Abstract
BACKGROUND & AIMS
Human primary liver cancer (PLC) is classified into biologically distinct subgroups, based on cellular origin. Liver cancer stem cells (CSCs) have been recently described. We investigated the ability of distinct lineages of hepatic cells to become liver CSCs and the phenotypic and genetic heterogeneity of PLC.
METHODS
We transduced mouse primary hepatic progenitor cells (HPC), lineage-committed hepatoblasts, and differentiated adult hepatocytes with transgenes encoding oncogenic H-Ras and simian virus 40 large-T antigen. The CSC properties of transduced cells and their ability to form tumors were tested by standard in vitro and in vivo assays and transcriptome profiling.
RESULTS
Irrespective of origin, all transduced cells acquired markers of CSC/progenitor cells, side populations, and self-renewal capacity in vitro. They also formed a broad spectrum of liver tumors, ranging from cholangiocarcinoma to hepatocellular carcinoma, which resembled human liver tumors, based on genomic and histologic analyses. The tumor cells co-expressed hepatocyte (HNF4A), biliary progenitor cell (keratin 19, EpCAM, A6), and mesenchyme (vimentin) markers and showed disregulation of genes that control the epithelial–mesenchymal transition. Gene expression analyses could distinguish tumors of different cellular origin, indicating the contribution of lineage-stage dependent genetic changes to malignant transformation. Activation of c-Myc and its target genes was required to reprogram adult hepatocytes into CSC and for tumors to develop. Stable knockdown of c-Myc in transformed adult hepatocytes reduced their CSC properties in vitro and suppressed growth of tumors in immunodeficient mice.
CONCLUSIONS
Any cell type in the mouse hepatic lineage can undergo oncogenic reprogramming into a CSC, by activating different cell type-specific pathways. Identification of common and cell-of-origin specific phenotypic and genetic changes could provide new therapeutic targets for liver cancer.
Keywords: Cell of origin, Cancer stem cell, HCC, c-MYC
The cardinal hallmark of cancer is a profound heterogeneity in cellular morphology, genetic landscape and response to therapeutic interventions. The nature of heterogeneity seen in tumors from the same and different organs remains an unresolved issue1. It is however recognized that activation of the same oncogenic processes at different lineage stages can affect both malignant potential and tumor morphology2,3.
Hepatocellular carcinoma (HCC) and cholangiocarcinoma (CCA) are the major primary adult human liver cancers. Both HCC and CCA are morphologically, genomically and clinically very heterogeneous with dismal clinical outcome4. In addition, a rare form of PLC, combined hepatocellular-cholangiocarcinoma (CHC) is now recognized which shares morphologic characteristics with HCC and CCA5. Although still debated, hepatocytes, cholangiocytes and adult liver stem/progenitor cell have been proposed as cells of origin for some or all PLC subtypes6. We have recently applied an integrative oncogenomic approach to address the clinical and functional implications of the overlapping phenotypes between HCC, CHC, and CCA, and identified a novel HCC subtype, CCA-like HCC (CLHCC), which expressed CCA traits7. Like CCA and CHC, CLHCC showed an aggressive behavior with shorter recurrence-free and overall survival as compared to HCC, and co-expressed embryonic stem cell (ESC)-like expression traits suggesting its derivation from bi-potential hepatic progenitor cells.
Many solid tumors contain a subset of cells that possess functional properties ascribed to normal stem cells, such as self-renewal, unlimited proliferative capacity and multi-potency (i.e. capacity of generating all tumorigenic and non-tumorigenic cell types in the tumor), leading to a hierarchical model of cancer with a CSC population at the apex of tumor formation8. The existence of CSCs (also known as tumor initiating cells) has been shown in a variety of solid tumors, including liver cancer8–10. Although it seems reasonable that evolution of CSCs from cells at different stages of differentiation may contribute, at least in part, to the phenotypic and genetic heterogeneity seen in liver cancer, whether a lineage stage may be a factor in acquisition of stemness properties at the cellular and molecular levels is not yet understood.
Here, we addressed whether the differentiation stage of distinct hepatic lineage cells (1) dictates the acquisition of CSC properties and (2) contributes to the phenotypic and genetic heterogeneity of PLC.
Materials and Methods
Isolation and Transduction of Hepatic Lineage Cells
All procedures were performed according to protocols approved by the Animal Care and Use Committee of the National Institutes of Health. C57BL/6NCr mice (National Laboratory for Cancer Research, NCI, Frederick, MD) were used for isolating HPCs, HBs and AHs. B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice (The Jackson Laboratory, Bar Harbor, ME) were used for isolating genetically labeled AHs. HPCs were activated with 0.1% 3,5-diethoxycarbonyl-1,4-dihydrocollidine (Bioserv, Frenchtown, NJ) diet and FACS sorted (FACS Vantage Cell Sorter, BD, San Jose, CA) as epithelial cell adhesion molecule (EpCAM)+ (a gift of Dr. Miyajima)/Lineage Cocktail− (BD) cells11. E-cadherin+ HBs were purified using MACS system (Miltenyi, Auburn, CA) from ED16.5 fetal livers12. AHs were isolated by a two-step collagenase (Worthington, Lakewood, NJ) perfusion method from 3-month-old male mice13. Construction and production of lentiviral vectors expressing oncogenic H-Ras-Luciferase/EGFP and SV40LT-mCherry is described in Supplementary Material. Cells were co-transduced with concentrated lentiviruses 24 hours after plating and cultured for 3 weeks to collect sufficient number of transduced cells from low frequency HPCs. H-Ras-EGFP+/SV40LT-mCherry+ HPCs, HBs and AHs were sorted using FACS Vantage Cell Sorter (BD).
