Keywords: Endemism, Genome size, Island flora, Low-copy nuclear markers, Molecular dating
Highlights
-
•
Analyses showed resolution among 1/3 of the Diospyros species form NC clade III.
-
•
Some morphological distinct species could not be discriminated with the markers used.
-
•
Diospyros arrived relative recently (5–20 mya) in New Caledonia.
-
•
Chromosome counts confirmed the investigated species to be diploid.
-
•
Genome sizes varied nearly fourfold among species examined.
Abstract
To clarify phylogenetic relationships among New Caledonian species of Diospyros, sequences of four plastid markers (atpB, rbcL, trnK–matK and trnS–trnG) and two low-copy nuclear markers (ncpGS and PHYA) were analysed. New Caledonian Diospyros species fall into three clades, two of which have only a few members (1 or 5 species); the third has 21 closely related species for which relationships among species have been mostly unresolved in a previous study. Although species of the third group (NC clade III) are morphologically distinct and largely occupy different habitats, they exhibit little molecular variability. Diospyros vieillardii is sister to the rest of the NC clade III, followed by D. umbrosa and D. flavocarpa, which are sister to the rest of this clade. Species from coastal habitats of western Grande Terre (D. cherrieri and D. veillonii) and some found on coralline substrates (D. calciphila and D. inexplorata) form two well-supported subgroups. The species of NC clade III have significantly larger genomes than found in diploid species of Diospyros from other parts of the world, but they all appear to be diploids. By applying a molecular clock, we infer that the ancestor of the NC clade III arrived in New Caledonia around 9 million years ago. The oldest species are around 7 million years old and the youngest ones probably much less than 1 million years.
1. Introduction
New Caledonia is an island group located in the southwestern Pacific about 1300 km east of Australia, ranging from around 19° to 23° south with an land area of ca. 19,000 km2. It consists of the main island Grande Terre (ca. 16,000 km2), Iles Belep (in the north), Ile des Pins (in the south), Loyalty Islands (in the east) and several other smaller islands. The continental part of New Caledonia (mainly Grande Terre) separated from Gondwanan during late Cretaceous (ca. 80 million years ago, mya; McLoughlin, 2001). During the Palaeocene to late Eocene, this continental sliver was submerged for at least 20 million years (myr), and a thick layer of oceanic mantle accumulated (Pelletier, 2006). After Grande Terre re-emerged in the late Eocene (37 mya), this heavy-metal rich oceanic material covered most of the land. Today, around 1/3 of the main island is still covered with ultramafic substrates. Because Grande Terre was totally submerged, it is highly unlikely that lineages that were already present in this area before the split from Gondwanan could have survived locally. Current hypotheses suggest that biota present today are derived from elements/ancestors that reached New Caledonia via long distance dispersal (e.g. Morat et al., 2012; Pillon, 2012; Grandcolas et al., 2008) mainly from Australia, New Guinea and Malaysia. Hypotheses of other islands between Australia and New Caledonia having served as stepping stones or refuges for Gondwanan taxa now endemic (e.g. Amborella) have been proposed by a few authors (Ladiges and Cantrill, 2007), but there is no consensus of when they existed or how large and numerous they might have been. The New Caledonian climate is tropical to subtropical. The main island is split by a mountain range into a humid eastern portion (2000–4000 mm precipitation per year) and a dry western part (1000 mm precipitation per year) with prevailing winds and rain coming from the south east. New Caledonia is one of the 34 biodiversity hotspots (Mittermeier et al., 2004; Myers et al., 2000), and nearly 75% of the native flora is endemic (Morat et al., 2012), which is the fourth highest for an island (Lowry, 1998). Among these endemic taxa are 98 genera and three families, Amborellaceae, Oncothecaceae and Phellinaceae (Morat et al., 2012). One of the reasons hypothesised for the high level of endemism found in New Caledonia is the ultramafic substrates, which have acted as a filter for colonising species that were already pre-adapted to this special soil (Pillon et al., 2010).
