Abstract
The cancer stem cell model was described for hematologic malignancies in 1997 and since then evidence has emerged to support it for many solid tumors as well, including colon cancer. This model proposes that certain cells within the tumor mass are pluripotent and capable of self-renewal and have an enhanced ability to initiate distant metastasis. The cancer stem cell model has important implications for cancer treatment, since most current therapies target actively proliferating cells and may not be effective against the cancer stem cells that are responsible for recurrence. In recent years great progress has been made in identifying markers of both normal and malignant colon stem cells. Proteins proposed as colon cancer stem cell markers include CD133, CD44, CD166, ALDH1A1, Lgr5, and several others. In this review we consider the evidence for these proteins as colon cancer stem cell markers and as prognostic indicators of colon cancer survival. Additionally, we discuss potential functions of these proteins and the implications this may have for development of therapies that target colon cancer stem cells.
Keywords: Beta-catenin, CD44, CD133, Crypt, Differentiation, Intestinal epithelial cell, Integrin, Review, Small intestine, Stem cell
2. INTRODUCTION
Colon cancer is the third most common cancer in the United States and the second leading cause of cancer deaths with an estimated 150,000 newly diagnosed cases and 50,000 deaths in 2008 (1). Most deaths from colon cancer occur from metastasis of the cancer to other tissues, most commonly the liver (2). Over the last decade the cancer stem cell (CSC) model, also known as the hierarchy model, has become increasingly accepted as an explanation for cancer spread and recurrence (3–5). Evidence for this model was first described for hematologic malignancies (6), and in recent years experimental support for it has emerged for many solid tumors as well. This model posits that a small subpopulation of tumor cells have an enhanced potency and ability to initiate distant metastases compared to the rest of the tumor cells, are capable of self-renewal, form metastatic tumors of heterogeneous cell types that resemble the primary tumor, and are more resistant to therapy (3, 5). The stochastic model of cancer progression, in contrast, holds that each cell within the tumor mass is equally capable of initiating distant metastases (3–5).
The biology of the normal intestinal epithelium has been described in several recent reviews (7–10) and is beyond the focus of this article. Before discussing colon CSC markers, however, it is necessary to briefly describe what is known of normal small intestinal and colon stem cells. In both the mouse small intestine and colon, the stem cells lie towards the bottom of the crypt in the proliferative zone, and are responsible for generating all epithelial cell types along the crypt-villus axis. It is thought that 4 to 6 stem cells are present in each crypt (11). In the small intestine two stem cell populations have been identified (Figure 1), the crypt base columnar cells, marked by expression of the G-protein receptor Lgr5 and positioned just above the Paneth cells at the bottom of the crypt (12), and cells at the +4 position from the bottom of the crypt (11), which recent research has shown are marked by expression of the polycomb group gene Bmi1 (13). Both cell types have been demonstrated to fulfill the criteria for stem cells (pluripotency and self-renewal capacity), though it is unclear at this point whether they represent the same or distinct cell populations. In the colon the precise position of the stem cells has not been determined, though they are known to lie at or near the bottom of the crypt. Colon stem cells are also marked by Lgr5 expression (12). In mice, stem cells marked by Bmi1 expression are only present in the small intestine (13).
Figure 1.
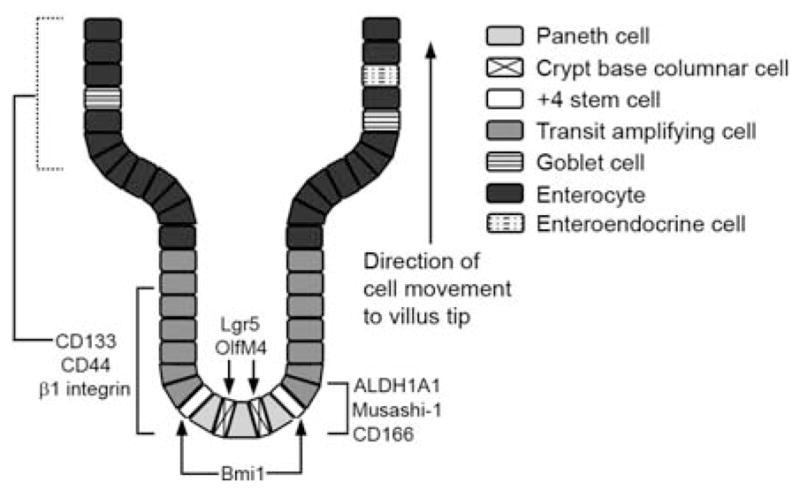
Schematic diagram showing expression patterns of some proposed normal intestinal stem cell and CSC markers in normal small intestine. In colon, the precise position of stem cells at the bottom of the crypt has not been determined. Additionally, Bmi1+ stem cells were detected in mouse small intestine but not mouse colon (13). Dotted line for CD133 expression in villus cells indicates differing results concerning its expression in these cells. The precise position of the cells in the bottom part of the crypt that express ALDH1A1 and CD166 has not been determined. Musashi-1 is not detected in Paneth cells in the mouse small intestine (103). Additional details are described in the text.
A stem cell origin for colon cancers is suggested by the observation of proliferating multipotent cell expansion at the intestinal epithelial crypt base that occurs during adenoma development in familial adenomatous polyposis (FAP) patients (14), who have mutations in the adenomatous polyposis coli (APC) gene that predispose them to gastrointestinal polyps and cancer. It has also long been known from histological examination of human colon adenocarcinomas that they contain multiple intestinal cell types of varying degrees of differentiation (15–17), and that single colon adenocarcinomas have variable expression of proteins such as MUC2 (18), neuroendocrine markers (19), and villin (20). The stochastic and CSC models differ in explaining how this arises. The stochastic model posits that this heterogeneity results from a multiclonal origin of the tumor. The CSC model, by contrast, proposes that colon adenocarcinomas arise from a single multipotent stem cell that gives rise to tumors containing multiple cell types. It predicts that a single CSC from the original tumor is capable of initiating metastatic tumors that are histopathologically similar to the tumor of origin. It is only in the last five years, however, that experimental evidence directly demonstrating the origin of colon CSCs from normal colon stem cells has been provided and that colon CSC molecular markers have been identified.
The majority of colon cancers arise through dysfunctional regulation of the Wnt/beta-catenin pathway, either through inactivating mutations in the beta–catenin negative regulator APC or activating mutations in beta-catenin. This leads to abnormal accumulation of a beta-catenin/transcription factor T-cell factor 4 (Tcf4) complex in the nucleus (21, 22), activation of the Tcf4 target c-MYC and subsequent suppression of differentiation through inhibition of p21CIP1/WAF1 expression (23). As will be discussed in more detail later, both Bmi1+ and Lgr5+ stem cell populations are capable of generating tumors in mice when beta-catenin signaling is activated specifically in these cells (12, 13). Broadly speaking, two different approaches have been used to identify potential markers of colon CSCs. First, the molecular profile of genes activated by the beta-catenin/Tcf4 complex has led to the identification of colon CSC markers such as Lgr5 (23, 24). In the other approach, CSC markers such as CD133 (25, 26), identified from cancers of other tissues, have been tested as colon CSC markers.