In Vivo Experiments
Immunodeficient NOD/SCID mice (National Laboratory for Cancer Research, NCI) were used for cell transplantation experiments. For limiting dilution assay, 10, 100 and 1000 H-Ras-EGFP+/SV40LT-mCherry+ cells were injected subcutaneously into the lower flanks (4 mice/group). Frequencies of tumor initiating cells were calculated by L-Calc software (Stemcell Technologies, Tukwila, WA) 5 weeks after transplantation. To assess orthotopic growth and establish tumor cell lines, 150,000 sorted cells were injected into the left liver lobe (5 mice/group). Mice were subjected to bioluminescent imaging using Xenogen-IVIS-200 Imaging System (Caliper Life Sciences, Hopkinton, MA) twice a week, and sacrificed when they displayed symptoms of disease (16–18 days after transplantation). Liver tumors were dissociated, and sorted H-Ras-EGFP+/SV40LT-mCherry+ cells were propagated to establish tumor cell lines. To assess the effect of c-Myc knockdown on tumor growth, 100 H-Ras-EGFP+/SV40LT-mCherry+ AHs expressing c-Myc shRNA (a gift of Dr. Manley)14 or scrambled shRNA were injected subcutaneously as described above. Tumor length (l) and width (w) were measured by external caliper once a week. Tumor volume (v) in mm3 was calculated using the formula: v = l × w2/2.
In Vitro Experiments
Analysis of side population (SP) was performed as described10. Expression of hepatic lineage and CSC markers was analyzed on LSRII Flow Cytometer (BD) using antibodies and corresponding isotype controls described in Supplementary Material. For spheroid formation assay, 500 cells/well were seeded in multiple wells of ultra-low attachment 96-well plates (Corning, Tewksbury, MA) in serum-free growth medium containing 1% methylcellulose (R&D Systems, Minneapolis, MN). Spheroids were dissociated and replated once a week for 6 weeks. Average number and diameters (d1; d2) of spheroids were calculated using ImageJ software (NIH, Bethesda, MD). Spheroid volumes were calculated in μm3 using the formula: v = d1 × d2 2/2, where d2 designates the shorter diameter. Spheroids with diameter less than 50 μm were excluded from analysis. Nuclear ploidy was determined using using Cycletest Plus DNA Reagent Kit (BD) according to the manufacturer's protocol. Quantitative reverse transcription polymerase chain reaction and western blotting was performed as described previously11,12. Primers are listed in Supplementary Table 1, antibodies are described in Supplementary Material.
Histology and Immunohistochemistry
Individual liver tumors (≥3mm diameter) were macrodissected 16–18 days after intrasplenic injection of H-Ras-EGFP+/SV40LT-mCherry+ cells and divided into two parts for immunohistochemical and microarray analyses. Mean percentage of tumor areas occupied by HCC-, CCA- and epithelial-mesenchymal transition (EMT)-like phenotypes was analyzed semiquantitatively on hematoxylin-eosin stained tumor sections by A.H. and V.M.F. Paraffin-embedded tumor sections were stained with anti-H-Ras (Life Technologies), anti-SV40LT (Abcam), anti-hepatocyte nuclear factor 4 alpha (HNF4A) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-keratin 19 (Developmental Studies Hybridoma Bank), anti-laminin (Abcam, Cambridge, MA), anti-vimentin (Abcam) and anti-A615.
Microarray
Total RNA of histologically confirmed liver tumors and primary HPCs, HBs and AHs were isolated by RNeasy Mini Kit (Qiagen, Valencia, CA). Linear amplification of 400 ng RNA was performed with Illumina TotalPrep RNA Amplification Kit (Life Technologies). A total of 750 ng cRNA of each sample were hybridized on Sentrix Mouse Expression BeadChips version 2 (Illumina, San Diego, CA). Data analysis is described in Supplementary Material. All microarray data were submitted to GEO (accession number GSE41312), and accessible at http://www.ncbi.nlm.nih.gov/geo/.
Statistics
Data were analyzed by Student's t-test, Mann-Whitney test, Poisson GLM test, 1-way ANOVA and Tukey's post hoc test as indicated. P values less than 0.05 were considered significant.
Results
Oncogenic H-Ras and SV40LT Reprogram Progenitor Cells and Mature Hepatocytes into Cancer Stem Cells
Activation of Ras pathway and disruption of p53 and Rb pathways are commonly found in rodent16 and human4,17 HCCs. To study the contribution of lineage stage to liver oncogenesis, we stably co-transduced primary cultures of murine HPCs, HBs and AHs with oncogenic H-Ras-Luciferase/EGFP and SV40LT-mCherry lentiviral vectors (a combination referred to as H-Ras/SV40LT). H-Ras-EGFP+/SV40LT-mCherry+ HPCs, HBs and HPCs were FACS sorted with the same gating parameters to ensure comparable viral load and transgene expression (Supplementary Figure 1A and B). All three cell types acquired CSC properties as defined by increase and/or acquisition of SP fraction18, CD133 expression9, and ability to grow as self-renewing spheres19 (Figure 1A–D). However, limiting dilution analysis revealed significantly higher frequency of tumor-initiating cells among transduced HPCs. As few as 10 transduced HPCs produced tumors in 6/8 injections compared to transduced HBs (2/8) and AHs (0/8) by 5 weeks after subcutaneous transplantation (Figure 2A). Ex vivo bioluminescence imaging revealed the highly metastatic nature of tumors (Supplementary Figure 2A and B). Similar results were obtained with orthotopic transplantation (Figure 2B–D). In contrast, subcutaneous injection of 3 million normal HPCs did not generate tumor after 6 months.
Figure 1.