Ebenaceae are pantropical and belong to the order Ericales (APG, 2009); the majority of species occur in Africa (incl. Madagascar) and the Indo-Pacific region. Duangjai et al. (2006) divided Ebenaceae into two sub families, Lissocarpoideae and Ebenoideae. Lissocarpoideae are monogeneric (Lissocarpa, 8 species in northwestern South America), and Ebenoideae include Diospyros, Euclea (18 species in Africa) and Royena (17 species in Africa). This classification of Ebenaceae in two subfamilies and four genera has been also supported by palynological data (Geeraerts et al., 2009).
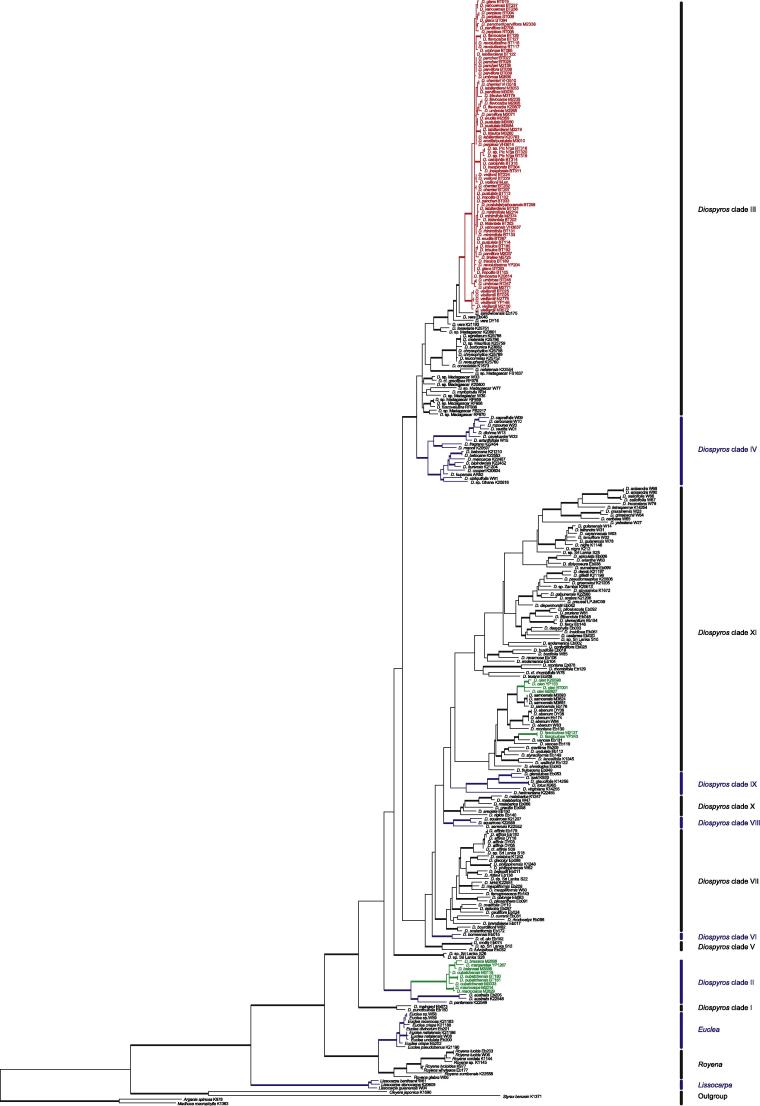
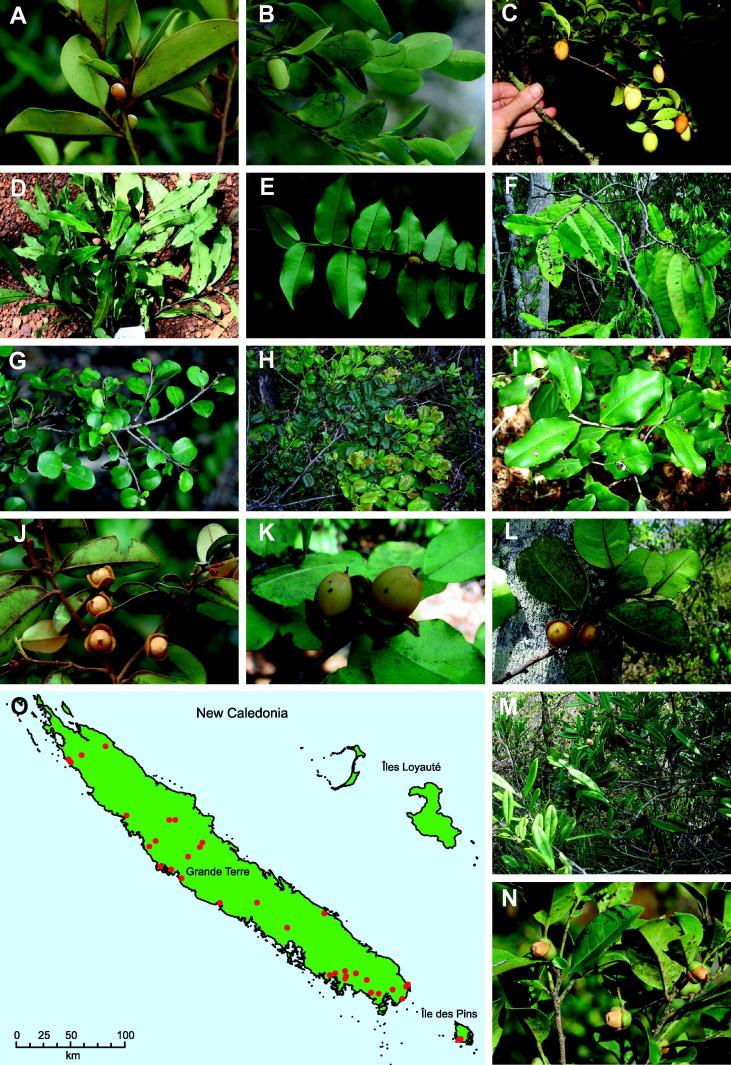
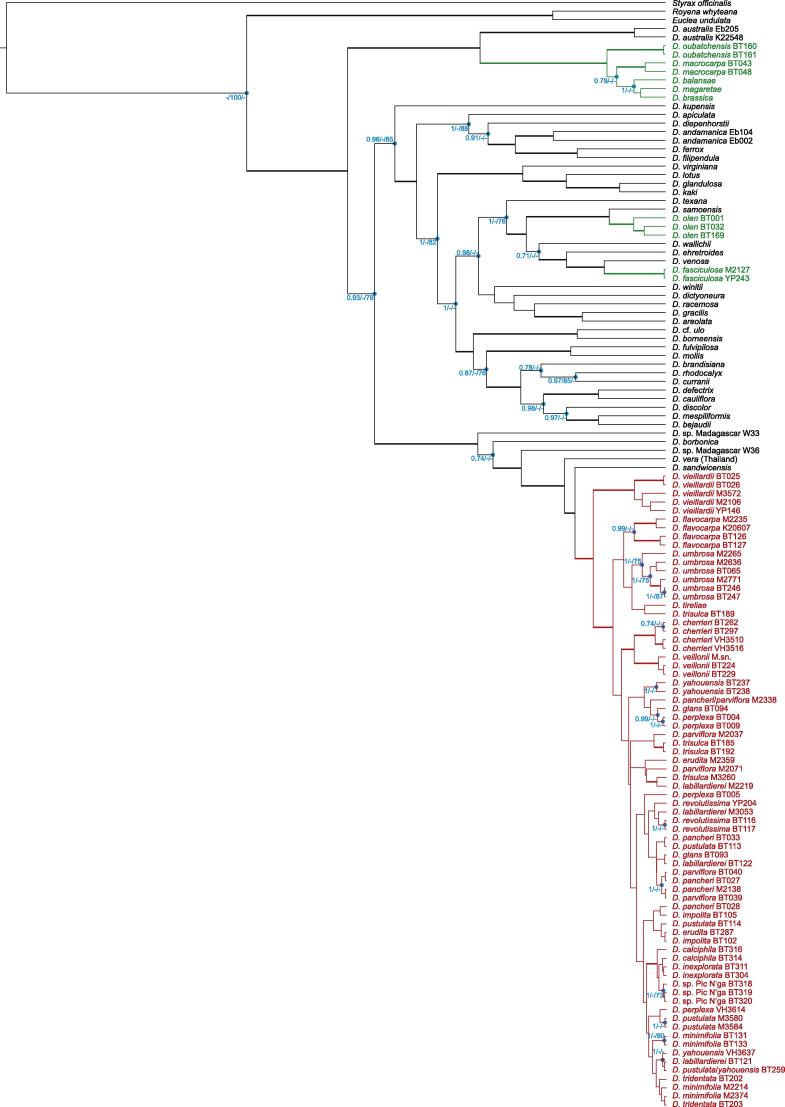
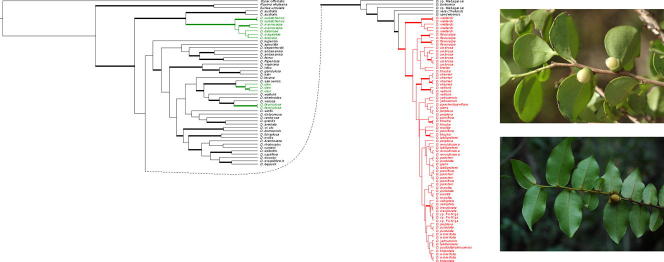
In this paper, we use the circumscription of Diospyros as proposed by Duangjai et al. (2006). Diospyros is the largest genus of Ebenaceae with more than 500 species, making it also one of the largest angiosperm genera. The greatest species of diversity is in Asia and the Pacific region (∼300 species). Fruits of some species (persimmons; e.g. D. kaki, D. lotus and D. virginiana) are edible, and ebony wood (e.g. D. ebenum) is one of the most expensive timbers. Species of Diospyros are shrubs or trees that occur in most tropical and subtropical habitats, where they are often important and characteristic elements. Duangjai et al. (2009) found 11 mostly well-resolved clades within Diospyros. In New Caledonia, there are 31 described Diospyros species, of which all but one are endemic, and they belong to three clades (Duangjai et al., 2009; Fig. 4, clades II, III and XI). The first clade (clade II) contains five species from New Caledonia that are related to Australian species of Diospyros. The second clade (clade III) includes species from Hawai‘i, Indian Ocean islands and 24 taxa from New Caledonia, within which the species from New Caledonia form a sublcade, here termed NC clade III. Although Duangjai et al. (2009) analysed more than 8000 base pairs of plastid DNA, low variability and little resolution was found among these endemic New Caledonian species. The third clade (clade XI), consisting of taxa from Asia, America, Pacific Islands and New Caledonia, includes two Diospyros species from New Caledonia, one endemic and the other found throughout the southern Pacific. These two species are not sister species, accounting for two more colonisations of New Caledonia (i.e. four in total). Similar, multiple colonisation events are also found among other organisms in New Caledonia (e.g. Murienne et al., 2005). Diospyros is observed in all types of New Caledonian vegetation except mangrove; the species range from sea level up to ca. 1250 m (New Caledonia’s highest point is 1628 m). There are several micro-endemics restricted to just a small area (White, 1992). Most New Caledonian Diospyros species from clade III are morphologically clearly defined and restricted by edaphic factors and occur on just one substrate type. For example, D. labillardierei (Fig. 1D) is distinctive with its long narrow leaves and Salix-like habit; it is a rheophyte on non-ultramafic substrates. Diospyros veillonii (Fig. 1F) is a remarkable species with coralloid inflorescence axes (unique among New Caledonian Diospyros) and large leaves, but is known from only a single locality in dry forest on black clay soil. Other species have broader distributions and ecologies, such as D. parviflora (Fig. 1J), which grows on both ultramafic and non-ultramafic substrates and is widespread throughout Grande Terre and Balabio Island in dense humid forests as well as in more open and dry vegetation. Some species can have similar ecological requirements, but are morphologically well differentiated; for example D. vieillardii (Fig. 1A) has a calyx narrower than its prune-like fruit, whereas D. glans (Fig. 1N) has a thick calyx much wider than its fruit, but both grow in maquis vegetation and co-occur at some sites.
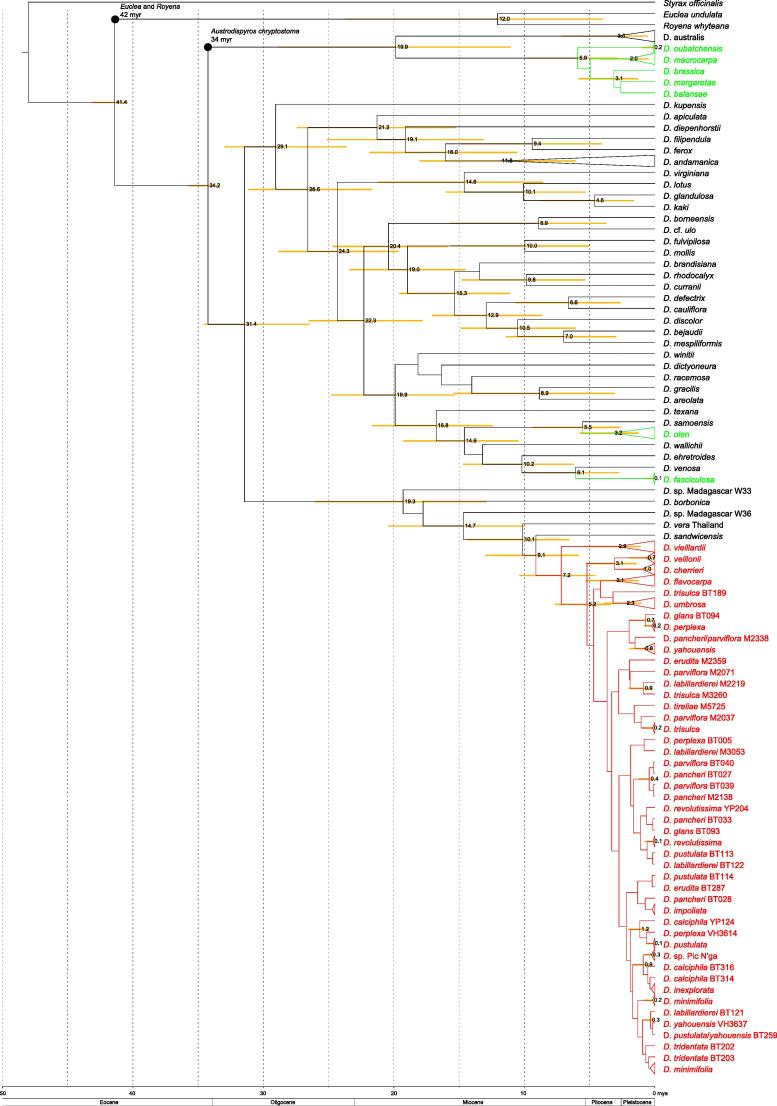
Fig. 4.
One of 210 equally parsimonius trees of the plastid data set. Clades are named according to Duangjai et al. (2009). Bold branches have more than 70% support in all three analysis. New Caledonian taxa are coloured, red represents clade III NC.
Fig. 1.
Examples of Diospyros species from New Caledonia (A–N) and Map of New Caledonia with collection points (O). A: D. vieillardii; B: D. umbrosa; C: D. flavocarpa, D: D. labillardierei; E: D. pancheri, F: D. veillonii; G: D. minimifolia; H: D. pustulata; I: D. cherrieri; J: D. parviflora; K: D.perplexa; L: D. yaouhensis; M: D. revolutissima; N: D. glans; O: Map of New Caledonia with sampling localities. Photographs taken by: C. Chambrey (I), V. Hequet (F, K, L), J. Munzinger (A, B, C, E, G, H, J, M, N) and B. Turner (D).