Markers for human CSCs have most commonly been identified based on their ability to form tumors when subcutaneously injected in immune-compromised (usually NOD/SCID) mice. Limiting dilution of tumor cells sorted for a particular molecular marker is performed to determine the number of cells required to initiate tumor formation compared to the number of unsorted tumor cells required for tumor formation. The ability to form spheroid cultures in vitro (27) in stem cell medium has also been used to test for CSC markers. This review will focus on proposed colon CSC markers, their expression in normal colonic mucosa and colon cancers, and their utility as prognostic indicators and targets for specifically eliminating colon CSCs.
3. COLON CANCER STEM CELL MARKERS
3.1. CD133
CD133 (Prominin-1), a transmembrane glycoprotein which was first identified as a potential CSC marker for brain tumors (28), was proposed as a colon CSC marker in 2007. Both O’Brien et al. and Ricci-Vitiani et al. found that the tumors formed from CD133 positive cells injected into SCID mice resembled the tumor from which they were taken and formed tumors of differentiated cell types that were mostly CD133 negative. The CD133 negative cells from these tumors did not form metastases in mice even when injected at much higher numbers than CD133 positive cells (25, 26). O’Brien et al. found that CD133 positive colon tumor cells were 200-fold enriched for CSCs compared to unsorted tumor cells and that colon tumors contained a higher percentage of CD133 positive cells than normal tissue (25).
Both reports also indicated that the vast majority of CD133 positive cells were not CSCs and thus that CD133 could not be relied on as a single marker of colon CSCs. O’Brien et al. calculated that 1 in 262 CD133 positive cells were cancer initiating cells (25), while Ricci-Vitiani et al. found that, for 4 of 13 tumors tested, injection of 10,000 CD133 positive cells was not sufficient to form tumors, though for each tumor, only two mice were injected (26). Additionally, Chu et al. did not observe an enhanced tumor initiating capacity for CD133 positive cells isolated from 3 different primary colon tumors and directly injected into mice (29), and Feng et al. did not observe any difference in the tumor initiating capacity of CD133 positive or negative HCT116 cells (30). The contrasting results of Chu et al. could be due to differences in methodology. Cells from primary tumors were used in this study, whereas 10 of 17 tumors tested by O’Brien et al. were metastatic. Chu et al. subcutaneously injected tumor cells in mice, whereas O’Brien et al. used renal capsule transplantation (25, 29). Chu et al. also directly injected cells into mice, while Ricci-Vitiani cultured cells in stem cell medium for 48 hours prior to injection (26, 29). It is more difficult, however, to explain the contrasting results of Shmelkov et al., who, like O’Brien et al., used cells from metastatic tumors, but observed that CD133 negative cells had the same tumor initiating capacity as CD133 positive cells and that CD133 was expressed equally in differentiated and undifferentiated cells in the normal human colon (31). Using a mouse knock-in model in which lacZ expression is driven by the CD133 promoter, this report also described heterogeneous expression of CD133 in undifferentiated crypt cells and differentiated villi (31). Using a similar method, however, Zhu et al. found that CD133 expression was enriched in the mouse small intestinal epithelial crypt, with 26% of crypt cells but only 3% of villi cells being positive for CD133 as indicated by expression of the lacZ reporter gene. In this report it was also found that expression of activated beta-catenin in CD133+ stem cells resulted in a disproportionate expansion of these cells at the crypt base (32). In another mouse study, Snippert et al. found that CD133 is expressed in both stem cells and the transit amplifying cells that lie above them in the small intestinal epithelial crypt (33), more in agreement with the results of Zhu et al. (32). One possible explanation for the differing results in humans is that the epitope recognized by the antibody used in these studies, AC133, is lost upon differentiation (34). Thus, it may be the particular glycosylated form of CD133 recognized by AC133 that is a marker of colon CSCs. The contrasting results of Shmelkov et al. (31) and Zhu et al. (32) in the mouse knock-in models in which lacZ was fused to the CD133 promoter, however, are more difficult to explain.
CD133 mRNA or protein expression in tumors has been found in most reports to correlate with a poor prognosis. Circulating levels (35) and tumor levels (36) of CD133 mRNA are associated with lower survival. Tumor levels of CD133 mRNA also correlate with distant metastasis formation for rectal tumors (37). CD133 protein expression assessed by immunohistochemistry in a colon adenocarcinoma microarray (38) and by percentage of CD133 expressing cells (39) significantly correlated with low survival, and in a case control study high CD133 protein expression levels are correlated with liver metastasis (40). Choi et al., however, did not find that CD133 expression assessed by immunohistochemistry correlated with survival (41).
Potential functions for CD133 in colon CSCs remain to be demonstrated. Zhu et al did not observe an overt phenotype in CD133 null mice (32), and reduction of CD133 expression by siRNA in two colon cancer cell lines, Caco-2 and LoVo, did not affect invasion, proliferation, or colony formation (40). While Feng et al. found that CD133 expression inversely correlated with the degree of differentiation of colon cancer cell lines, siRNA knockdown of CD133 had no effect on differentiation (30). In two independent studies, siRNA reduction of CD133 in primary human colon tumor cells does not affect colony formation (30, 42), and one of these studies additionally demonstrated that tumorigenicity of these cells injected subcutaneously in SCID mice is not affected (42). In total, the evidence suggests that CD133 has value as a marker of colon CSCs and as an indicator of the aggressiveness of colonic tumors, but that it is not likely to be a useful therapeutic target for elimination of colon CSCs.
3.2. CD44
CD44, a receptor for the extracellular matrix protein hyaluranon, was first identified as a potential CSC marker for breast cancers (43) and is, like CD133, a transmembrane glycoprotein. Expression of CD44 is also activated by the beta-catenin/Tcf-4 complex in colon cancer cells (23). CD44 expression is confined to the crypt epithelium in non-neoplastic mucosa of Apc+/Apc1638 mice and increases with progression of cancer (44). CD44 has many variant splice forms, and prior to its identification as a potential colon CSC marker, it had been demonstrated that expression of several CD44 splice variants correlates with colon cancer progression (45–50). Thus when evaluating studies examining CD44 as a potential colon CSC marker it must be taken into account that the methods employed may not distinguish between different CD44 splice variants. Dalerba et al. first identified CD44 as a potential CSC marker (51). In this study, 2 of 5 mice developed tumors when injected with 10,000 CD44+ cells from human colon tumor cells that had been serially passaged in NOD/SCID mice, compared to 0 of 5 mice injected with CD44− cells. Though a direct comparison with tumor initiating capacity of CD133+ cells was not performed, CD133 was expressed in a larger percentage of cells than CD44, suggesting that CD44 is a more selective marker for colon CSCs. Du et al. also found that CD44 is more selective as a colon CSC marker than CD133, since as few as 1 of 100 CD44+ cells from human tumors were capable of initiating tumor formation in SCID mice. Additionally, CD44+ cells were found to have greater tumor initiating potential than CD133+ cells from the same tumor, and 24% of CD44+ cells, but only 1.8% of CD133+ cells, were capable of forming tumor spheroid holoclones (42).