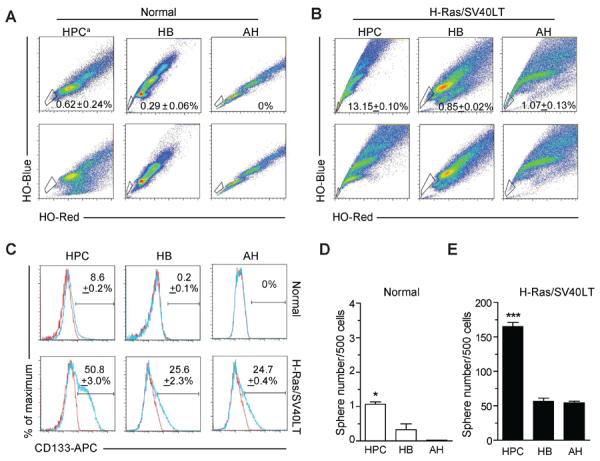
H-Ras/SV40LT-transduced HPCs, HBs and AHs acquire cancer stem cell properties in vitro. (A and B) Analysis of side population (SP) by flow cytometry in freshly isolated normal (A) and transduced (B) hepatic lineage cells. SP cells were identified by Hoechst 33342 (HO) staining. Fumitremorgin was used to set up the SP gate (FACS plots at the bottom). HPC: hepatic progenitor cell; HB: hepatoblast; AH: adult hepatocyte; a cultured HPCs at passage 5. Numbers represent mean ± SD of three experiments. (C) Analysis of CD133 expression by flow cytometry. Blue line: CD133-APC; red line: isotype control. Numbers represent mean ± SD of three experiments. (D–E) Spheroid forming ability. Freshly isolated normal (D) and transduced (E) hepatic lineage cells were cultured at low density in 1% methylcellulose. Sphere number was quantified after 7 days. Data represent mean ± SD of four experiments. Significant differences were evaluated by ANOVA and Poisson GLM. * P < 0.05; *** P < 0.001.
Figure 2.
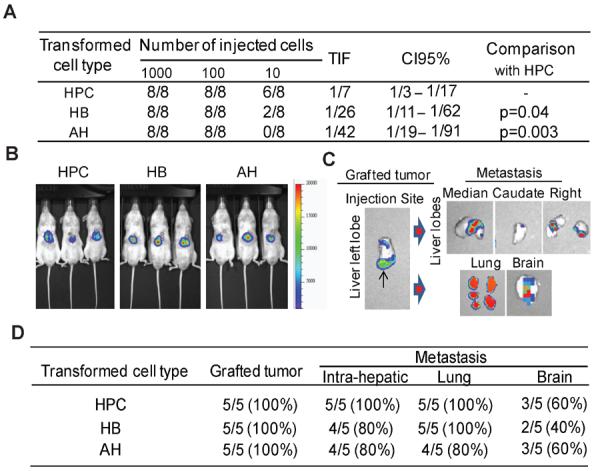
H-Ras/SV40LT-transduced HPCs, HBs and AHs give rise to fast growing tumors in two models of transplantation. (A) Limiting dilution analysis. H-Ras-Luciferase/EGFP and SV40LT-mCherry double positive cells were FACS sorted and injected subcutaneously in lower flanks of NOD/SCID mice. The frequency of tumor initiating cells (TIF) and confidence interval (CI95%) were calculated based on the number of resulting tumors/injection after 5 weeks. (B–D) Orthotopic tumor growth and metastatic ability. (B) Representative bioluminescence images of mice at 11 days after transplantation of 150,000 cells of each type. (C) Ex vivo bioluminescence imaging of liver, lung and brain 16 days after transplantation. (D) Incidence of primary grafted tumors and intrahepatic, lung and brain metastases.
To gain more insight into the tumorigenicity of the transformed cells, we established four clonal cell lines from each HPC-, HB- and AH-derived liver tumors. Irrespective of tumor cell-of-origin, all cell lines expressed hepatic progenitor/biliary (keratin 19, EpCAM, A6)6,15,20 and CSC-associated (CD133, CD44, CD29, CD49f, CD90, Sca-1)9,18,21 markers, had comparable size of SP fraction, and possessed high self-renewal capacity through 6 passages (Supplementary Figure 3A–D). This indicates that any hepatic lineage cell was susceptible to oncogene-driven transformation, and could acquire similar attributes of liver CSC producing aggressive liver cancer. However, primitive HPCs were more susceptible to transformation than more differentiated HBs and AHs.
Unambiguous in Vitro Transformation of Terminally Differentiated Hepatocytes
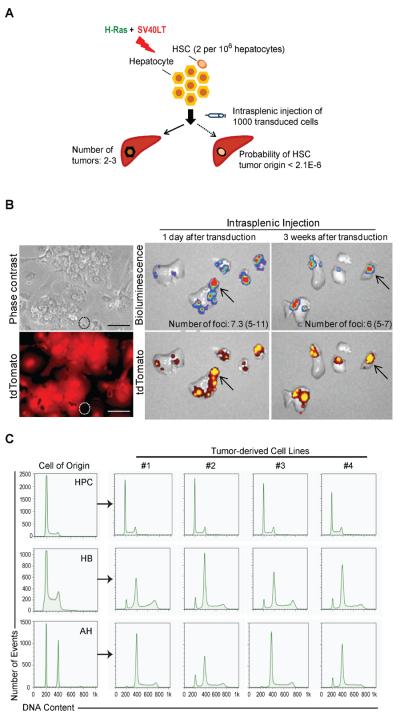
Hepatic stem cells (HSC) are extremely rare in normal adult mouse liver22,23. Although we optimized primary AH isolation to obtain hepatocytes of high purity, it could not be ruled out that contaminating HSCs were targeted by the transforming oncogenes and selectively amplified during 3-week-growth in culture. To test this possibility, we first compared the estimated number of HSCs in normal adult mouse liver with the frequency of liver tumors initiated after intrasplenic transplantation of low number (1000) of H-Ras/SV40LT-transduced AHs. Given that the frequency of EpCAM+ non-parenchymal cells in primary AH culture was on average 0.13%, and only 0.16% of this fraction possessed sphere-forming potential and ability to differentiate along hepatocytic or biliary epithelial lineages20, the estimated number of HSCs did not exceed 2 per 106 hepatocytes. To avoid in vitro selection bias, transduced AHs were maintained in culture for only one day before transplantation. Transplanted cells produced 2–3 liver tumors per mouse by 18 days after injection. Assuming 100% efficiency of transduction and transformation, the probability that the tumors were derived from transduced HSCs is negligible (≤ 2.1×10−6) (Figure 3A).