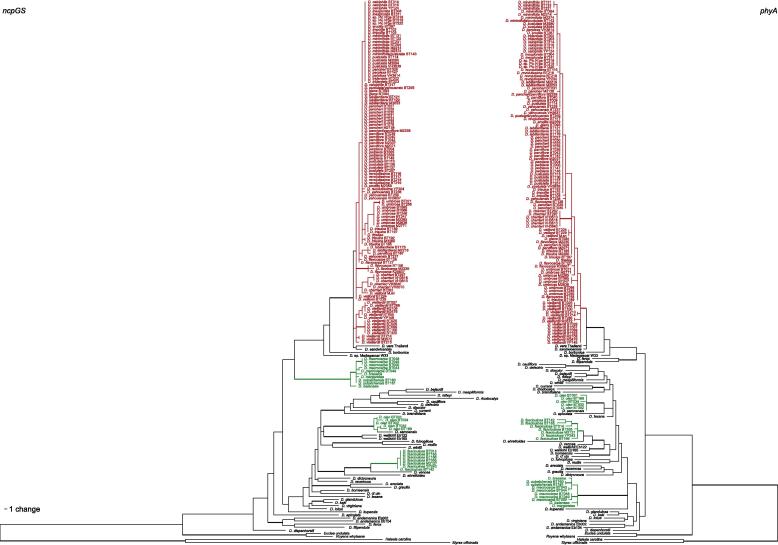
For establishing phylogenetic relationships, sequences of low-copy nuclear genes are not as often used as regions from the plastid genome, often due to methodological difficulties. Low-copy genes are present in one or few copies in the genome, and primers are often highly specific for individual groups, requiring them therefore to be newly designed for each study. On the other hand, low-copy nuclear markers are normally highly informative and as they are biparentally inherited they may also help detect recent hybridization (e.g. Moody and Rieseberg, 2012). However, in a study of Hawaiian endemics in two unrelated genera, Pillon et al. (2013) found that although two low-copy nuclear loci displayed a high level of variability, they also exhibited heterozygosity, intraspecific variation, and retention of ancient alleles; allele coalescence was older than the species under study. Nonetheless, we hoped that inclusion of low-copy nuclear genes might provide additional insight into species relationships and thus included two such loci. Phytochrome A (PHYA) belongs to the gene family of the phytochromes, which has eight members across the seed plants (PHYA–PHYE in angiosperms and PHYN–PHYP in gymnosperms); PHYN/PHYA, PHYO/PHYC and PHYP/PHYBDE are orthologs, the rest being paralogs of the others (Mathews et al., 2010). Genes of this family encode photoreceptor proteins that mediate developmental responses to red and far red light. The three main paralogs (PHYA, PHYB and PHYC) are different enough to be amplified with specific primers (Zimmer and Wen, 2012). Sequences of phy genes have been used successfully across the flowering plants (e.g. Mathews et al., 2010; Nie et al., 2008; Bennett and Mathews, 2006) for phylogenetic reconstruction. The gene PHYA used in this study consists of four exons and three introns. Glutamine synthetase (GS), codes for a protein involved in nitrogen assimilation. There are two main types of GS genes, cytosolic- and chloroplast-expressed. Chloroplast-expressed glutamine synthetase (ncpGS) consists of 12 exons and 11 introns and has been shown to be a single-copy gene in plants (Emshwiller and Doyle, 1999). This combination of coding and non-coding regions has been shown to be highly informative for inferring phylogenetic trees of various groups (e.g. Oxalidaceae, Emshwiller and Doyle, 1999; Passiflora, Yockteng and Nadot, 2004; Spiraeanthemum, Pillon et al., 2009a; Codia, Pillon et al., 2009b; Achillea millefolium, Guo et al., 2012).
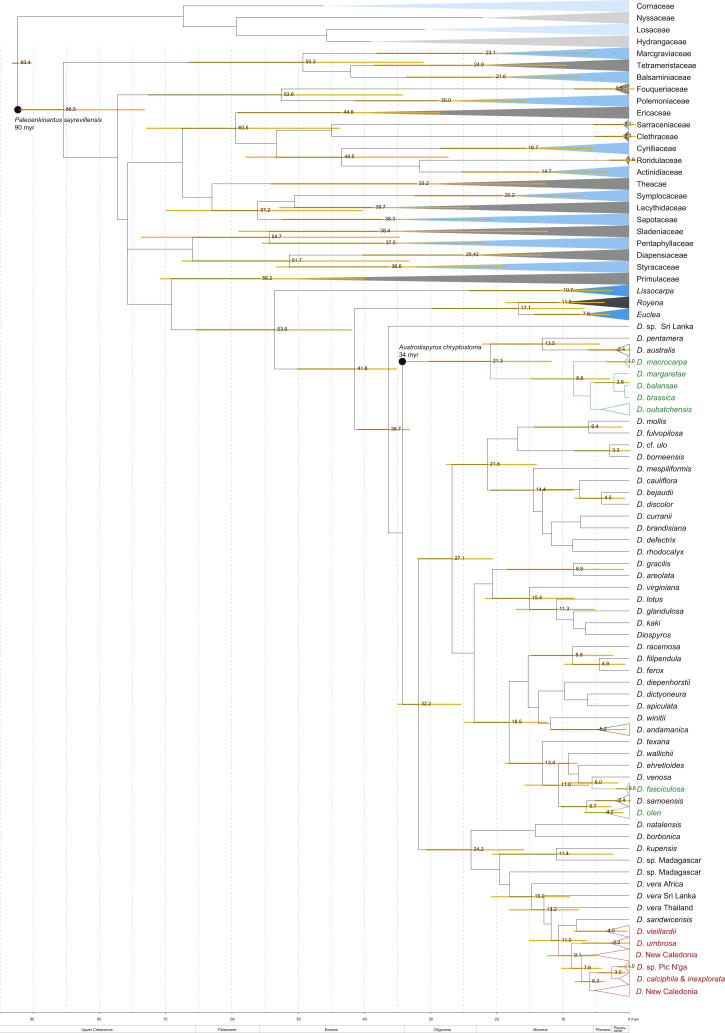
Beside phylogenetic relationships, the age of clades is of interest. In many cases, there are no fossils available for direct dating of a group of interest in a particular region, which is often the case for islands and is certainly true for New Caledonia (the few fossils recorded to date are older than the last emergence of the island and are not certain to be angiosperms; Salard and Avias, 1968). Rates of DNA divergence are generally consistent with a molecular clock (Zuckerkandl and Pauling, 1965), and therefore DNA data contain information about the relative ages of taxa. When substitution rates (e.g. Silvestro et al., 2011; Alba et al., 2000) or fossils belonging to defined clades (e.g. Pirie and Doyle, 2012; Magallón, 2010) are taken into consideration, the relative ages obtained can be transformed into absolute ages. Placement of fossils in the correct position in the phylogenetic tree is crucial for accurate interpretation (Forest, 2009). Some previous studies have has been published on the subject of the age of asterids (e.g. Millán-Martínez, 2010; Bell et al., 2010; Bremer et al., 2004) to which Ericales belong, and fossil Diospyros are known from some localities (mainly in India and North America), but none has been found in New Caledonia. Austrodiospyros cryptostoma (Basinger and Christophel, 1985), a fossil from Australia has many morphological similarities to D. australis of clade II (Duangjai et al., 2009). It is thus far the only fossil belonging to a clade that includes Diospyros species from New Caledonia. We treat A. cryptostoma as member of clade II in this study.
Genome sizes vary nearly 2400-fold across angiosperms (Pellicer et al., 2010). Most variation in DNA amount is caused by different amounts of non-coding, repetitive DNA, such as pseudogenes, retrotransposons, transposons and satellite repeats (Leitch, 2007; Bennett and Leitch, 2005; Parisod et al., 2009; Petrov, 2001). Genome sizes and chromosome numbers of Diospyros are within the range of those of other members of Ericales (Bennett and Leitch, 2010). Nuclear DNA amounts in Diospyros range from 0.78 pg (1C-value) in diploid D. rhodocalyx up to 4.06 pg in nonaploid D. kaki cultivars (Tamura et al., 1998). The basic chromosome number in Diospyros is 2n = 2x = 30, and most species seem to be diploid (e.g. Tamura et al., 1998; White, 1992). There are some reports of polyploid Diospyros, mostly from cultivated species (e.g. D. rhombifolia 4x, D. ebenum 6x, D. kaki 6x and 9x, D. virginiana 6x and 9x; Tamura et al., 1998). White (1992) provided chromosome counts for nine New Caledonian species of Diospyros (D. calciphila, D. fasciculosa, D. flavocarpa, D. minimifolia, D. olen, D. parviflora, D. umbrosa, D. vieillardii and D. yaouhensis), all of which are diploid.
Duangjai et al. (2009) found little sequence variation in the markers investigated among many species from NC clade III, which could indicate recent diversification. White (1992), who described most the New Caledonian Diospyros species, suspected some hybridization was taking place. The main aim of this study was to clarify relationships among New Caledonian Diospyros species, especially of those belonging to clade III (Duangjai et al., 2009). Furthermore, if we were able to find more variable than those previously studied, we wanted to elucidate potential factors underlying speciation (e.g. ecological speciation, hybrid speciation and introgression) and understand better differences in speciation rates of the clades that reached New Caledonia independently. We used low-copy nuclear markers, PHYA and ncpGS because they offered the prospect of resolving relationships within this clade and detecting possible hybrid species. We also included samples from nine additional species that were not available for the study of Duangjai et al. (2009). Moreover, we conducted dating analyses to obtain estimates of the ages for the lineages to which New Caledonian Diospyros species belong. We also present chromosome numbers and genome sizes of some additional New Caledonian species of Diospyros; we wished to examine further the hypothesis that polyploidy (perhaps involving hybridization) might have played a role in producing diversity in this comparatively species-rich clade.