Unlike CD133 (30, 42), siRNA reduction of CD44 in primary human colon cancer cells reduces colony formation and inhibits tumor formation from cells injected subcutaneously in SCID mice (42). HT29 cells transfected with CD44 siRNA also exhibit reduced colony formation and tumor formation in SCID mice (52) and reduced migration (53). Additionally, shRNA specifically targeting the CD44v6 splice variant inhibits adenoma formation in APC Min/+ mice (54). In normal human colon expression of CD44 is found at the crypt base where stem cells are located (figure 1; (42)). Patel et al. found increased expression of CD44 in normal appearing intestinal mucosa from patients with adenomatous polyps, and that increased expression of CD44 in the normal appearing tissue correlated with increasing age and the number of polyps in the patient (55). Additionally, expression of CD44 in rat colon increased following exposure to the carcinogen dimethylhydrazine (56), and increased expression of CD44 appears to precede K-ras and p53 gene alterations (45), suggesting that increased expression of CD44 may be an early indicator for the presence of colon CSCs. Taken together, this evidence suggests that CD44 may be a useful therapeutic target for elimination of colon CSCs and that it is a more specific marker for colon CSCs than CD133. As noted above, however, many different splice variants of CD44 exist. Therefore, an important subject for future studies will be to determine whether colon CSCs express particular splice variants of CD44. If so, this may allow the design of targeted therapies with greater efficacy and specificity.
3.3. CD166
CD166/activated leukocyte cell adhesion molecule (ALCAM) belongs to the immunoglobulin superfamily of cell adhesion molecules, with five extracellular immunoglobulin domains, a transmembrane region and a short cytoplasmic C-terminal region. It is found at sites of cell-cell contact and is involved in both homotypic and heterotypic (to CD6) adhesion. CD166 was observed by Weichert et al. in 2004 to have increased and heterogeneous expression in colon carcinoma (57) and was first identified as a potential colon CSC marker by Dalerba et al. in 2007 (51). While it was not examined as an individual stem cell marker in this study, sorting for CD166 in addition to CD44 or in addition to both CD44 and epithelial cell adhesion molecule (EpCAM) identified a population of colon CSCs with increased tumor initiating potential. Vermeulen et al. observed that sorting dissociated colonic adenocarcinomas for CD166 in addition to CD133 did not increase the frequency of colonosphere formation from single cells compared to sorting for CD133 alone (58), although CD166 expression is enriched in tumor spheroids derived from CD133-sorted primary colon cancer cells (59).
The function of CD166 both in normal colonic mucosa and in colon CSCs is unclear. Weichert et al. observed expression of CD166 to be confined to a few cells at the crypt base in normal human colonic epithelium (57), while Levi et al. (56) observed similar limited expression in young rat colonic mucosa (figure 1). Knockout of CD166 in mice does not cause overt phenotypes (60), suggesting it is not essential for intestinal stem cell function, and the effect of CD166 knockdown in colon cancer cells on tumor initiating potential and colonosphere formation does not appear to have been determined. Colon expression of CD166 in macroscopically normal mucosa increases with age and with the number of polyps present (55) and increases in rat colon following exposure to dimethylhydrazine (56), suggesting, as with CD44, that increased expression of CD166 may be an early indicator of the presence of CSCs. Weichert et al. observed a significant correlation between CD166 protein expression in primary colon adenocarcinomas and shortened patient survival (57). CD166 protein expression in a colonic adenocarcinoma tissue sample microarray also correlated somewhat (p=0.07) with low survival, but CD133 expression was found to be a stronger prognostic indicator of low survival in this study (61). Notably, however, the value of CD166 as a single prognostic marker for colon CSCs is unclear, since in sorting studies of colon cancer cells it has mainly been examined in combination with other stem cell markers. Its value as a potential colon CSC therapeutic target is also not clear, since siRNA knockdown studies in colon CSC cells or colon cancer-derived cell lines do not appear to have been performed.
3.4. Aldehyde dehydrogenase 1A1 (ALDH1A1)
ALDH1A1 was first identified as a marker for normal and malignant breast stem cells in 2007 by Ginestier et al. (62), and was subsequently identified as a marker for normal and malignant colon stem cells in 2009 by Huang et al. (63). In this study, direct comparison of ALDH1A1, CD133, and CD44 expression in normal colon cells indicated that ALDH1A1 was restricted to fewer cells at the crypt base than either CD133 or CD44 (figure 1), indicating it is a more selective marker for the least differentiated cells. Cells at the crypt base expressing ALDH1A1 were a subset of those expressing the other two markers, and in colon tumors ALDH1A1 was expressed in a smaller proportion of cells. In adenomatous crypts the proportion of cells expressing ALDH1A1 increases by fivefold (63). Based on this and the observation that ALDH1A1+/CD44− and ALDH1A1+/CD133− cells initiate tumors in SCID mice, the authors conclude that ALDH1A1 is a selective marker for colon CSCs. Compared to selection for ALDH1A1 expression alone, combined selection for ALDH1A1 and CD133 somewhat increases the proportion of tumor initiating cells and the size of tumors, and combined selection for ALDH1A1 and CD44 does not affect the rate of tumor initiation but generates tumors of somewhat larger size (63). The findings on ALDH1A1 expression in normal colon and colon cancer were confirmed by immunohistochemical staining of normal human colon and a tissue array of 67 colon cancers, though it should be noted that a small percentage of cancers in this study were negative for ALDH1A1 (64). Carpentino et al. also observed an expansion of crypt cells expressing ALDH1A1 in patients with chronic colitis, an important risk factor for CRC (65).
ALDH1A1 is of particular interest because of its role in detoxifying cyclophosphamide class chemotherapeutic agents (reviewed in (66)). Thus, it is conceivable that disabling ALDH1A1 could make colon CSCs more sensitive to these drugs. It should be noted, however, that ALDH1A1 is one of 19 members of the ALDH superfamily (reviewed in (67)), at least one of which, ALDH3A1, also confers resistance to cyclophosphamide-derived drugs (68, 69) and has been detected by activity assays in colon cancers (70). The detailed expression pattern of ALDH3A1, however, in the normal colon and in colon adenocarcinomas has not yet been characterized.