Figure 3.
Tumors are derived from transformed AHs but not from HSCs. (A) Schematic overview of the approach used to compare the number of resulting tumors with the estimated frequency of hepatic stem cells (HSCs). Primary AH culture was transduced and cultured for only 1 day to exclude the possibility of selective overgrowth of HSCs. One thousand transduced cells were injected via spleen into NOD/SCID mice and liver tumors were counted after 18 days. Probability of tumor initiation by transduced HSCs was calculated using binomial distribution. (B) Left panel: phase contrast and fluorescence images of 24-hour primary hepatocyte culture established from Rosa26-CAG-stop-tdTomato mouse one week after i.v. injection of Ad-Cre virus. Dotted circle marks tdTomato-negative non-parenchymal cells. Scale bar: 100 μm. Right panel: Ex vivo bioluminescence and fluorescence images of livers 18 days after injection of 105 hepatocytes co-transduced with H-Ras/SV40LT and cultured for 1 or 21 days prior to intrasplenic transplanatation. (C) Flow cytometry analysis of DNA content in tumor cell lines (#1–4) established from liver tumors initiated by H-Ras/SV40LT-transduced HPCs, HBs and AHs.
We next isolated and transduced genetically labeled primary AHs from ROSA26-CAG-stop-tdTomato reporter mice24. AHs showed strong, homogeneous expression of tdTomato upon Adeno-Cre-mediated recombination (Figure 3B, left panel). Regardless of the duration of in vitro culture (1 day versus 21 days), transduced AHs produced tumors with comparable frequencies and displayed overlapping luciferin and tdTomato signals indicating that the tumors originated from AHs (Figure 3B, right panel).
Lastly, we found a significant increase in nuclear ploidy in AH tumors, a characteristic of differentiated hepatocytes25. In contrast, cells isolated from HPC tumors were diploid (Figure 3C). These results demonstrate that the terminally differentiated AHs but not contaminating HSCs were targets of oncogenic transformation.
H-Ras/SV40LT Induce Liver Cancer of Multilineage Differentiation
To examine the impact of cell-of-origin on tumor histopathology, we subjected 14 HPC-, 28 HB- and 28 AH-derived liver tumors to histopathological analyses. Tumor cells retained strong sub-membranous H-Ras and nuclear SV40LT staining confirming their origin from H-Ras/SV40LT-expressing cells (Supplementary Figure 4). Irrespective of cell-of-origin, tumors were moderately to poorly differentiated with varying contribution of HCC-, CCA- and EMT-like phenotypes supporting the concept of continuous spectrum of human PLC26 (Figure 4A and Supplementary Figure 5). AH tumors showed a predominant HCC-like phenotype (on average 60% of the tumor cross-section areas) characterized by polygonal, hepatocyte-like tumor cells arranged in solid pattern. HB tumors displayed mostly CCA-like phenotype (53%) composed of columnar or cuboid cholangiocyte-like tumor cells arranged in glandular structures surrounded by abundant fibrous stroma. HPC tumors had mostly EMT-like phenotype (85%) characterized by sheets of spindle-shaped, mesenchymal-like cancer cells. Majority of HCC-like tumor cells expressed HNF4A, a central mediator of hepatocyte differentiation27. HNF4A was also detected in CCA- and EMT-like tumor cells albeit with lower frequency. We observed strong, uniform expression of progenitor/biliary markers keratin 19 and A6 (Figure 4A) regardless of tumor cell-of-origin. Furthermore, EMT- and HCC-like tumor cells showed intense cytoplasmic and extracellular staining for laminin, a component of the hepatic progenitor cell niche in rodent and human livers23, and were uniformly positive for mesenchymal marker vimentin.
Figure 4.
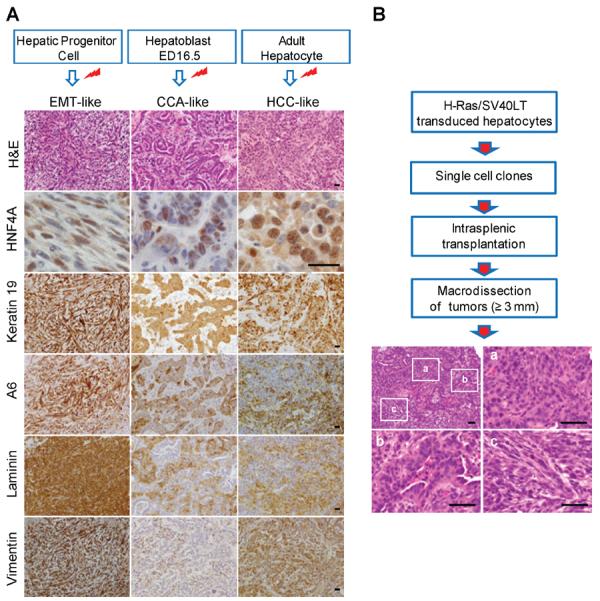
(A) Representative H&E images and immunostaining of HCC-, CCA- and EMT-like tumor phenotypes. Paraffin-embedded sections were counterstained with haematoxilin. Red marks indicate transduction with H-Ras/SV40LT. EMT: epithelial-mesenchymal transition; CCA: cholangiocarcinoma; HCC: hepatocellular carcinoma; H&E: hematoxylin-eosin; HNF4A: hepatocyte nuclear factor 4 alpha. Scale bar: 25 μm. (B) Schematic overview of the approach and representative H&E staining of 1/42 tumors derived from a single cell clone of HRas/SV40LT-transduced AHs. Letters a, b, and c denote HCC-, CCA- and EMT-like areas within the same tumor shown. Scale bar: 50 μm.