2. Materials and methods
2.1. Material
Material from New Caledonian Diospyros species was collected by B. Turner (BT), J. Munzinger (JM), Yohan Pillon (YP) or Vanessa Hequet (VH). When fertile, a voucher was made with several duplicates sent to various herbaria. When sterile, one voucher per population was taken; this was compared to already existing collections in Noumea Herbarium (NOU) from the same location and referred to that species if similar. One putatively new species was detected while doing fieldwork for this project, here called D. sp. Pic N’ga. Other Ebenaceae samples are from previous studies (Duangjai et al., 2009). Outgroup taxa and a few Diospyros samples were taken from the Royal Botanic Gardens, Kew, DNA Bank (http://apps.kew.org/dnabank/homepage.html). Compared to the sampling of Duangjai et al. (2009), we added material of the following New Caledonian species: D. erudita, D. glans, D. impolita, D. inexplorata, D. margaretae, D. tireliae, D. tridentata, D. trisulca and D. veillonii (for details see Table 1). The three un-sampled species from New Caledonia (D. fastidiosa, D. nebulosa and D. neglecta) are rare and have not been seen after their description.
Table 1.
Table of accessions; showing all individuals used in this study. Sequences provided by S. Duangjai are indicated.
| Taxon | Acc.-nr. | Origin | Voucher | Herbarium | atpB | rbcL | matK & trnK intron | trnS–trnG | ncpGS | PHYA |
|---|---|---|---|---|---|---|---|---|---|---|
| D. abyssinica (Hiern) F. White | K1672 | Africa | Gilbert & Sebseke 8803 | K | DQ923883 | EU980646 | DQ923990 | EU981061 | ||
| D. affinis Thwaites | DY03 | Sri Lanka | Yakandawala 03 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291310 | ||
| D. affinis | DY05 | Sri Lanka | Yakandawala 05 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291311 | ||
| D. affinis | DY18 | Sri Lanka | Yakandawala 18 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291312 | ||
| D. affinis | Eb179 | Sri Lanka | Samuel s.n. | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291313 | ||
| D. affinis | Eb180 | Sri Lanka | Samuel s.n. | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291314 | ||
| D. cf. affinis | S09 | Sri Lanka | Samuel 09 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291315 | ||
| D. andamanica (Kurz) Bakh. | Eb002 | Thailand | Duangjai 068 | KUFF, W | DQ923884 | EU980645 | DQ923991 | EU981060 | KF291447 | KF291624 |
| D. andamanica | Eb104 | Thailand | Duangjai 162 | KUFF, W | DQ923950 | EU980755 | DQ924057 | EU981170 | KF291448 | KF291625 |
| D. anisandra S.F. Blake | W68 | Guatemala | Wallnöfer 6012 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291316 | ||
| D. anisandra | W80 | Guatemala | Frisch 2006-1 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291317 | ||
| D. apiculata Hiern | Eb006 | Thailand | Duangjai 072 | KUFF | EU980813 | EU980647 | EU980936 | EU981062 | KF291449 | KF291626 |
| D. areolata King & Gamble | Eb160 | Brunei | Duangjai et al. 33 | BRUN, W, WU | Duangjai unpubl | Duangjai unpubl | Duangjai unpubl | KF291318 | KF291450 | KF291627 |
| D. artanthifolia Mart. ex Miq. | W15 | Peru | Pirie 62 | W | DQ923885 | EU980648 | DQ923992 | EU981063 | ||
| D. australis (R.Br) Hiern | Eb205 | Australia | Wallnöfer & Duangjai 13944 | WU | DQ923887 | EU980650 | DQ923994 | EU981065 | ||
| D. australis | K22548 | Australia | Forster 7848 | K | DQ923886 | EU980649 | DQ923993 | EU981064 | ||
| D. balansae Guillaumin | M3556 | New Caledonia | Munzinger 3556 | NOU015466 | EU980814 | EU980651 | EU980937 | EU981066 | KF291451 | KF291628 |
| D. batocana Hiern | K21210 | Namibia | Steyl 88 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291319 | ||
| D. batocana | K22553 | Zambia | Pope et al. 2196 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291320 | ||
| D. bejaudii Lecomte | Eb011 | Thailand | Duangjai 075 | KUFF, W | DQ923888 | EU980652 | DQ923995 | EU981067 | KF291452 | KF291629 |
| D. bipindensis Gürke | K22452 | Gabon | Stone & Niangadouma 3554 | MO | DQ923889 | EU980653 | DQ923996 | EU981068 | ||
| D. borbonica I. Richardson | K23682 | Reunion | Chase REU10042 | REU, WU | EU980815 | EU980654 | EU980938 | EU981069 | KF291453 | KF291630 |
| D. borneensis Hiern | Eb015 | Thailand | Duangjai 079 | KUFF, W | DQ923890 | EU980655 | DQ923997 | EU981070 | KF291454 | KF291631 |
| D. bourdillonii Brandis | W82 | India | DeFranceschi 18.12.2006 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291321 | ||
| D. brandisiana Kurz | Eb017 | Thailand | Duangjai & Sinbumrung 007 | KUFF, W | DQ923891 | EU980656 | DQ923998 | EU981071 | KF291455 | KF291632 |
| D. brassica F. White | M2898 | New Caledonia | Munzinger 2898 | NOU007949 | DQ923892 | EU980657 | DQ923999 | EU981072 | KF291456 | KF291633 |
| D. buxifolia (Blume) Hiern | Eb018 | Thailand | Duangjai 081 | KUFF, W | EU980816 | EU980658 | EU980939 | EU981073 | ||
| D. buxifolia | W85 | India | DeFranceschi 18.12.2006 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291322 | ||
| D. calciphila F. White | BT314 | New Caledonia | Munziner et al. 6650 | MPU, NOU, P | KF291801 | KF291860 | KF291919 | KF291323 | KF291457 | KF291634 |
| D. calciphila | BT316 | New Caledonia | Munziner et al. 6650 | MPU, NOU, P | KF291802 | KF291861 | KF291920 | KF291324 | KF291458 | KF291635 |
| D. calciphila | BT317 | New Caledonia | Munziner et al. 6653 | MPU, NOU, P | KF291459 | KF291636 | ||||
| D. calciphila | YP124 | New Caledonia | Pillon 124 | NOU006325 | KF291460 | KF291637 | ||||
| D. capreifolia Mart. ex Hiern | W09 | French Guiana | Prévost & Sabatier 3476 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291325 | ||
| D. carbonaria Benoist | W10 | French Guiana | Prévost & Sabatier 3470 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291326 | ||
| D. caribaea (A.DC.) Standl. | W65 | Cuba | Abbott 19004 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291327 | ||
| D. castanea (Craib) Fletcher | Eb020 | Thailand | Duangjai 083 | KUFF, W | DQ923893 | EU980660 | DQ924000 | EU981075 | ||
| D. cauliflora Blume | Eb024 | Thailand | Duangjai 087 | KUFF, W | DQ923894 | EU980661 | DQ924001 | EU981076 | KF291461 | KF291638 |
| D. cavalcantei Sothers | W22 | French Guiana | Prévost et al. 4671 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291328 | ||
| D. cayennensis A.DC. | W03 | French Guiana | Prévost 3430 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291329 | ||
| D. celebica Bakh. | K1242 | Indonesia | Chase 1242 | K | DQ923897 | EU980664 | DQ924004 | EU981079 | ||
| D. cherrieri F. White | BT262 | New Caledonia | Chambrey & Turner 16 | NOU079551, WU062860 | KF291803 | KF291862 | KF291921 | KF291330 | KF291463 | KF291640 |
| D. cherrieri | BT297 | New Caledonia | Chambrey & Turner 17 | NOU079547 | KF291804 | KF291863 | KF291922 | KF291331 | KF291464 | KF291641 |
| D. cherrieri | VH3510 | New Caledonia | Hequet 3510 | NOU015245 | EU980818 | EU980665 | EU980941 | EU981080 | KF291465 | KF291642 |
| D. cherrieri | VH3516 | New Caledonia | Hequet 3516 | NOU015251 | EU980819 | EU980666 | EU980942 | EU981081 | KF291466 | KF291643 |
| D. cherrieri | VH3610 | New Caledonia | Hequet 3610 | NOU016962 | KF291467 | KF291644 | ||||
| D. cherrieri | VH3640 | New Caledonia | Hequet 3640 | NOU017014 | KF291468 | KF291645 | ||||
| D. chrysophyllos Poir. | K25758 | Mauritius | Page 45 | MAU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291332 | ||
| D. chrysophyllos | K25769 | Mauritius | Page 71 | MAU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291333 | ||
| D. clementium Bakh. | Eb154 | Brunei | Duangjai et al. 24 | BRUN, W, WU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291334 | ||
| D. confertiflora (Hiern) Bakh. | Eb028 | Thailand | Duangjai 091 | KUFF, W | DQ923898 | EU980667 | DQ924005 | EU981082 | ||
| D. consolatae Chiov. | K1673 | Africa | Beentje 2168 | K | DQ923899 | EU980668 | DQ924006 | EU981083 | ||