3.5. ATP binding cassette proteins
ATP binding cassette proteins (ABC) use cellular ATP to drive the transport of drugs, metabolites, and other compounds across cell membranes. This protein family consists of about 50 members, two of which, MDR1 (multi-drug resistance protein 1; also known as ABCB1 or p-glycoprotein) and ABCG2 have been identified as responsible for resistance to chemotherapeutic agents. ABCG2 was cloned from human breast cancer MCF-7 cells and demonstrated to be responsible for resistance to doxorubicin and related compounds (71). Since that time it has been demonstrated to be capable of effluxing a wide variety of chemotherapeutic drugs (reviewed in (72)). In 2001, Zhou et al. (73) demonstrated that ABCG2 conferred the side population (SP) phenotype, defined by the ability to efflux the fluorescent dye Hoechst 33342, which is a property of normal hematopoietic stem cells. Additionally, they demonstrated that it was expressed by normal stem cells from a variety of tissue types. As drug resistance is a characteristic of CSCs, it was logical to investigate ABCG2 as a CSC marker. Subsequently, SP cells were shown to be present in human cell lines derived from a variety of tumor types (74). Studies in colon cancer cell lines have yielded differing results. Haraguchi et al. demonstrated the presence of SP cells in six different colon cancer cell lines (75). Burkert et al., however, did not find any difference in tumorigenic potential of SP and non-SP Caco-2 and HT29 cells, and also observed that expression of CD44 and CD133 was similar in SP and non-SP cells. Interestingly, ABCG2 was only expressed in less than 5% of SP cells, and expression did not differ between SP and non-SP cells, suggesting that other membrane transporters may be responsible for the SP phenotype in colon cancer cells (76). Other methods, such as direct flow cytometry sorting for ABCG2, may need to be utilized to determine if ABCG2 is a colon CSC marker.
A direct assessment of MDR1 as a colon CSC marker has not been performed. In HT29 cells selected for CD133 expression, siRNA targeting MDR1 increases sensitivity of the cells to paclitaxel (77), suggesting that MDR1 is present in colon CSCs. Immunohistochemical staining for MDR1 expression in ileum and colon, however, indicates that MDR1 expression may not be limited to crypt stem cells (78). Salinomycin, recently demonstrated to specifically target breast CSCs (79), may act in part by inhibiting MDR1 function (80). It will obviously be of great interest to determine whether colon CSCs are also sensitive to salinomycin.
3.6. Lgr5
Barker et al. identified Lgr5 (Leucine-rich G-protein coupled receptor 5) as an intestinal epithelial stem cell marker in 2007 (12). It is one of about eighty genes demonstrated to be a target of the Wnt-regulated transcription factor T-cell factor 4 (23, 24). Histological examination of its expression (figure 1) determined that it is present only in the small number of cycling columnar cells at the crypt base in the small intestine and colon (12, 81). Proof of Lgr5 as a marker of normal small intestinal and colon stem cells came from lineage tracing studies in mice in which the Lgr5 promoter-mediated activation of Cre recombinase induces irreversible activation of a lacZ reporter gene in Lgr5+ cells. Expression of lacZ gene is retained indefinitely in the crypt bottom, lacZ+ cells emanate from the crypt bottom and move up the crypt, and, after 60 days, lacZ+ cells of all cell types present in the small intestine and colon are observed. In this study, Lgr5 is expressed in fewer cells per crypt than CD133 (about 4 Lgr5+ cells compared to about 30 CD133+ cells), indicating it is a more selective marker for normal stem cells than CD133 (12). Additionally, Sato et al. demonstrated in an in vitro culture system that a single Lgr5+ mouse small intestinal crypt cell could generate villus- like domains which contained all of the differentiated cell types found in vivo (82).
The low level of Lgr5 expression has made it difficult to purify Lgr5+ cells from colon tumors by flow cytometry and perform tests on their tumorigenicity in SCID mice. Evidence that Lgr5 is a marker for colon CSCs has come from mouse studies in which APC is selectively knocked out in Lgr5+ cells. In both small intestine and colon this led to formation of transformed beta-catenin high/Lgr5+ cells at the bottom of the crypt, formation of micro-adenomas within 3 weeks, and adenoma formation by 36 days. In contrast, conditional deletion of APC in the transit-amplifying cells above the Lgr5+ stem cells very rarely led to adenoma formation (83). In humans, McClanahan et al. found by gene expression profiling that Lgr5 is overexpressed in 10 of 15 primary human colon tumors (84). Becker et al. observed expansion in the number of Lgr5+ cells in premalignant lesions in the human small intestine and colon, indicating that expansion in the number of Lgr5+ cells is an early event in premalignant transformation of colon CSCs (85). Additionally, Uchida et al. observed Lgr5 mRNA overexpression in 35 of 50 human colon cancers and in 7 of 7 sporadic adenomas, and that expression of Lgr5 significantly increased with cancer stage (81).
Lgr5 is an orphan receptor for which the ligand and downstream G-protein effectors have not been determined (reviewed in (7)). Lgr5 null mice exhibit 100% neonatal lethality with gastrointestinal distension (86), suggesting an essential function, but the phenotype of conditional intestinal epithelial cell Lgr5 null mice does not appear to have been determined. Thus it is not known whether Lgr5 is essential for colon CSC function, and thus possibly could be a specific colon CSC therapeutic target, or may only serve as a marker for colon CSCs.
3.7. Other potential colon CSC markers
Other proteins identified as potential colon CSC markers include CD24, beta1 integrin (CD29), EpCAM, OlfM4, Bmi1 and Musashi-1. In most cases these proteins have not been tested as single markers for selection of tumor-initiating colon CSCs in NOD/SCID mice. Low CD24 expression in combination with high CD44 expression was identified as a selection marker for breast CSCs in 2003 (43). Interestingly, CD24+/CD133+ colon cancer cells have enhanced capacity for forming in vitro colonospheres compared to CD24−/CD133+ cells (58). Contrasting results have been obtained on its expression in colon cancer cells, with Vermeulen et al. observing decreased expression (58) and Choi et al. increased expression (41) with increased differentiation. Weichert et al. observed that increased cytoplasmic expression of CD24 correlated with shortened survival in colon cancer patients (87).
Beta1 integrin expression is enriched in the proliferative zone of colonic crypts (figure 1), and selects for intestinal epithelial cells with enhanced clonogenic capacity as measured in a soft agar assay (88). Disruption of beta1 integrin heterodimer interaction with extracellular matrix leads to rapid onset of apoptosis of colonic crypt cells (89), and the alpha2 beta1 integrin ligand collagen I promotes a stem-cell like phenotype of colon cancer cell lines with increased expression of Bmi1 and CD133 (90). Beta1 integrin expression is enriched in colonospheres derived from CD133+ primary colon tumor cells (59), but selection of primary colon cancer cells for beta1 integrin expression in addition to CD133 expression does not select for more clonogenic cells than selection for CD133 alone (58). It does not appear, however, that beta1 integrin has been tested as a single selection marker for colon CSCs.