To provide additional evidence that all three tumor phenotypes were initiated by a single cell, we transplanted via spleen 15 single cell-derived clonal lines established from H-Ras/SV0LT-transduced AHs. Fourteen out of 15 clones (93.3%) showed comparable frequency of engraftment and kinetics of tumor growth. More significantly, all examined tumors (n=42) displayed overlapping HCC-CCA-EMT-like phenotypes indistinguishable from the tumors initiated by a bulk of transduced AHs (Figure 4B). We concluded that upon oncogenic transformation, murine HPCs, HBs and AHs were capable of initiating liver cancers of multilineage differentiation closely resembling human PLCs, and the differentiation stage of cell-of-origin had a profound impact on tumor phenotype.
Common Activation of EMT-Related Pathways in Distinct Hepatic Lineage Cells during Oncogenic Reprogramming
Next, we analyzed the transcriptome of tumors described above (n=50) to define key molecular similarities/differences between tumors and corresponding cell-of-origin (4 samples each). Tumor groups displayed higher degree of similarity to each other than to their cell-of-origin by bioequivalence test28 (Figure 5A). We identified 590 genes with significant common dysregulation among the three tumor groups (Figure 5B and Supplementary Table 2). Hierarchical clustering of common genes separated tumors according to their cell-of-origin (Figure 5C). A significant proportion of common genes were associated with EMT consistent with the highly metastatic nature of all three tumor groups (Supplementary Figure 6A). Gene set enrichment analysis (GSEA)29 showed significant enrichment of a 35-gene EMT-signature30 in HPC, HB and AH tumors (Supplementary Figure 6B).
Figure 5.
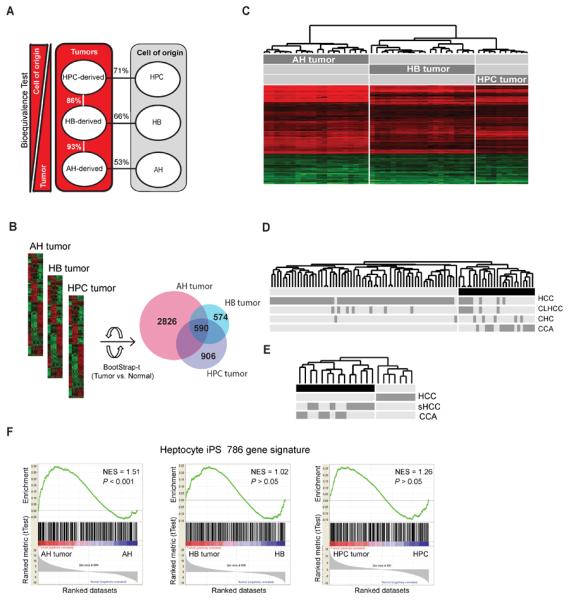
Transcriptomic characteristics of liver tumors derived from distinct hepatic lineage cells. (A) Bioequivalence test of similarities between HPC-, HB- and AH-derived tumors and their respective cell-of-origin. Data were evaluated at fold change >1.5 and P < 0.05. (B) Venn diagram of differentially expressed genes in HPC, HB and AH tumors after normalization to corresponding normal cells. Bootstrap-t test: P < 0.001; fold change (C) Supervised hierarchical clustering of tumors based on 590 commonly differentially expressed genes. (D and E) Hierarchical clustering of human PLC data sets including HCC, CCA, CCA-like HCC (CLHCC), combined hepatocellular-cholangiocarcinoma (CHC) and scirrhous HCC (sHCC) from Woo et al.7 (D) and Seok et al.26 (E) using murine to human homologue genes comprised within the 590-gene common gene signature. (F) Gene set enrichment analysis using a hepatocyte iPSC gene signature (786 genes)32. iPSC: induced pluripotent stem cell; NES: normalized enrichment score, P < 0.05 was considered significant.
Common Gene Signature Identifies Human PLC Subtypes
Human PLCs are pheno- and genotypically highly heterogeneous4,7. In this study we generated a mouse model that mimics subtypes of PLC and tested the utility of our signature to classify distinct PLC subtypes. When the 590-gene common gene signature was applied to a data set of 70 human HCCs, 13 CCAs and 7 CHCs7 using cross-species comparison, it correctly predicted 100% CCs, 71% CHCs, 89% HCCs and identified 7/8 misclassified HCCs as HCCs with CCA-like genomic traits (CLHCC) (Figure 5D). To avoid classification-bias due to the predominance of HCCs in the first data set, we then applied the signature to gene expression data of 8 HCCs, 6 CCAs and 7 scirrhous HCCs (sHCC)26 (Figure 5E). Common gene signature correctly predicted all HCCs confirming the anticipated common traits between CCA and sHCC (Figure 5E).
Lineage Stage Determines the Transcriptional Programs Required for Transformation
Clear separation of the tumors based on their cell-of-origin by hierarchical clustering suggested that distinct hepatic lineage cells dysregulate cell type-specific transcriptional programs in response to the same oncogenic stimuli. AH tumors showed the largest number of differentially expressed genes compared to their cell-of-origin (2826 versus 574 and 906 genes in HB and HPC tumors, respectively) by Bootstrap simulation (Figure 5B). Network analysis of tumor group-specific genes identified more significantly changed transcription factors in AH tumors (i.e. E2f1, Klf6, Myc) compared to HB (i.e. Sp1, Foxo1) and HPC tumors (i.e. Cebpb, Esrrb).