| D. cooperi (Hutchinson & Dalziel) F. White | K20604 | Ghana | Merello et al. 1350 | MO | DQ923900 | EU980669 | DQ924007 | EU981084 | ||
| D. crassinervis (Krug & Urb.) Standl. | W23 | Cuba | Rainer s.n. | W | DQ923901 | EU980670 | DQ924008 | EU981085 | ||
| D. curranii Merr. | Eb031 | Thailand | Duangjai 094 | KUFF, W, WU | DQ923902 | EU980671 | DQ924009 | EU981086 | KF291469 | KF291646 |
| D. dasyphylla Kurz | Eb033 | Thailand | Duangjai 096 | KUFF, W | DQ923903 | EU980672 | DQ924010 | EU981087 | ||
| D. defectrix Fletcher | Eb097 | Thailand | Duangjai 155 | KUFF, WU | KF291805 | KF291864 | KF291923 | KF291335 | KF291470 | KF291647 |
| D. dendo Welw. ex Hiern | K21197 | Central African Republic | Harris & Fay 1594 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291336 | ||
| D. dichroa Sandwith | W13 | French Guiana | Sabatier et al. 4457 | W | DQ923904 | EU980673 | DQ924011 | EU981088 | ||
| D. dictyoneura Hiern | Eb038 | Thailand | Duangjai 100 | KUFF, W | EU980674 | EU980820 | EU980943 | EU981089 | KF291471 | KF291648 |
| D. diepenhorstii Miq. | Eb042 | Thailand | Duangjai 103 | KUFF, W | DQ923905 | EU980675 | DQ924012 | EU981090 | KF291472 | KF291649 |
| D. discolor Willd. | Eb088 | Thailand | Duangjai 146 | KUFF, WU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291337 | KF291473 | KF291650 |
| D. ebenum J. Koenig ex Retz | DY06 | Sri Lanka | Yakandawala 06 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291338 | ||
| D. ebenum | DY08 | Sri Lanka | Yakandawala 08 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291339 | ||
| D. ebenum | Eb174 | Sri Lanka | Samuel s.n. | WU | EU980677 | EU980821 | EU980944 | EU981092 | ||
| D. ebenum | W83 | India | Ramesh Diosass-2 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291340 | ||
| D. ebenum | W84 | India | DeFranceschi 21.12.2006 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291341 | ||
| D. egrettarum I. Richardson | K25788 | Mauritius | Page 122 | MAU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291342 | ||
| D. ehretioides Wall. ex G. Don | Eb043 | Thailand | Duangjai 104 | KUFF, W | DQ923907 | EU980678 | DQ924014 | EU981093 | KF291474 | KF291651 |
| D. eriantha Charmp. ex Benth | W63 | Taiwan | Chung & Anderberg 1401 | HAST | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291343 | ||
| D. erudita F. White | BT287 | New Caledonia | Chambrey & Turner 20 | NOU | KF291806 | KF291865 | KF291924 | KF291344 | KF291475 | KF291652 |
| D. erudita | M2359 | New Caledonia | Munzinger et al. 2359 | NOU003840 | EU980845 | EU980739 | EU980968 | EU981154 | KF291476 | KF291653 |
| D. erudita/pustulata | M3010 | New Caledonia | Munziner et al. 3010 | NOU008358 | EU980841 | EU980735 | EU980964 | EU981150 | ||
| D. fasciculosa (F. Muell.) F. Muell. | BT014 | New Caledonia | Munzinger et al. 6617 | NOU | KF291477 | KF291654 | ||||
| D. fasciculosa | BT142 | New Caledonia | MacKee 27341 | NOU022840 | KF291478 | KF291655 | ||||
| D. fasciculosa | BT165 | New Caledonia | KF291479 | KF291656 | ||||||
| D. fasciculosa | BT166 | New Caledonia | KF291480 | KF291657 | ||||||
| D. fasciculosa | BT335 | New Caledonia | KF291481 | KF291658 | ||||||
| D. fasciculosa | M2127 | New Caledonia | Munzinger 2127 | NOU003604 | DQ923908 | EU980679 | DQ924015 | EU981094 | KF291482 | KF291659 |
| D. fasciculosa | YP243 | New Caledonia | Pillon et al. 243 | NOU010096 | EU980822 | EU980680 | EU980945 | EU981095 | KF291483 | KF291660 |
| D. ferox Bakh. | Eb146 | Brunei | Duangjai et al. 012 | BRUN, W, WU | DQ923909 | EU980681 | DQ924016 | EU981096 | KF291484 | KF291661 |
| D. ferruginescens Bakh. | Eb143 | Brunei | Duangjai et al. 007 | BRUN, W, WU | DQ923911 | EU980685 | DQ924018 | EU981100 | ||
| D. filipendula Pierre ex Lecomte | Eb048 | Thailand | Duangjai 109 | KUFF | DQ923912 | EU980686 | DQ924019 | EU981101 | KF291485 | KF291662 |
| D. flavocarpa (Vieill. ex P. Parm.) F. White | BT126 | New Caledonia | Munzinger et al. 6625 | NOU | KF291807 | KF291866 | KF291925 | KF291345 | KF291486 | KF291663 |
| D. flavocarpa | BT127 | New Caledonia | Munzinger et al. 6625 | NOU | KF291808 | KF291867 | KF291926 | KF291346 | KF291487 | KF291664 |
| D. flavocarpa | BT156 | New Caledonia | Munzinger et al. 6632 | NOU | KF291488 | KF291665 | ||||
| D. flavocarpa | K20607 | New Caledonia | McPherson & Lowry 18563 | NOU022877 | DQ923913 | EU980687 | DQ924020 | EU981102 | KF291489 | KF291666 |
| D. flavocarpa | K20614 | New Caledonia | Lowry et al. 5783 | NOU023319 | EU980870 | EU980782 | EU980993 | EU981197 | ||
| D. flavocarpa | M2235 | New Caledonia | Munzinger 2235 | NOU006659 | EU980825 | EU980688 | EU980948 | EU981103 | KF291490 | KF291667 |
| D. flavocarpa | M2905 | New Caledonia | Munzinger et al. 2905 | NOU007977 | EU980826 | EU980689 | EU980949 | EU981104 | ||
| D. fragrans Gürke | K22454 | Gabon | SIMAB 010610 | MO | DQ923914 | EU980690 | DQ924021 | EU981105 | ||
| D. frutescens Blume | Eb049 | Thailand | Duangjai 110 | KUFF, W | EU980827 | EU980691 | EU980950 | EU981106 | ||
| D. fulvopilosa Fletcher | Eb052 | Thailand | Duangjai 113 | KUFF, W | DQ923915 | EU980692 | DQ924022 | EU981107 | KF291491 | KF291668 |
| D. fuscovelutina Baker | RF938 | Madagascar | RF 938 | W | DQ923979 | EU980803 | DQ924088 | EU981218 | ||
| D. gabunensis Gürke | K22560 | Tanzania | Bidgood et al. 2890 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291347 | ||
| D. gilletii De Wild | K21198 | Cameroon | Harris & Fay 884 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291348 | ||
| D. glandulosa Lace | Eb053 | Thailand | Duangjai 114 | KUFF, W | DQ923916 | EU980693 | DQ924023 | EU981108 | KF291492 | KF291669 |
| D. glans F. White | BT019 | New Caledonia | KF291809 | KF291868 | KF291927 | KF291349 | ||||
| D. glans | BT093 | New Caledonia | Turner et al. 093 | MPU | KF291810 | KF291869 | KF291928 | KF291350 | KF291493 | KF291670 |
| D. glans | BT094 | New Caledonia | Turner et al. 094 | MPU | KF291811 | KF291870 | KF291929 | KF291351 | KF291494 | KF291671 |
| D. glaucifolia Metcalf | K14256 | China | Chase 14256 | K | DQ923917 | EU980694 | DQ924024 | EU981109 | ||
| D. cf. gracilipes Hiern | RF978 | Madagascar | RNF 978 | W | DQ923918 | EU980695 | DQ924025 | EU981110 | ||
| D. gracilis Fletcher | Eb058 | Thailand | Duangjai 019 | BK, BKF, KUFF, WU | KF291812 | KF291871 | KF291930 | KF291352 | KF291495 | KF291672 |
| D. greenweyi F. White | K21205 | Somalia | Friis et al. 4991 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291353 | ||
| D. grisebachii (Heirn) Standl. | W64 | Cuba | Abbott 18937 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291354 | ||
| D. guianensis (Aubl.) Gürke | W14 | French Guiana | Prévost & Sabatier 4029 | W | DQ923919 | EU980696 | DQ924026 | EU981111 | ||
| D. guianensis | W78 | French Guiana | Mori 25921 | NY, W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291355 | ||
| D. hartmaniana S. Knapp | K22455 | Panama | McPherson & Richardson 15959 | MO | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291356 | ||
| D. impolita F. White | BT102 | New Caledonia | Schmid 5010 | NOU019538 | KF291813 | KF291872 | KF291931 | KF291357 | KF291496 | KF291673 |
| D. impolita | BT105 | New Caledonia | Schmid 5010 | NOU019538 | KF291814 | KF291873 | KF291932 | KF291358 | KF291497 | KF291674 |
| D. inconstans Jacq. | W79 | Ecuador | Rainer 1682 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291359 | ||
| D. inexplorata F. White | BT304 | New Caledonia | MacKee 22791 | NOU005818 | KF291815 | KF291874 | KF291933 | KF291360 | KF291498 | KF291675 |
| D. inexplorata | BT311 | New Caledonia | MacKee 22791 | NOU005818 | KF291816 | KF291875 | KF291934 | KF291361 | KF291499 | KF291676 |
| D. insidiosa Bakh. | Eb061 | Thailand | Duangjai 120 | KUFF, W | DQ923920 | EU980697 | DQ924027 | EU981112 | ||
| D. iturensis (Gürke) Letouzey & F. White | K21204 | Cameroon | Harris & Fay 1513 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291362 | ||
| D. kaki L.f. | K920 | Japan | Chase 920 | K | DQ923921 | EU980698 | DQ924028 | EU981113 | KF291500 | KF291677 |