EpCAM expression in colon adenocaracinomas was first described more than 30 years ago (91), and overexpression of EpCAM was found in 98% of primary human colon cancers as assessed by immunohistochemical staining (92). Colon cancer cells selected for EpCAM expression in addition to CD44 expression had greatly enhanced tumor initiating capacity compared to cells selected only for CD44. 21 of 28 mice developed tumors when injected with 200 EpCAM+/CD44+ colon cancer cells, while only 2 of 5 mice injected with 10,000 singly-selected CD44+ cells developed tumors (51). EpCAM is cleaved by TACE and pre-senilin 2, resulting in release of the extracellular domain and release of the intracellular domain into the cytoplasm. The released extracellular domain appears to act as an autocrine stimulator of EpCAM cleavage (93). The EpCAM intracellular domain is a coactivator of the Wnt/beta-catenin pathway through its association with the beta-catenin/Tcf4 transcriptional activation complex in the nucleus. The EpCAM intracellular domain is detectable in the nuclei of colon carcinoma cells, but not of normal cells, and siRNA knockdown increases apoptosis and decreases proliferation of HCT-8 colon cancer cells (93). Consistent with these observations, membranous EpCAM expression is observed to be lost in budding colon cancer cells (94). These studies indicate that EpCAM may have value as both a prognostic indicator of colon CSCs and a potential therapeutic target.
OlfM4, a member of the olfactomedin glycoprotein family, is specifically expressed in Lgr5+ stem cells in the mouse small intestine (figure 1; (95)) and in both the human small intestine and colon (96). It is expressed in a subset of cells in colorectal carcinoma as detected by immunohistochemistry (96), and additional studies indicate it is overexpressed in colon cancer (97, 98). Another study, while observing OlfM4 mRNA overexpression in well-differentiated colon tumors compared to normal tissue, found that expression decreased in poorly differentiated tumors (99). Since OlfM4 is a secreted protein, it has been difficult to test it as a colon CSC marker.
The polycomb group gene Bmi1 is a marker of small intestinal stem cells at the +4 position from the crypt bottom (figure 1). As observed for Lgr5+ stem cells, the Bmi1+ cells are pluripotent, capable of self-renewal, and form adenomas when the Wnt/beta-catenin signaling pathway is activated within them (13). Bmi1 is overexpressed in precancerous colonic lesions and is more highly overexpressed in cancerous tissues (100). High Bmi1 expression in tumors also predicts poor survival for colon cancer patients (101).
Two different research groups identified the RNA binding protein Musashi-1 as a marker for normal small intestinal stem cells in 2003 (102, 103). Both reports describe Musashi-1 expression in the crypt base columnar cells and in cells at the +4 position (figure 1). Potten et al. also detect Musashi-1 in mouse large intestinal crypt cells and increased Musashi-1 expression in adenomas of Min mice compared to adjacent normal tissue (103). Musashi-1 is also overexpressed in human colonic adenocarcinomas, and injection of Musashi-1 siRNA into xenografted tumors derived from HCT116 cells resulted in a greater than 75% reduction in tumor volume. Musashi-1 knockdown by siRNA also reduced proliferation, stimulated apoptosis, and caused an increase in expression of the differentiation marker p21WAF1 in HCT116 cells (104). This suggests Musashi-1 may have value both as a colon CSC marker and as a therapeutic target.
4. SUMMARY AND PERSPECTIVE
Although tremendous progress has been made in identifying the molecular characteristics of colon CSCs, there is still an urgent need for therapeutic strategies that specifically target them. Therapies targeted to the epidermal growth factor receptor and the vascular endothelial growth factor receptor have proven only marginally effective against metastatic colon cancer (105), and it is likely that complete tumor eradication will require multiple therapeutic approaches. The CSCs that appear to be responsible for cancer recurrence may lie dormant for many years (106, 107), and thus are resistant to many current therapies that target actively proliferating cells. Exposure of colon cancer cell lines to colon cancer chemotherapeutic agents oxaliplatin, 5-FU, or the combination of the two (FOLFOX), selects for cells that have higher CD133 expression (108, 109). Exposure of colon CSC-derived xenografts in mice to FOLFOX, while effective at reducing tumor size, also significantly increased the proportion of CD133+ cells (110), indicating a higher percentage of the surviving cells are colon CSCs. One therapeutic approach being actively investigated is to stimulate differentiation of colon CSCs, thus making them more vulnerable to existing therapeutic agents. Salinomycin, recently identified to target breast CSCs, appears to operate in part by this mechanism (79). Among the colon CSC markers that have been identified so far, the cell surface proteins CD44 and Lgr5 appear to have potential as therapeutic targets since they are cell surface proteins with restricted expression, and both may be important to colon CSC function.