To assess lineage stage-specific transcriptional memory31, we performed GSEA using a hepatocyte-derived induced pluripotent stem cell (iPSC) signature of 786 genes32. The signature was significantly enriched in AH (P < 0.001) but not in HB or HPC tumors (Figure 5E). This suggests that induction of ESC-related genes is indispensable for oncogenic reprogramming of AHs. AH tumors demonstrated the highest overlap with a published module map of ESC genes33 (42.1% versus 19.7% and 22.7% in HB and HPC tumors, respectively) (Supplementary Figure 7A and 7B–D). Significantly, AH tumors showed a strong upregulation (21.1-fold) of Myc, a major link between ESCs and cancer34 (Figure 6A and Supplementary Figure 7B). GSEA using a list of 229 Myc E-box target genes35 confirmed a significant enrichment in AH (P < 0.0001) but not in HPC or HB tumors (Supplementary Figure 7E).
Figure 6.
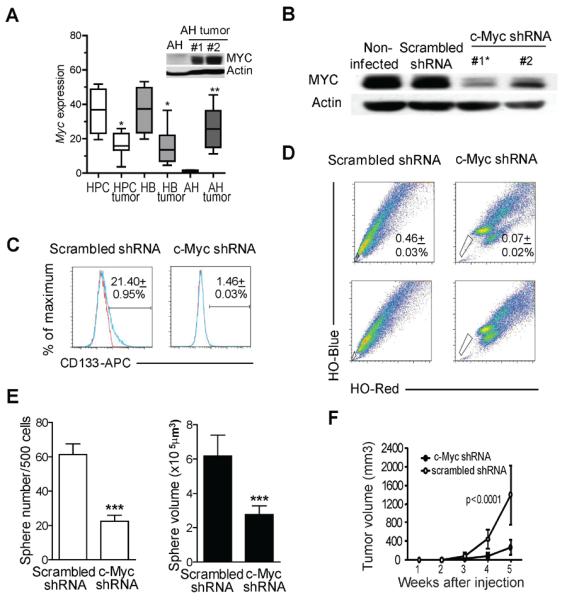
Myc is required for oncogenic reprogramming of AHs. (A) Box-plot analysis of Myc expression in HPC, HB and AH tumors and their normal counterparts based on microarray data. Significant differences were calculated by Mann-Whitney test. *P < 0.05; **P < 0.01. Inset: western blot analysis of c-Myc and actin in AHs and AH tumors. (B) Western blot analysis of c-Myc protein in H-Ras/SV40LT-transduced AHs infected with c-Myc shRNA or scrambled shRNA retroviruses. Actin was used as loading control. Asterisk marks the clone used for functional assays. (C) Analysis of CD133 expression by flow cytometry. Blue line: CD133-APC; red line: isotype control. Numbers show the mean ± SD of three experiments. (D) FACS analysis of side population identified by Hoechst 33342 (HO) staining. Numbers show the mean ± SD of three experiments. (E) Effects of c-Myc knockdown on spheroid forming ability. Cells expressing c-Myc shRNA or scrambled shRNA were seeded at low density in ultra-low attachment 96-well plates in 1% methylcellulose. Spheroids were counted after 7 days. White bars: sphere number; black bars: sphere volume. Data represent the mean ± SD of four experiments. Significant differences were evaluated by Poisson GLM and Student's t-test. *** P < 0.001. (F) Effects of c-Myc knockdown on tumor growth. One hundred cells expressing c-Myc shRNA or scrambled shRNA were injected in lower flanks of NOD/SCID mice (n=5 for each cell type). Graph shows the kinetics of subcutaneous tumor growth. Significant differences were evaluated by Student's t-test.
Myc is Required for H-Ras/SV40LT-Mediated Oncogenic Reprogramming of Adult Hepatocytes
To corroborate the role of c-Myc in transformation of AHs, we stably knocked down c-Myc in H-Ras/SV40LT-transduced AHs using shRNA-expressing retroviral vectors14 (Figure 6B). Functional relationships between c-Myc signaling and CSC properties were tested by standard in vitro and in vivo assays (Figure 6C–F). Knockdown of c-Myc significantly reduced the number of CD133+ cells (1.5% compared to 21.4% in control cells transduced with scrambled shRNA) (Figure 6C), decreased the size of SP population (Figure 6D), and diminished the sphere forming capacity and sphere size (Figure 6E). Subcutaneous tumor growth was also significantly reduced in c-Myc shRNA-expressing cells compared to control cells (Figure 6F).
Discussion
In this study, we used a mosaic model of genetically defined liver cancer36 initiated from distinct hepatic lineage cells to address the cellular origin of PLC. We targeted the cells with the same oncogenes, oncogenic H-Ras and SV40LT. Our results show that any hepatic lineage cell can be target of oncogenic transformation and acquire common CSC mode of tumorigenesis via activation of diverse cell-specific pathways.
Despite extensive efforts, the origin of CSCs in liver cancer is not fully elucidated. Hepatic stem/progenitor cells, terminally differentiated hepatocytes and cholangiocytes have been implicated as potential cells-of-origin of PLC. Expansion of progenitor cells from terminal branches of the biliary tree in rodent hepatocarcinogenesis models and frequent expression of stem/progenitor cell markers in experimental and human HCCs favor the hypothesis of progenitor cell origin at least for some HCCs6, whereas sequential phenotypic changes in diseased liver, such as emergence of dysplastic foci, nodules and finally HCC37, support oncogenic transformation of mature hepatocytes. Human CHC which displays phenotypic and gene expression traits of hepatic progenitor cells is regarded as the best example of hepatic stem/progenitor cell-derived tumor5–7.