| D. kirkii Hiern | K22551 | Zimbabwe | Poilecot 7650 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291363 | ||
| D. kupensis Gosline | AR62 | Cameroon | Russell 62 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291501 | KF291678 | |
| D. labillardierei F. White | BT121 | New Caledonia | Munzinger et al. 6624 | NOU | KF291817 | KF291876 | KF291935 | KF291364 | KF291502 | KF291679 |
| D. labillardierei | BT122 | New Caledonia | Munzinger et al. 6624 | NOU | KF291818 | KF291877 | KF291936 | KF291365 | KF291503 | KF291680 |
| D. labillardierei | BT179 | New Caledonia | KF291504 | KF291681 | ||||||
| D. labillardierei | K20763 | New Caledonia | McPherson & Munzinger 18038 | MO | DQ923922 | EU980699 | DQ924029 | EU981114 | ||
| D. labillardierei | M2219 | New Caledonia | Munzinger 2219 | NOU006657 | EU980828 | EU980700 | EU980951 | EU981115 | KF291505 | KF291682 |
| D. labillardierei | M3053 | New Caledonia | Munzinger 3053 | NOU008407 | EU980829 | EU980701 | EU980952 | EU981116 | KF291506 | KF291683 |
| D. lanceifolia Roxb. | K1245 | Indonesia | Chase 1245 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291366 | ||
| D. leucomelas Poir. | K25752 | Mauritius | Page 16 | MAU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291367 | ||
| D. lotus L. | D16 | Living coll. HBV | Turner D16 | Living coll. HBV | KF291507 | KF291684 | ||||
| D. lotus | K965 | Living coll. Kew 1882-3501 | Chase 965 | K | DQ923924 | EU980703 | DQ924031 | EU981118 | ||
| D. macrocarpa (Vieill.) Hiern | BT043 | New Caledonia | KF291508 | KF291685 | ||||||
| D. macrocarpa | BT044 | New Caledonia | KF291509 | KF291686 | ||||||
| D. macrocarpa | BT048 | New Caledonia | KF291510 | KF291687 | ||||||
| D. macrocarpa | BT049 | New Caledonia | KF291511 | KF291688 | ||||||
| D. macrocarpa | BT050 | New Caledonia | KF291512 | KF291689 | ||||||
| D. macrocarpa | M2014 | New Caledonia | Munzinger 2014 | NOU003637 | EU980830 | EU980704 | EU980953 | EU981119 | ||
| D. macrocarpa | M2829 | New Caledonia | Munzinger 2829 | NOU008233 | DQ923925 | EU980705 | DQ924032 | EU981120 | ||
| D. maingayi (Hiern) Bakh. | Eb073 | Thailand | Duangjai 131 | KUFF, W | DQ923926 | EU980706 | DQ924033 | EU981121 | ||
| D. malabarica (Desr.) Kostel. | Eb066 | Thailand | Duangjai 006 | KUFF, W | EU980708 | DQ923928 | DQ924035 | EU981123 | ||
| D. malabarica | K1247 | Indonesia | Chase 1247 | K | DQ923927 | EU980707 | DQ924034 | EU981122 | ||
| D. malabarica | W47 | South East Asia, cult. USA | Abbott 14325 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291368 | ||
| D. mannii Hiern | K20597 | Ghana | Merello et al. 1348 | MO | DQ923929 | EU980709 | DQ924036 | EU981124 | ||
| D. margaretae F. White | YP1267 | New Caledonia | Pillon 1267 | NOU049432, WU062863 | KF291819 | KF291878 | KF291937 | KF291369 | KF291513 | KF291690 |
| D. maritima Blume | Eb209 | Malaysia | Wallnöfer 13948 | W | DQ923930 | EU980710 | DQ924037 | EU981125 | ||
| D. melanida Poir. | K25786 | Mauritius | Page 112 | MAU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291370 | ||
| D. melocarpa F. White | K22457 | Gabon | SIMAB 012319 | MO | DQ923931 | EU980711 | DQ924038 | EU981126 | ||
| D. mespiliformis Hochst. Ex A.DC. | Eb206 | Tropical Africa | Wallnöfer & Duangjai 13945 | W | DQ923932 | EU980712 | DQ924039 | EU981127 | KF291514 | KF291691 |
| D. mespiliformis | W60 | Senegal | Prinz 2005-5 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291371 | ||
| D. minimifolia F. White | BT131 | New Caledonia | Dagostini 203 | NOU019556 | KF291820 | KF291879 | KF291938 | KF291372 | KF291515 | KF291692 |
| D. minimifolia | BT133 | New Caledonia | Dagostini 203 | NOU019556 | KF291821 | KF291880 | KF291939 | KF291373 | KF291516 | KF291693 |
| D. minimifolia | BT231 | New Caledonia | Veillon 7206 | NOU019554 | KF291517 | KF291694 | ||||
| D. minimifolia | BT264 | New Caledonia | Chambrey & Turner 24 | NOU079549, WU062872 | KF291518 | KF291695 | ||||
| D. minimifolia | M2214 | New Caledonia | Munzinger 2214 | NOU006263 | EU980831 | EU980714 | EU980954 | EU981129 | KF291519 | KF291696 |
| D. minimifolia | M2374 | New Caledonia | Munzinger 2374 | NOU006677 | EU980832 | EU980715 | EU980955 | EU981130 | KF291520 | KF291697 |
| D. minimifolia/pustulata | BT143 | New Caledonia | KF291521 | KF291698 | ||||||
| D. mollis Griff. | Eb074 | Thailand | Duangjai 132 | KUFF, W | DQ923934 | EU980716 | DQ924041 | EU981131 | KF291522 | KF291699 |
| D. montana Roxb. | Eb078 | Thailand | Duangjai 136 | KUFF, W | DQ923935 | EU980717 | DQ924042 | EU981132 | ||
| D. montana | Eb130 | Thailand | Duangjai & Sinbumrung 017 | KUFF, W | DQ923943 | EU980733 | DQ924050 | EU981148 | ||
| D. myriophylla (H. Perrier) G.E. Schatz & Lowry | W34 | Madagascar | Sieder 209 | W | DQ923974 | EU980797 | DQ924083 | EU981212 | ||
| D. natalensis (Harv.) Brenan | K22554 | Zambia | Bingham 10635 | K | DQ923936 | EU980718 | DQ924043 | EU981133 | ||
| D. nigra (J.F. Gmel.) Perrier | K212 | Cult. Mexico | Chase 212 | NCU | DQ923906 | EU980676 | DQ924013 | EU981091 | ||
| D. nigra | K1146 | Cult. Mexico | Chase 1146 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291374 | ||
| D. obliquifolia (Hiern ex Gürke) F. White | W91 | Cameroon | Rainer 6.3.2007 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291375 | ||
| D. oblonga Wall. Ex G. Don. | Eb083 | Thailand | Duangjai 141 | KUFF, W | DQ923937 | EU980719 | DQ924044 | EU981134 | ||
| D. olen Hiern | BT001 | New Caledonia | Munzinger et al. 6609 | NOU | KF291822 | KF291881 | KF291940 | KF291376 | KF291523 | KF291700 |
| D. olen | BT032 | New Caledonia | KF291524 | KF291701 | ||||||
| D. olen | BT034 | New Caledonia | KF291525 | KF291702 | ||||||
| D. olen | BT169 | New Caledonia | Munzinger et al. 6634 | NOU | KF291526 | KF291703 | ||||
| D. olen | BT302 | New Caledonia | KF291527 | KF291704 | ||||||
| D. olen | K20598 | New Caledonia | Lowry et al. 5628 | MO, NOU004840 | DQ923938 | EU980720 | DQ924045 | EU981135 | ||
| D. olen | M2827 | New Caledonia | Munzinger 2827 | NOU008235 | EU980833 | EU980721 | EU980956 | EU981136 | ||
| D. olen | YP153 | New Caledonia | Pillon 153 | NOU006438 | EU980834 | EU980722 | EU980957 | EU981137 | ||
| D. oubatchensis Kosterm. | BT160 | New Caledonia | LeCore et al. 768 | NOU079472 | KF291823 | KF291882 | KF291941 | KF291377 | KF291528 | KF291705 |
| D. oubatchensis | BT161 | New Caledonia | LeCore et al. 768 | NOU079472 | KF291824 | KF291883 | KF291942 | KF291378 | KF291529 | KF291706 |
| D. oubatchensis | M3118 | New Caledonia | Munzinger 3118 | NOU009675 | EU980835 | EU980723 | EU980958 | EU981138 | ||
| D. oubatchensis | M3333 | New Caledonia | Munzinger 3333 | NOU011201 | EU980836 | EU980724 | EU980959 | EU981139 | ||
| D. ovalifolia Wight | DY10 | Sri Lanka | Yakandawala 10 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291379 | ||
| D. pancheri Kosterm. | BT027 | New Caledonia | Munzinger et al. 6619 | NOU | KF291825 | KF291884 | KF291943 | KF291380 | KF291530 | KF291707 |
| D. pancheri | BT028 | New Caledonia | Munzinger et al. 6619 | NOU | KF291826 | KF291885 | KF291944 | KF291381 | KF291531 | KF291708 |
| D. pancheri | BT029 | New Caledonia | Munzinger et al. 6619 | NOU | KF291532 | KF291709 | ||||
| D. pancheri | BT030 | New Caledonia | Munzinger et al. 6620 | NOU | KF291533 | KF291710 | ||||
| D. pancheri | BT031 | New Caledonia | Munzinger et al. 6620 | NOU | KF291534 | KF291711 | ||||
| D. pancheri | BT033 | New Caledonia | Munzinger et al. 6620 | NOU | KF291827 | KF291886 | KF291945 | KF291382 | KF291535 | KF291712 |
| D. pancheri | BT035 | New Caledonia | Munzinger et al. 6620 | NOU | KF291536 | KF291713 | ||||
| D. pancheri | BT076 | New Caledonia | KF291537 | KF291714 | ||||||
| D. pancheri | M2138 | New Caledonia | Munzinger 2138 | NOU003868 | EU980837 | EU980725 | EU980960 | EU981140 | KF291538 | KF291715 |
| D. pancheri/parviflora | M2338 | New Caledonia | Munzinger 2338 | NOU006586 | EU980838 | EU980726 | EU980961 | EU981141 | KF291539 | KF291716 |