Acknowledgments
A part of the work presented in this communication has been supported by grants to Dr Majumdar by NIH/NIA (AG014343) and the Department of Veterans Affairs (VA Merit Review)
Abbreviations
- CSC
cancer stem cell
- FAP
familial fdenomatous polyposis
- APC
adenomatous polyposis coli
- NOD/SCID
non-obese diabetic/severe combined immunodeficiency
- ALDH
aldehyde dehydrogenase
- ABC
ATP binding cassette protein
- Lgr5
leucine-rich G-protein coupled receptor 5
- EpCAM
epithelial cell adhesion molecule
References
- 1.Jemal A, Center MM, Ward E, Thun MJ. Cancer occurrence. Methods Mol Biol. 2009;471:3–29. doi: 10.1007/978-1-59745-416-2_1. [DOI] [PubMed] [Google Scholar]
- 2.Welch JP, Donaldson GA. The clinical correlation of an autopsy study of recurrent colorectal cancer. Ann Surg. 1979;189:496–502. doi: 10.1097/00000658-197904000-00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 7.Barker N, Clevers H. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology. 2010;138:1681–96. doi: 10.1053/j.gastro.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Ricci-Vitiani L, Fabrizi E, Palio E, De Maria R. Colon cancer stem cells. J Mol Med. 2009;87:1097–104. doi: 10.1007/s00109-009-0518-4. [DOI] [PubMed] [Google Scholar]
- 9.Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138:2151–62. doi: 10.1053/j.gastro.2009.12.063. [DOI] [PubMed] [Google Scholar]
- 10.Potten CS, Gandara R, Mahida YR, Loeffler M, Wright NA. The stem cells of small intestinal crypts: where are they? Cell Prolif. 2009;42:731–50. doi: 10.1111/j.1365-2184.2009.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjerknes M, Cheng H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- 12.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 13.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boman BM, Walters R, Fields JZ, Kovatich AJ, Zhang T, Isenberg GA, Goldstein SD, Palazzo JP. Colonic crypt changes during adenoma development in familial adenomatous polyposis: immunohistochemical evidence for expansion of the crypt base cell population. Am J Pathol. 2004;165:1489–98. doi: 10.1016/S0002-9440(10)63407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dexter DL, Spremulli EN, Fligiel Z, Barbosa JA, Vogel R, VanVoorhees A, Calabresi P. Heterogeneity of cancer cells from a single human colon carcinoma. Am J Med. 1981;71:949–56. doi: 10.1016/0002-9343(81)90312-0. [DOI] [PubMed] [Google Scholar]
- 16.Marsh KA, Stamp GW, Kirkland SC. Isolation and characterization of multiple cell types from a single human colonic carcinoma: tumourigenicity of these cell types in a xenograft system. J Pathol. 1993;170:441–50. doi: 10.1002/path.1711700407. [DOI] [PubMed] [Google Scholar]
- 17.Pierce GB, Nakane PK, Martinez-Hernandez A, Ward JM. Ultrastructural comparison of differentiation of stem cells of murine adenocarcinomas of colon and breast with their normal counterparts. J Natl Cancer Inst. 1977;58:1329–45. doi: 10.1093/jnci/58.5.1329. [DOI] [PubMed] [Google Scholar]
- 18.Blank M, Klussmann E, Kruger-Krasagakes S, Schmitt-Graff A, Stolte M, Bornhoeft G, Stein H, Xing PX, McKenzie IF, Verstijnen CP, et al. Expression of MUC2-mucin in colorectal adenomas and carcinomas of different histological types. Int J Cancer. 1994;59:301–6. doi: 10.1002/ijc.2910590302. [DOI] [PubMed] [Google Scholar]
- 19.Grabowski P, Schonfelder J, Ahnert-Hilger G, Foss HD, Stein H, Berger G, Zeitz M, Scherubl H. Heterogeneous expression of neuroendocrine marker proteins in human undifferentiated carcinoma of the colon and rectum. Ann N Y Acad Sci. 2004;1014:270–4. doi: 10.1196/annals.1294.030. [DOI] [PubMed] [Google Scholar]
- 20.West AB, Isaac CA, Carboni JM, Morrow JS, Mooseker MS, Barwick KW. Localization of villin, a cytoskeletal protein specific to microvilli, in human ileum and colon and in colonic neoplasms. Gastroenterology. 1988;94:343–52. doi: 10.1016/0016-5085(88)90421-0. [DOI] [PubMed] [Google Scholar]
- 21.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 22.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 23.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 24.Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M, Van Gijn ME, Suijkerbuijk S, Van de Wetering M, Marra G, Clevers H. The Intestinal Wnt/TCF Signature. Gastroenterology. 2007;132:628–32. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 25.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 26.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 27.Kunz-Schughart LA, Kreutz M, Knuechel R. Multicellular spheroids: a three-dimensional in vitro culture system to study tumour biology. Int J Exp Pathol. 1998;79:1–23. doi: 10.1046/j.1365-2613.1998.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 29.Chu P, Clanton DJ, Snipas TS, Lee J, Mitchell E, Nguyen ML, Hare E, Peach RJ. Characterization of a subpopulation of colon cancer cells with stem cell-like properties. Int J Cancer. 2009;124:1312–21. doi: 10.1002/ijc.24061. [DOI] [PubMed] [Google Scholar]
- 30.Feng HL, Liu YQ, Yang LJ, Bian XC, Yang ZL, Gu B, Zhang H, Wang CJ, Su XL, Zhao XM. Expression of CD133 correlates with differentiation of human colon cancer cells. Cancer Biol Ther. 2010;9:216–23. doi: 10.4161/cbt.9.3.10664. [DOI] [PubMed] [Google Scholar]
- 31.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, Chadburn A, Murphy AJ, Valenzuela DM, Gale NW, Thurston G, Yancopoulos GD, D’Angelica M, Kemeny N, Lyden D, Rafii S. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–7. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snippert HJ, van Es JH, van den Born M, Begthel H, Stange DE, Barker N, Clevers H. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136:2187–2194. doi: 10.1053/j.gastro.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Kemper K, Sprick MR, de Bree M, Scopelliti A, Vermeulen L, Hoek M, Zeilstra J, Pals ST, Mehmet H, Stassi G, Medema JP. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010;70:719–29. doi: 10.1158/0008-5472.CAN-09-1820. [DOI] [PubMed] [Google Scholar]
- 35.Lin EH, Hassan M, Li Y, Zhao H, Nooka A, Sorenson E, Xie K, Champlin R, Wu X, Li D. Elevated circulating endothelial progenitor marker CD133 messenger RNA levels predict colon cancer recurrence. Cancer. 2007;110:534–42. doi: 10.1002/cncr.22774. [DOI] [PubMed] [Google Scholar]
- 36.Artells R, Moreno I, Diaz T, Martinez F, Gel B, Navarro A, Ibeas R, Moreno J, Monzo M. Tumour CD133 mRNA expression and clinical outcome in surgically resected colorectal cancer patients. Eur J Cancer. 2010;46:642–9. doi: 10.1016/j.ejca.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Saigusa S, Tanaka K, Toiyama Y, Yokoe T, Okugawa Y, Ioue Y, Miki C, Kusunoki M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann Surg Oncol. 2009;16:3488–98. doi: 10.1245/s10434-009-0617-z. [DOI] [PubMed] [Google Scholar]
- 38.Horst D, Kriegl L, Engel J, Kirchner T, Jung A. CD133 expression is an independent prognostic marker for low survival in colorectal cancer. Br J Cancer. 2008;99:1285–9. doi: 10.1038/sj.bjc.6604664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li CY, Li BX, Liang Y, Peng RQ, Ding Y, Xu DZ, Zhang X, Pan ZZ, Wan DS, Zeng YX, Zhu XF, Zhang XS. Higher percentage of CD133+ cells is associated with poor prognosis in colon carcinoma patients with stage IIIB. J Transl Med. 2009;7:56. doi: 10.1186/1479-5876-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horst D, Scheel SK, Liebmann S, Neumann J, Maatz S, Kirchner T, Jung A. The cancer stem cell marker CD133 has high prognostic impact but unknown functional relevance for the metastasis of human colon cancer. J Pathol. 2009;219:427–34. doi: 10.1002/path.2597. [DOI] [PubMed] [Google Scholar]