Here, we provide conclusive evidence that acquisition of CSC properties is independent of the cell-of-origin in PLC. Forced expression of oncogenic H-Ras/SV40LT reprogrammed diverse hepatic lineage cells into CSCs as judged by an increase or acquisition of (i) expression of CSC/progenitor markers (e.g. keratin 19, A6, EpCAM, CD133), EMT- and ESC-like transcriptional programs, (ii) side population and long-term self-renewal in vitro, (iii) high tumorigenicity and (iv) metastatic capacity in various tumorigenicity assays. Furthermore, irrespective of the hepatic lineage hierarchy, transduced cells were capable of multilineage differentiation and gave rise to a continuum of liver cancers from HCC to CCA indicating that any hepatic lineage cell can be cell-of-origin of PLC. In concordance with recent findings which associate EMT, stem cell traits and cancer26,38, our genome-wide expression analysis revealed a significant upregulation of EMT- and ESC-related genes in HPC, HB and AH tumors compared to their respective cell-of-origin. Similarly, human mammary epithelial cells were reported to spontaneously dedifferentiate into stem-like cells, a process that was enhanced by oncogenic transformation39. Recent work has demonstrated generation of CSCs by oncogenic reprogramming of human fibroblasts40. Together these studies suggest that CSCs may evolve de novo from non-tumorigenic progeny during tumor progression, which holds important implications for cancer therapy.
Nonetheless, the nature of target cells may have a profound impact on susceptibility to transformation, tumor histopathology and global gene expression profiles. Thus, the same oncogenic alterations yielded higher frequency of tumor initiating cells among transduced HPCs compared to HBs and AHs. More striking disparity was described in acute myeloid leukemia where only common myeloid but not committed progenitors could be transformed by meningioma 141. The relatively small differences in the frequency of tumor initiating cells among transduced HPCs, HBs and AHs may be attributed to our choice of transforming onocogenes. Active Ras is known to promote undifferentiated state42, whereas SV40LT-mediated inhibition of p53 could contribute to acquisition and maintenance of CSC properties43, thereby diminishing the differences in the susceptibility for transformation among diverse hepatic lineage cells.
Likewise, differentiation state of the cell of origin affected the histopathology of the resulting tumors. Even though all transformed hepatic lineage cells initiated liver cancer with HCC-, CCA- and EMT-like phenotypes, the frequency of each phenotype was very variable in tumors with different cell-of-origin. Tumors initiated by mature AHs were predominantly of HCC-like pattern indicating that tumorigenic cells retained at least part of the differentiation program typical of the original cells. Tumors derived from committed HBs displayed a prominent presence of CCA-like phenotype, while HPC tumors adopted a more primitive mesenchymal-like state. This is consistent with recent findings that histological diversity in human CCA may reflect the differences in cholangiocyte phenotypes that initiate the corresponding tumors44. In contrast to our findings, overexpression of Notch receptor together with Akt in adult mouse hepatocytes resulted in the formation of only cholangiocarcinomas45. As Notch is a major regulator of biliary differentiation, the described prevalence of cholangiocarcinomas could be related to the nature of the transforming agent, suggesting that different transforming stimuli may define directions of differentiation in the same target cell. These data suggest that both the cell-of-origin and type of cancer-predisposing genetic alterations could contribute to the phenotypic and molecular diversity of PLC.
Consistent with this, expression analysis clearly distinguished tumors of different cell-of-origin indicating that distinct genetic changes are needed for oncogenic transformation of diverse hepatic lineage cells. Notably, comparison of gene expression profiles among HPC, HB and AH tumors and freshly isolated normal counterparts revealed that AH tumors displayed drastically more differentially expressed genes than HB or HPC tumors and activated the highest number of ESC-related genes. Among these was Myc with a remarkable 21-fold upregulation, which was associated with coordinated activation of Myc-centered interaction networks. Although the significance of c-Myc in liver cancer biology is widely described in rodents46 and human47,48, this is the first study which identified c-Myc as a key element of ESC-related genes activated during oncogenic reprogramming of AHs. We validated these findings by knockdown of c-Myc in H-Ras/SV40LT-expressing AHs which significantly reduced the frequency of CSCs and delayed tumor development in immunocompromised mice.
In conclusion, our study provides the first comprehensive and systematic comparison of hepatocarcinogenesis initiated by controlled oncogenic transformation of cells at specific stages of hepatic lineage. Differentiated hepatocytes, hepatoblasts and adult hepatic progenitor cells were isolated at high purity and efficiently transduced with the same combination of H-Ras and SV40LT oncogenes. This permitted a unique and direct side-by-side comparison of cellular and molecular characteristics of transformed cells both in vitro and in vivo. We formally demonstrated that any hepatic lineage cell can be reprogrammed into CSC by activating diverse cell type-specific pathways. Furthermore, we described common and cell-of-origin specific phenotypic and genetic changes which accurately differentiated murine tumors according to their origin providing an important tool to phenotypically classify morphologically diverse human PLC. Thus, identification of cells that are susceptible to oncogenic transformation and relevant molecular pathways is essential for a deeper understanding the origin of liver cancer and development of more effective therapeutic strategies.
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Program of the National Cancer Institute, NIH. We thank S. Arya and CP. Day for advices on lentiviral vectors; E. A. Conner for help with animal procedures; B. Taylor and S. Banerjee for assistance with flow cytometry, and T. Hoang for help with immunohistochemistry.
Abbreviations
- AH
adult hepatocyte
- CCA
cholangiocarcinoma
- CHC
combined hepatocellular-cholangiocarcinoma
- CLHCC
CCA-like hepatocellular carcinoma
- CSC
cancer stem cell
- EGFP
enhanced green fluorescent protein
- EMT
epithelial-mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- ESC
embryonic stem cell
- HB
hepatoblast
- HCC
hepatocellular carcinoma
- HNF4A
hepatocyte nuclear factor 4 alpha
- HPC
hepatic progenitor cell
- HSC
hepatic stem cell
- NOD/SCID
Nonobese diabetic/severe combined immunodeficiency
- PLC
primary liver cancer
- sHCC
scirrhous HCC
- SV40LT
simian virus 40 large T antigen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors disclose no conflicts.
Authors contribution: Study concept and design: SST, AH. Acquisition of data: AH, VMF, JBA, JUM, CR, MK, HA, MD. Analysis and interpretation of data: AH, VMF, JBA, DK. Drafting of the manuscript: AH, VMF. Critical revision of the manuscript for important intellectual content: SST, VMF, JAB, DK, JUM. Statistical analysis: DS. Study supervision: SST, VMF
All microarray data were submitted to GEO (accession number GSE41312), and accessible at http://www.ncbi.nlm.nih.gov/geo/.