| D. parviflora (Schltr.) Bakh. | BT038 | New Caledonia | KF291828 | KF291887 | KF291946 | KF291383 | ||||
| D. parviflora | BT039 | New Caledonia | KF291829 | KF291888 | KF291947 | KF291384 | KF291540 | KF291717 | ||
| D. parviflora | BT040 | New Caledonia | KF291541 | KF291718 | ||||||
| D. parviflora | BT042 | New Caledonia | KF291542 | KF291719 | ||||||
| D. parviflora | BT187 | New Caledonia | Munzinger et al. 6636 | NOU | KF291543 | KF291720 | ||||
| D. parviflora | M2037 | New Caledonia | Munzinger 2037 | NOU002519 | EU980839 | EU980727 | EU980962 | EU981142 | KF291544 | KF291721 |
| D. parviflora | M2071 | New Caledonia | Munzinger 2071 | NOU002608 | EU980869 | EU980776 | EU980992 | EU981191 | KF291545 | KF291722 |
| D. parviflora | M2708 | New Caledonia | Munzinger 2708 | NOU006658 | EU980728 | EU980840 | EU980963 | EU981143 | ||
| D. parviflora | M3035 | New Caledonia | Munzinger 3035 | NOU008397 | EU980842 | EU980736 | EU980965 | EU981151 | ||
| D. pentamera (Woolls & F. Muell.) F. Muell. | K22549 | Australia | Forster & Booth 25525 | K | DQ923939 | EU980729 | DQ924046 | EU981144 | ||
| D. perplexa F. White | BT004 | New Caledonia | Munzinger et al. 6611 | NOU | KF291830 | KF291889 | KF291948 | KF291385 | KF291546 | KF291723 |
| D. perplexa | BT005 | New Caledonia | Munzinger et al. 6611 | NOU | KF291831 | KF291890 | KF291949 | KF291386 | KF291547 | KF291724 |
| D. perplexa | BT009 | New Caledonia | Munzinger et al. 6611 | NOU | KF291832 | KF291891 | KF291950 | KF291387 | KF291548 | KF291725 |
| D. perplexa | BT147 | New Caledonia | Munzinger et al. 6630 | NOU | KF291549 | KF291726 | ||||
| D. perplexa | BT148 | New Caledonia | Munzinger et al. 6630 | NOU | KF291550 | KF291727 | ||||
| D. perplexa | VH3614 | New Caledonia | Hequet et al. 3614 | NOU016957 | EU980873 | EU980786 | EU980996 | EU981201 | KF291551 | KF291728 |
| D. philippinensis A.DC. | K1248 | Indonesia | Chase 1248 | K | DQ923940 | EU980730 | DQ924047 | EU981145 | ||
| D. philippinensis | W62 | Taiwan | Chung & Anderberg 1400 | HAST | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291388 | ||
| D. pilosanthera Blanco | Eb091 | Thailand | Duangjai 149 | KUFF, W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291389 | ||
| D. pilosiuscula G. Don | Eb092 | Thailand | Duangjai 150 | KUFF, W | DQ923941 | EU980731 | DQ924048 | EU981146 | ||
| D. preussii Gürke | LPJMO39 | Cameroon | LPJMO39 | YA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291390 | ||
| D. pruriens Dalzell | W81 | India | DeFranceschi 18.12.2006 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291391 | ||
| D. pseudomespilus Mildbr. | K20606 | Gabon | Walters et al. 956 | MO | DQ923942 | EU980732 | DQ924049 | EU981147 | ||
| D. puncticulosa Bakh. | Eb150 | Brunei | Duangjai et al. 018 | BRUN, W, WU | DQ923944 | EU980734 | DQ924051 | EU981149 | ||
| D. pustulata F. White | BT113 | New Caledonia | KF291833 | KF291892 | KF291951 | KF291392 | KF291552 | KF291729 | ||
| D. pustulata | BT114 | New Caledonia | KF291834 | KF291893 | KF291952 | KF291393 | KF291553 | KF291730 | ||
| D. pustulata | BT136 | New Caledonia | Munzinger et al. 6629 | NOU | KF291554 | KF291731 | ||||
| D. pustulata | BT137 | New Caledonia | Munzinger et al. 6629 | NOU | KF291555 | KF291732 | ||||
| D. pustulata | BT257 | New Caledonia | Cambrey & Turner 21 | NOU079548, WU062871 | KF291556 | KF291733 | ||||
| D. pustulata | M3580 | New Caledonia | Munzinger 3580 | NOU016720 | EU980843 | EU980737 | EU980966 | EU981152 | KF291557 | KF291734 |
| D. pustulata | M3584 | New Caledonia | Munzinger 3584 | NOU016734 | EU980844 | EU980738 | EU980967 | EU981153 | KF291558 | KF291735 |
| D. pustulata | VH3638 | New Caledonia | Hequet et al. 3638 | NOU017016 | KF291559 | KF291736 | ||||
| D. pustulata/yahouensis | BT259 | New Caledonia | Chambrey & Turner 26 | WU062855 | KF291835 | KF291894 | KF291953 | KF291394 | KF291560 | KF291737 |
| D. racemosa Roxb. | Eb106 | Thailand | Duangjai 164 | KUFF | EU980856 | EU980759 | EU980979 | EU981174 | KF291561 | KF291738 |
| D. revaughanii I. Richardson | K25760 | Mauritius | Page 47 | MAU | Duangjai unpubl | Duangjai unpubl | Duangjai unpubl | KF291395 | ||
| D. revolutissima F. White | BT116 | New Caledonia | MacKee 22382 | NOU023189 | KF291836 | KF291895 | KF291954 | KF291396 | KF291562 | KF291739 |
| D. revolutissima | BT117 | New Caledonia | MacKee 22382 | NOU023189 | KF291837 | KF291896 | KF291955 | KF291397 | KF291563 | KF291740 |
| D. revolutissima | BT218 | New Caledonia | Munzinger et al. 6640 | NOU | KF291564 | KF291741 | ||||
| D. revolutissima | BT219 | New Caledonia | Munzinger et al. 6640 | NOU | KF291565 | KF291742 | ||||
| D. revolutissima | YP204 | New Caledonia | Pillon 204 | NOU009155 | EU980846 | EU980740 | EU980969 | EU981155 | KF291566 | KF291743 |
| D. rhodocalyx Kurz | Eb096 | Thailand | Duangjai 154 | KUFF, WU | KF291838 | KF291897 | KF291956 | KF291398 | KF291567 | KF291744 |
| D. rhombifolia Hemsl. | Eb129 | Thailand | Duangjai & Sinbumrung 016 | KUFF, W | DQ923945 | EU980741 | DQ924052 | EU981156 | ||
| D. cf. rhombifolia | W76 | Cult. USA, (South East Asia) | Abbott 20824 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291399 | ||
| D. ridleyi Bakh. | Eb138 | Brunei | Duangjai et al. 002 | BRUN, W, WU | DQ923946 | EU980742 | DQ924053 | EU981157 | KF291568 | KF291745 |
| D. rigida Hiern | Eb140 | Brunei | Duangjai et al. 004 | BRUN, W, WU | DQ923947 | EU980743 | DQ924054 | EU981158 | ||
| D. ropourea B. Walln. | W20 | French Guiana | Wallnöfer 13459 | W | DQ923948 | EU980744 | DQ924055 | EU981159 | ||
| D. salicifolia Humb. & Bonpl. ex Willd. | W66 | Guatemala | Abbott 19765 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291400 | ||
| D. salicifolia | W67 | Guatemala | Abbott 19777 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291401 | ||
| D. samoensis A. Gray | Eb176 | Cult. Hawaii Bot Garden | Kiehn s.n. | WU | EU980745 | EU980847 | EU980970 | EU981160 | ||
| D. samoensis | M3593 | Vanuatu | Munzinger 3593 | NOU080070 | EU980848 | EU980746 | EU980971 | EU981161 | ||
| D. samoensis | M3624 | Vanuatu | Munzinger 3624 | NOU080138, NOU080139 | EU980849 | EU980747 | EU980972 | EU981162 | KF291569 | KF291746 |
| D. samoensis | M3691 | Vanuatu | Munzinger 3691 | NOU | EU980850 | EU980748 | EU980973 | EU981163 | ||
| D. sandwicensis (A.DC.) Fosberg | Eb175 | Cult. Hawaii Bot Garden | Kiehn s.n. | WU | EU980851 | EU980749 | EU980974 | EU981164 | KF291570 | KF291747 |
| D. scabra (Chiov.) Cufod. | K21206 | Ethiopia | Wondefrash & Tefera 9622 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291402 | ||
| D. scalariformis Fletcher | Eb172 | Thailand | Duangjai & Sinbumrung s.n. | KUFF, W | EU980750 | EU980852 | EU980975 | EU981165 | ||
| D. senensis Klotzsch | K22552 | Zambia | Bingham 11092 | K | EU980853 | EU980751 | EU980976 | EU981166 | ||
| D. squarrosa Klotzsch | K21207 | Somalia | Friis et al. 4894 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291417 | ||
| D. squarrosa | K22555 | Zambia | Bingham & Downie 11465 | K | EU980854 | EU980752 | EU980977 | EU981167 | ||
| D. styraciformis King & Gamble | Eb149 | Brunei | Duangjai et al. 017 | BRUN, W, WU | DQ923949 | EU980753 | DQ924056 | EU981168 | ||
| D. sumatrana Miq. | Eb099 | Thailand | Duangjai 157 | KUFF, W | EU980855 | EU980754 | EU980978 | EU981169 | ||
| D. tenuiflora A.C.Sm. | W32 | Brazil | Maas et al. 9186 | NY, W | DQ923923 | EU980702 | DQ924030 | EU981117 | ||
| D. tesselaria Poir. | K25751 | Mauritius | Page 15 | MAU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291418 | ||
| D. tetrandra Hiern | W31 | French Guiana | Prévost & Sabatier 4713 | W | DQ923951 | EU980756 | DQ924058 | EU981171 | ||
| D. tetrasperma Sw. | K14254 | Mexico | Chase 14254 | K, W | DQ923952 | EU980757 | DQ924059 | EU981172 | ||
| D. texana Scheele | Eb208 | Middle America | Wallnöfer & Duangjai 13946 | W | DQ923953 | EU980758 | DQ924060 | EU981173 | KF291575 | KF291752 |