- 41.Choi D, Lee HW, Hur KY, Kim JJ, Park GS, Jang SH, Song YS, Jang KS, Paik SS. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J Gastroenterol. 2009;15:2258–64. doi: 10.3748/wjg.15.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du L, Wang H, He L, Zhang J, Ni B, Wang X, Jin H, Cahuzac N, Mehrpour M, Lu Y, Chen Q. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–60. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 43.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, Clevers H, Pals ST. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154:515–23. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H, Yang XL, Rosada C, Hamilton SR, August JT. CD44 expression in colorectal adenomas is an early event occurring prior to K-ras and p53 gene mutation. Arch Biochem Biophys. 1994;310:504–7. doi: 10.1006/abbi.1994.1199. [DOI] [PubMed] [Google Scholar]
- 46.Kopp R, Fichter M, Schalhorn G, Danescu J, Classen S. Frequent expression of the high molecular, 673-bp CD44v3, v8–10 variant in colorectal adenomas and carcinomas. Int J Mol Med. 2009;24:677–83. doi: 10.3892/ijmm_00000279. [DOI] [PubMed] [Google Scholar]
- 47.Mulder JW, V, Wielenga J, Polak MM, van den Berg FM, Adolf GR, Herrlich P, Pals ST, Offerhaus GJ. Expression of mutant p53 protein and CD44 variant proteins in colorectal tumorigenesis. Gut. 1995;36:76–80. doi: 10.1136/gut.36.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ropponen KM, Eskelinen MJ, Lipponen PK, Alhava E, Kosma VM. Expression of CD44 and variant proteins in human colorectal cancer and its relevance for prognosis. Scand J Gastroenterol. 1998;33:301–9. doi: 10.1080/00365529850170900. [DOI] [PubMed] [Google Scholar]
- 49.Suh SI, Baek WK, Park JW, Bae OS, Suh MH, Choe BK. Identification of CD44 splice variant in Korean colorectal cancers and cell lines. J Korean Med Sci. 1995;10:169–75. doi: 10.3346/jkms.1995.10.3.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi K, Yamaguchi A, Urano T, Goi T, Nakagawara G, Shiku H. Expression of CD44 variant exons 8–10 in colorectal cancer and its relationship to metastasis. Jpn J Cancer Res. 1995;86:292–7. doi: 10.1111/j.1349-7006.1995.tb03053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramaniam V, I, Vincent R, Gilakjan M, Jothy S. Suppression of human colon cancer tumors in nude mice by siRNA CD44 gene therapy. Exp Mol Pathol. 2007;83:332–40. doi: 10.1016/j.yexmp.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Subramaniam V, I, Vincent R, Gardner H, Chan E, Dhamko H, Jothy S. CD44 regulates cell migration in human colon cancer cells via Lyn kinase and AKT phosphorylation. Exp Mol Pathol. 2007;83:207–15. doi: 10.1016/j.yexmp.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Misra S, V, Hascall C, De Giovanni C, Markwald RR, Ghatak S. Delivery of CD44 shRNA/nanoparticles within cancer cells: perturbation of hyaluronan/CD44v6 interactions and reduction in adenoma growth in Apc Min/+ MICE. J Biol Chem. 2009;284:12432–46. doi: 10.1074/jbc.M806772200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel BB, Yu Y, Du J, Levi E, Phillip PA, Majumdar AP. Age-related increase in colorectal cancer stem cells in macroscopically normal mucosa of patients with adenomas: a risk factor for colon cancer. Biochem Biophys Res Commun. 2009;378:344–7. doi: 10.1016/j.bbrc.2008.10.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levi E, Misra S, Du J, Patel BB, Majumdar AP. Combination of aging and dimethylhydrazine treatment causes an increase in cancer-stem cell population of rat colonic crypts. Biochem Biophys Res Commun. 2009;385:430–3. doi: 10.1016/j.bbrc.2009.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weichert W, Knosel T, Bellach J, Dietel M, Kristiansen G. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol. 2004;57:1160–4. doi: 10.1136/jcp.2004.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105:13427–32. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang DD, Kim YJ, Lee CN, Aggarwal S, McKinnon K, Mesmer D, Norton J, Birse CE, He T, Ruben SM, Moore PA. Expansion of CD133 (+) colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br J Cancer. 2010;102:1265–75. doi: 10.1038/sj.bjc.6605610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKinnon PJ, McLaughlin SK, Kapsetaki M, Margolskee RF. Extracellular matrix-associated protein Sc1 is not essential for mouse development. Mol Cell Biol. 2000;20:656–60. doi: 10.1128/mcb.20.2.656-660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horst D, Kriegl L, Engel J, Kirchner T, Jung A. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Invest. 2009;27:844–50. doi: 10.1080/07357900902744502. [DOI] [PubMed] [Google Scholar]
- 62.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–9. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, Li C, Wang LP, Roby KF, Orsulic S, Connolly DC, Zhang Y, Montone K, Butzow R, Coukos G, Zhang L. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69:8208–15. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sladek NE. Aldehyde dehydrogenase-mediated cellular relative insensitivity to the oxazaphosphorines. Curr Pharm Des. 1999;5:607–25. [PubMed] [Google Scholar]
- 67.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moreb JS, Mohuczy D, Ostmark B, Zucali JR. RNAi-mediated knockdown of aldehyde dehydrogenase class-1A1 and class-3A1 is specific and reveals that each contributes equally to the resistance against 4-hydroperoxycyclophosphamide. Cancer Chemother Pharmacol. 2007;59:127–36. doi: 10.1007/s00280-006-0233-6. [DOI] [PubMed] [Google Scholar]
- 69.Sreerama L, Sladek NE. Identification and characterization of a novel class 3 aldehyde dehydrogenase overexpressed in a human breast adenocarcinoma cell line exhibiting oxazaphosphorine-specific acquired resistance. Biochem Pharmacol. 1993;45:2487–505. doi: 10.1016/0006-2952(93)90231-k. [DOI] [PubMed] [Google Scholar]
- 70.Wroczynski P, Nowak M, Wierzchowski J, Szubert A, Polanski J. Activities of cytosolic aldehyde dehydrogenase isozymes in colon cancer: determination using selective, fluorimetric assays. Acta Pol Pharm. 2005;62:427–33. [PubMed] [Google Scholar]
- 71.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998;95:15665–70. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding XW, Wu JH, Jiang CP. ABCG2: a potential marker of stem cells and novel target in stem cell and cancer therapy. Life Sci. 2010;86:631–7. doi: 10.1016/j.lfs.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 73.Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, Sorrentino BP. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–34. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 74.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–33. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haraguchi N, Utsunomiya T, Inoue H, Tanaka F, Mimori K, Barnard GF, Mori M. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–13. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 76.Burkert J, Otto WR, Wright NA. Side populations of gastrointestinal cancers are not enriched in stem cells. J Pathol. 2008;214:564–73. doi: 10.1002/path.2307. [DOI] [PubMed] [Google Scholar]