References
- 1.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–22. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 2.Ince TA, Richardson AL, Bell GW, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–70. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland KD, Proost N, Brouns I, et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–64. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis. Semin Liver Dis. 2011;31:173–87. doi: 10.1055/s-0031-1276646. [DOI] [PubMed] [Google Scholar]
- 5.Yeh MM. Pathology of combined hepatocellular-cholangiocarcinoma. J Gastroenterol Hepatol. 2010;25:1485–92. doi: 10.1111/j.1440-1746.2010.06430.x. [DOI] [PubMed] [Google Scholar]
- 6.Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818–22. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- 7.Woo HG, Lee JH, Yoon JH, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–41. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–96. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–56. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 10.Marquardt JU, Raggi C, Andersen JB, et al. Human hepatic cancer stem cells are characterized by common stemness traits and diverse oncogenic pathways. Hepatology. 2011;54:1031–42. doi: 10.1002/hep.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa T, Factor VM, Marquardt JU, et al. Hepatocyte growth factor/c-met signaling is required for stem-cell-mediated liver regeneration in mice. Hepatology. 2012;55:1215–26. doi: 10.1002/hep.24796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SB, Seo D, Choi D, et al. Contribution of hepatic lineage stage-specific donor memory to the differential potential of induced mouse pluripotent stem cells. Stem Cells. 2012;30:997–1007. doi: 10.1002/stem.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao CY, Factor VM, Thorgeirsson SS. Reduced growth capacity of hepatocytes from c-myc and c-myc/TGF-alpha transgenic mice in primary culture. Biochem Biophys Res Commun. 1996;222:64–70. doi: 10.1006/bbrc.1996.0698. [DOI] [PubMed] [Google Scholar]
- 14.David CJ, Chen M, Assanah M, et al. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Factor VM, Radaeva SA, Thorgeirsson SS. Origin and fate of oval cells in dipin-induced hepatocarcinogenesis in the mouse. Am J Pathol. 1994;145:409–22. [PMC free article] [PubMed] [Google Scholar]
- 16.Yaswen P, Goyette M, Shank PR, et al. Expression of c-Ki-ras, c-Ha-ras, and c-myc in specific cell types during hepatocarcinogenesis. Mol Cell Biol. 1985;5:780–6. doi: 10.1128/mcb.5.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvisi DF, Ladu S, Gorden A, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–28. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Chiba T, Kita K, Zheng YW, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–51. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 19.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okabe M, Tsukahara Y, Tanaka M, et al. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development. 2009;136:1951–60. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- 21.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci U S A. 2005;102:6942–7. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paku S, Schnur J, Nagy P, et al. Origin and structural evolution of the early proliferating oval cells in rat liver. Am J Pathol. 2001;158:1313–23. doi: 10.1016/S0002-9440(10)64082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzini S, Bird TG, Boulter L, et al. Characterisation of a stereotypical cellular and extracellular adult liver progenitor cell niche in rodents and diseased human liver. Gut. 2010;59:645–54. doi: 10.1136/gut.2009.182345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta S. Hepatic polyploidy and liver growth control. Semin Cancer Biol. 2000;10:161–71. doi: 10.1006/scbi.2000.0317. [DOI] [PubMed] [Google Scholar]
- 26.Seok JY, Na DC, Woo HG, et al. A fibrous stromal component in hepatocellular carcinoma reveals a cholangiocarcinoma-like gene expression trait and epithelial-mesenchymal transition. Hepatology. 2012;55:1776–86. doi: 10.1002/hep.25570. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–74. [PMC free article] [PubMed] [Google Scholar]
- 28.Wellek S. Testing Stastistical Hypothesis of Equivalence. Chapman & Hall/crc Press LLC; Boca Raton, FL: 2003. [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alonso SR, Tracey L, Ortiz P, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–60. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 31.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohi Y, Qin H, Hong C, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–9. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong DJ, Liu H, Ridky TW, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–24. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zender L, Xue W, Cordon-Cardo C, et al. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251–61. doi: 10.1101/sqb.2005.70.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hytiroglou P. Morphological changes of early human hepatocarcinogenesis. Semin Liver Dis. 2004;24:65–75. doi: 10.1055/s-2004-823097. [DOI] [PubMed] [Google Scholar]
- 38.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 39.Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–5. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scaffidi P, Misteli T. In vitro generation of human cells with cancer stem cell properties. Nat Cell Biol. 2011;13:1051–61. doi: 10.1038/ncb2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heuser M, Yun H, Berg T, et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer Cell. 2011;20:39–52. doi: 10.1016/j.ccr.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Vita G, Bauer L, da Costa VM, et al. Dose-dependent inhibition of thyroid differentiation by RAS oncogenes. Mol Endocrinol. 2005;19:76–89. doi: 10.1210/me.2004-0172. [DOI] [PubMed] [Google Scholar]
- 43.Yi L, Lu C, Hu W, et al. Multiple roles of p53 related pathways in somatic cell reprogramming and stem cell differentiation. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-1451. [DOI] [PubMed] [Google Scholar]
- 44.Komuta M, Govaere O, Vandecaveye V, et al. Histological diversity in cholangiocellular carcinoma reflects the different cholangiocyte phenotypes. Hepatology. 2012;55:1876–88. doi: 10.1002/hep.25595. [DOI] [PubMed] [Google Scholar]
- 45.Fan B, Malato Y, Calvisi DF, et al. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest. 2012;122:2911–5. doi: 10.1172/JCI63212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Seminars in Cancer Biology. 2006;16:313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Kaposi-Novak P, Libbrecht L, Woo HG, et al. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775–82. doi: 10.1158/0008-5472.CAN-08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shachaf CM, Kopelman AM, Arvanitis C, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–7. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.