman PA, Bennett CP, Woods CG, Roume J, Lyonnet S, Génin E, Le Merrer M, Munnich A, Gubler MC, Cox P, Macdonald F, Vekemans M, Johnson CA, Attié-Bitach T SOFFOET (Société Française de Foetopathologie) Spectrum of MKS1 and MKS3 mutations in Meckel syndrome: a genotype-phenotype correlation. Hum Mutat. 2007;28:523–524. doi: 10.1002/humu.9489. [DOI] [PubMed] [Google Scholar]
- 38.Brancati F, Iannicelli M, Travaglini L, Mazzotta A, Bertini E, Boltshauser E, D’Arrigo S, Emma F, Fazzi E, Gallizzi R, Gentile M, Loncarevic D, Mejaski-Bosnjak V, Pantaleoni C, Rigoli L, Salpietro CD, Signorini S, Stringini GR, Verloes A, Zabloka D, Dallapiccola B, Gleeson JG, Valente EM International JSRD Study Group. MKS3/TMEM67 mutations are a major cause of COACH Syndrome, a Joubert Syndrome related disorder with liver involvement. Hum Mutat. 2009;30:E432–E442. doi: 10.1002/humu.20924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consugar MB, Kubly VJ, Lager DJ, Hommerding CJ, Wong WC, Bakker E, Gattone VH2nd, Torres VE, Breuning MH, Harris PC. Molecular diagnostics of Meckel-Gruber syndrome highlights phenotypic differences between MKS1 and MKS3. Hum Genet. 2007;121:591–599. doi: 10.1007/s00439-007-0341-3. [DOI] [PubMed] [Google Scholar]
- 40.Mougou-Zerelli S, Thomas S, Szenker E, Audollent S, Elkhartoufi N, Babarit C, Romano S, Salomon R, Amiel J, Esculpavit C, Gonzales M, Escudier E, Leheup B, Loget P, Odent S, Roume J, Gérard M, Delezoide AL, Khung S, Patrier S, Cordier MP, Bouvier R, Martinovic J, Gubler MC, Boddaert N, Munnich A, Encha-Razavi F, Valente EM, Saad A, Saunier S, Vekemans M, Attié-Bitach T. CC2D2A mutations in Meckel and Joubert syndromes indicate a genotype-phenotype correlation. Hum Mutat. 2009;30:1574–1582. doi: 10.1002/humu.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoefele J, Sudbrak R, Reinhardt R, Lehrack S, Hennig S, Imm A, Muerb U, Utsch B, Attanasio M, O’Toole JF, Otto E, Hildebrandt F. Mutational analysis of the NPHP4 gene in 250 patients with nephronophthisis. Hum Mutat. 2005;4:411. doi: 10.1002/humu.9326. [DOI] [PubMed] [Google Scholar]
- 42.Wolf MT, Saunier S, O’Toole JF, Wanner N, Groshong T, Attanasio M, Salomon R, Stallmach T, Sayer JA, Waldherr R, Griebel M, Oh J, Neuhaus TJ, Josefiak U, Antignac C, Otto EA, Hildebrandt F. Mutational analysis of the RPGRIP1L gene in patients with Joubert syndrome and nephronophthisis. Kidney Int. 2007;12:1520–1526. doi: 10.1038/sj.ki.5002630. [DOI] [PubMed] [Google Scholar]
- 43.Varley KE, Mitra RD. Nested Patch PCR enables highly multiplexed mutation discovery in candidate genes. Genome Res. 2008;11:1844–1850. doi: 10.1101/gr.078204.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.