- 77.Liu C, Zhao G, Liu J, Ma N, Chivukula P, Perelman L, Okada K, Chen Z, Gough D, Yu L. Novel biodegradable lipid nano complex for siRNA delivery significantly improving the chemosensitivity of human colon cancer stem cells to paclitaxel. J Control Release. 2009;140:277–83. doi: 10.1016/j.jconrel.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 78.Blokzijl H, Vander Borght S, Bok LI, Libbrecht L, Geuken M, van den Heuvel FA, Dijkstra G, Roskams TA, Moshage H, Jansen PL, Faber KN. Decreased P-glycoprotein (P-gp/MDR1) expression in inflamed human intestinal epithelium is independent of PXR protein levels. Inflamm Bowel Dis. 2007;13:710–20. doi: 10.1002/ibd.20088. [DOI] [PubMed] [Google Scholar]
- 79.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riccioni R, Dupuis ML, Bernabei M, Petrucci E, Pasquini L, Mariani G, Cianfriglia M, Testa U. The cancer stem cell selective inhibitor salinomycin is a p-glycoprotein inhibitor. Blood Cells Mol Dis. 2010;45:86–92. doi: 10.1016/j.bcmd.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 81.Uchida H, Yamazaki K, Fukuma M, Yamada T, Hayashida T, Hasegawa H, Kitajima M, Kitagawa Y, Sakamoto M. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci. 2010;101:1731–1737. doi: 10.1111/j.1349-7006.2010.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 83.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 84.McClanahan T, Koseoglu S, Smith K, Grein J, Gustafson E, Black S, Kirschmeier P, Samatar AA. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–26. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- 85.Becker L, Huang Q, Mashimo H. Immunostaining of Lgr5, an intestinal stem cell marker, in normal and premalignant human gastrointestinal tissue. Scientific World Journal. 2008;8:1168–76. doi: 10.1100/tsw.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morita H, Mazerbourg S, Bouley DM, Luo CW, Kawamura K, Kuwabara Y, Baribault H, Tian H, Hsueh AJ. Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol. 2004;24:9736–43. doi: 10.1128/MCB.24.22.9736-9743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weichert W, Denkert C, Burkhardt M, Gansukh T, Bellach J, Altevogt P, Dietel M, Kristiansen G. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin Cancer Res. 2005;11:6574–81. doi: 10.1158/1078-0432.CCR-05-0606. [DOI] [PubMed] [Google Scholar]
- 88.Fujimoto K, Beauchamp RD, Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123:1941–8. doi: 10.1053/gast.2002.37065. [DOI] [PubMed] [Google Scholar]
- 89.Strater J, Wedding U, Barth TF, Koretz K, Elsing C, Moller P. Rapid onset of apoptosis in vitro follows disruption of beta 1-integrin/matrix interactions in human colonic crypt cells. Gastroenterology. 1996;110:1776–84. doi: 10.1053/gast.1996.v110.pm8964403. [DOI] [PubMed] [Google Scholar]
- 90.Kirkland SC. Type I collagen inhibits differentiation and promotes a stem cell-like phenotype in human colorectal carcinoma cells. Br J Cancer. 2009;101:320–6. doi: 10.1038/sj.bjc.6605143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herlyn D, Herlyn M, Steplewski Z, Koprowski H. Monoclonal antibodies in cell-mediated cytotoxicity against human melanoma and colorectal carcinoma. Eur J Immunol. 1979;9:657–9. doi: 10.1002/eji.1830090817. [DOI] [PubMed] [Google Scholar]
- 92.Went P, Vasei M, Bubendorf L, Terracciano L, Tornillo L, Riede U, Kononen J, Simon R, Sauter G, Baeuerle PA. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer. 2006;94:128–35. doi: 10.1038/sj.bjc.6602924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 94.Gosens MJ, van Kempen LC, van de Velde CJ, van Krieken JH, Nagtegaal ID. Loss of membranous Ep-CAM in budding colorectal carcinoma cells. Mod Pathol. 2007;20:221–32. doi: 10.1038/modpathol.3800733. [DOI] [PubMed] [Google Scholar]
- 95.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M, Clevers H. Transcription factor achaete scutelike 2 controls intestinal stem cell fate. Cell. 2009;136:903–12. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 96.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–7. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 97.Conrotto P, Roesli C, Rybak J, Kischel P, Waltregny D, Neri D, Castronovo V. Identification of new accessible tumor antigens in human colon cancer by ex vivo protein biotinylation and comparative mass spectrometry analysis. Int J Cancer. 2008;123:2856–64. doi: 10.1002/ijc.23861. [DOI] [PubMed] [Google Scholar]
- 98.Koshida S, Kobayashi D, Moriai R, Tsuji N, Watanabe N. Specific overexpression of OLFM4 (GW112/HGC-1) mRNA in colon, breast and lung cancer tissues detected using quantitative analysis. Cancer Sci. 2007;98:315–20. doi: 10.1111/j.1349-7006.2006.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu W, Liu Y, Zhu J, Wright E, Ding I, Rodgers GP. Reduced hGC-1 protein expression is associated with malignant progression of colon carcinoma. Clin Cancer Res. 2008;14:1041–9. doi: 10.1158/1078-0432.CCR-07-4125. [DOI] [PubMed] [Google Scholar]
- 100.Tateishi K, Ohta M, Kanai F, Guleng B, Tanaka Y, Asaoka Y, Tada M, Seto M, Jazag A, Lianjie L, Okamoto M, Isayama H, Yoshida H, Kawabe T, Omata M. Dysregulated expression of stem cell factor Bmi1 in precancerous lesions of the gastrointestinal tract. Clin Cancer Res. 2006;12:6960–6. doi: 10.1158/1078-0432.CCR-06-0449. [DOI] [PubMed] [Google Scholar]
- 101.Du J, Li Y, Li J, Zheng J. Polycomb group protein Bmi1 expression in colon cancers predicts the survival. Med Oncol. 2009 doi: 10.1007/s12032-009-9373-y. [DOI] [PubMed] [Google Scholar]
- 102.Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H, Chiba T. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett 2003. 2003;535:131–5. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- 103.Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, Ashton G, Clarke R, Sakakibara S, Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 104.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–58. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 105.Segal NH, Saltz LB. Evolving treatment of advanced colon cancer. Annu Rev Med. 2009;60:207–19. doi: 10.1146/annurev.med.60.041807.132435. [DOI] [PubMed] [Google Scholar]
- 106.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–46. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Quesnel B. Tumor dormancy and immunoescape. APMIS. 2008;116:685–94. doi: 10.1111/j.1600-0463.2008.01163.x. [DOI] [PubMed] [Google Scholar]
- 108.Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G, 2nd, Samuel S, Kim MP, Lim SJ, Ellis LM. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–7. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu Y, Kanwar SS, Patel BB, Nautiyal J, Sarkar FH, Majumdar AP. Elimination of Colon Cancer Stem-Like Cells by the Combination of Curcumin and FOLFOX. Transl Oncol. 2009;2:321–8. doi: 10.1593/tlo.09193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, Stassi G. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]