| D. tireliae F. White | M5725 | New Caledonia | Munzinger 5725 | NOU051026 | KF291843 | KF291902 | KF291961 | KF291419 | KF291576 | KF291753 |
| D. tridentata F. White | BT202 | New Caledonia | Munzinger et al. 6639 | NOU | KF291844 | KF291903 | KF291962 | KF291420 | KF291577 | KF291754 |
| D. tridentata | BT203 | New Caledonia | Munzinger et al. 6639 | NOU | KF291845 | KF291904 | KF291963 | KF291421 | KF291578 | KF291755 |
| D. trisulca F. White | BT185 | New Caledonia | Hequet (leg. Butin) 3820 | NOU031344 | KF291846 | KF291905 | KF291964 | KF291422 | KF291579 | KF291756 |
| D. trisulca | BT189 | New Caledonia | Hequet (leg. Butin) 3820 | NOU031344 | KF291847 | KF291906 | KF291965 | KF291423 | KF291580 | KF291757 |
| D. trisulca | BT192 | New Caledonia | Hequet (leg. Butin) 3820 | NOU031344 | KF291848 | KF291907 | KF291966 | KF291424 | KF291581 | KF291758 |
| D. trisulca | BT197 | New Caledonia | Munzinger et al. 6637 | NOU | KF291582 | KF291759 | ||||
| D. trisulca | M3179 | New Caledonia | Munzinger 3179 | NOU016896 | EU980871 | EU980784 | EU980994 | EU981199 | ||
| D. trisulca | M3260 | New Caledonia | Munzinger 3260 | NOU016891, WU062868 | EU980872 | EU980785 | EU980995 | EU981200 | KF291583 | KF291760 |
| D. cf. ulo Merr. | Eb152 | Brunei | Duangjai et al. 021 | BRUN, W, WU | EU980857 | EU980760 | EU980980 | EU981175 | KF291462 | KF291639 |
| D. umbrosa F. White | BT065 | New Caledonia | KF291849 | KF291908 | KF291967 | KF291425 | KF291584 | KF291761 | ||
| D. umbrosa | BT066 | New Caledonia | KF291585 | KF291762 | ||||||
| D. umbrosa | BT071 | New Caledonia | KF291586 | KF291763 | ||||||
| D. umbrosa | BT246 | New Caledonia | McPherson 2144 | NOU023234 | KF291850 | KF291909 | KF291968 | KF291426 | KF291587 | KF291764 |
| D. umbrosa | BT247 | New Caledonia | McPherson 2144 | NOU023234 | KF291851 | KF291910 | KF291969 | KF291427 | KF291588 | KF291765 |
| D. umbrosa | BT256 | New Caledonia | McPherson 2144 | NOU023234 | KF291589 | KF291766 | ||||
| D. umbrosa | M2265 | New Caledonia | Munzinger 2265 | NOU006679 | EU980858 | EU980761 | EU980981 | EU981176 | KF291590 | KF291767 |
| D. umbrosa | M2636 | New Caledonia | Munzinger 2636 | NOU006678 | EU980859 | EU980762 | EU980982 | EU981177 | KF291591 | KF291768 |
| D. umbrosa | M2771 | New Caledonia | Munzinger 2771 | NOU007912 | EU980860 | EU980763 | EU980983 | EU981178 | KF291592 | KF291769 |
| D. undulata Wall. Ex G. Don | Eb112 | Thailand | Duangjai 170 | KUFF, W | DQ923954 | EU980764 | DQ924061 | EU981179 | ||
| D. veillonii F. White | BT224 | New Caledonia | Veillon 7919 | NOU019582 | KF291852 | KF291911 | KF291970 | KF291428 | KF291593 | KF291770 |
| D. veillonii | BT229 | New Caledonia | Veillon 7919 | NOU019582 | KF291853 | KF291912 | KF291971 | KF291429 | KF291594 | KF291771 |
| D. veillonii | M.sn. | New Caledonia | Munzinger s.n. | Living coll. Hortus Veillonii | EU980861 | EU980765 | EU980984 | EU981180 | KF291595 | KF291772 |
| D. venosa Wall ex A.DC | Eb119 | Thailand | Duangjai 177 | KUFF, W | DQ923955 | EU980767 | DQ924062 | EU981182 | KF291596 | KF291773 |
| D. venosa | Eb131 | Thailand | Duangjai 059 | KUFF, W | EU980862 | EU980766 | EU980985 | EU981181 | ||
| D. vera (Lour.) A. Chev. | DY16 | Sri Lanka | Yakandawala 16 | PDA | EU980823 | EU980682 | EU980946 | EU981097 | ||
| D. vera | Eb045 | Thailand | Duangjai 106 | KUFF | DQ923910 | EU980683 | DQ924017 | EU981098 | KF291597 | KF291774 |
| D. vera | K21193 | Central African Republic | Harris & Fay 2032 | K | EU980824 | EU980684 | EU980947 | EU981099 | ||
| D. vestita Benoist | W01 | French Guiana | Molino 1849 | W | DQ923956 | EU980768 | DQ924063 | EU981183 | ||
| D. vieillardii (Hiern) Kosterm. | BT025 | New Caledonia | Munzinger et al. 6618 | NOU | KF291854 | KF291913 | KF291972 | KF291430 | KF291598 | KF291775 |
| D. vieillardii | BT026 | New Caledonia | Munzinger et al. 6618 | NOU | KF291855 | KF291914 | KF291973 | KF291431 | KF291599 | KF291776 |
| D. vieillardii | BT055 | New Caledonia | KF291600 | KF291777 | ||||||
| D. vieillardii | BT057 | New Caledonia | KF291601 | KF291778 | ||||||
| D. vieillardii | BT099 | New Caledonia | KF291602 | KF291779 | ||||||
| D. vieillardii | BT100 | New Caledonia | KF291603 | KF291780 | ||||||
| D. vieillardii | BT213 | New Caledonia | MacKee 25141 | NOU023242 | KF291604 | KF291781 | ||||
| D. vieillardii | BT214 | New Caledonia | MacKee 25141 | NOU023242 | KF291605 | KF291782 | ||||
| D. vieillardii | BT286 | New Caledonia | Chambrey & Turner 13 | NOU054004, WU062859 | KF291606 | KF291783 | ||||
| D. vieillardii | BT325 | New Caledonia | Munzinger et al. 6657 | NOU, P | KF291607 | KF291784 | ||||
| D. vieillardii | M2106 | New Caledonia | Munzinger 2106 | NOU006676 | EU980863 | EU980769 | EU980986 | EU981184 | KF291608 | KF291785 |
| D. vieillardii | M2776 | New Caledonia | Munzinger 2776 | NOU008207 | EU980864 | EU980770 | EU980987 | EU981185 | ||
| D. vieillardii | M3476 | New Caledonia | Munzinger 3476 | NOU012947 | KF291609 | KF291786 | ||||
| D. vieillardii | M3572 | New Caledonia | Munzinger 3572 | NOU016733 | EU980866 | EU980772 | EU980989 | EU981187 | KF291610 | KF291787 |
| D. vieillardii | YP146 | New Caledonia | Pillon 146 | NOU006400 | EU980867 | EU980773 | EU980990 | EU981052 | KF291611 | KF291788 |
| D. virginiana L. | K14255 | USA | Chase 14255 | K | DQ923957 | EU980774 | DQ924064 | EU981189 | KF291612 | KF291789 |
| D. wallichii King & Gamble ex King | Eb122 | Thailand | Duangjai 180 | KUFF, W | EU980868 | EU980775 | EU980991 | EU981190 | KF291613 | KF291790 |
| D. wallichii | Eb165 | Brunei | Duangjai et al. 41 | BRUN, W, WU | KF291614 | KF291791 | ||||
| D. winitii Fletcher | Eb123 | Thailand | Duangjai 181 | KUFF, WU | KF291615 | KF291792 | ||||
| D. yahouensis (Schltr.) Kosterm. | BT237 | New Caledonia | Schlechter 15059 | P00057340 | KF291856 | KF291915 | KF291974 | KF291432 | KF291616 | KF291793 |
| D. yahouensis | BT238 | New Caledonia | Schlechter 15059 | P00057340 | KF291857 | KF291916 | KF291975 | KF291433 | KF291617 | KF291794 |
| D. yahouensis | BT239 | New Caledonia | Schlechter 15059 | P00057340 | KF291618 | KF291795 | ||||
| D. yahouensis | VH3637 | New Caledonia | Hequet et al. 3637 | NOU017017 | KF291858 | KF291917 | KF291976 | KF291434 | KF291619 | KF291796 |
| D. yatesiana Standl. | W27 | Guatemala | Frisch s.n. | W | DQ923958 | EU980777 | DQ924065 | EU981192 | ||
| D. sp. Pic N’ga | BT318 | New Caledonia | Munzinger 6065 | NOU | KF291839 | KF291898 | KF291957 | KF291404 | KF291572 | KF291749 |
| D. sp. Pic N’ga | BT319 | New Caledonia | Munzinger 6065 | NOU | KF291840 | KF291899 | KF291958 | KF291405 | KF291573 | KF291750 |
| D. sp. Pic N’ga | BT320 | New Caledonia | Munzinger 6065 | NOU | KF291841 | KF291900 | KF291959 | KF291406 | KF291574 | KF291751 |
| D. sp. | FS1637 | Madagascar | Fischer & Sieder 1637 | W | DQ923959 | EU980778 | DQ924066 | EU981193 | ||
| D. sp. | FS2217 | Madagascar | Fischer & Sieder 2217 | W | DQ923960 | EU980779 | DQ924067 | EU981194 | ||
| D. sp. | K20600 | Madagascar | Rabenantoandro et al. 1246 | MO | DQ923961 | EU980780 | DQ924068 | EU981195 | ||
| D. sp. | K20601 | Madagascar | Rabevohitra et al. 3660 | MO | DQ923973 | EU980796 | DQ924082 | EU981211 | ||
| D. sp. | K20613 | Zambia | Zimba et al. 893 | MO | DQ923962 | EU980781 | DQ924069 | EU981196 | ||
| D. sp. | K20616 | Ghana | Schmidt et al. 2207 | MO | DQ923963 | EU980783 | DQ924070 | EU981198 | ||
| D. sp. | K25759 | Mauritius | Page 46 | MAU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291403 | ||
| D. sp. | RF958 | Madagascar | RNF 958 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291407 | ||
| D. sp. | RF959 | Madagascar | RNF 959 | W | DQ923980 | EU980804 | DQ924089 | EU981219 | ||
| D. sp. | RF970 | Madagascar | RNF 970 | W | DQ923964 | EU980787 | DQ924071 | EU981202 | ||
| D. sp. | S10 | Sri Lanka | Samuel 10 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291408 | ||
| D. sp. | S12 | Sri Lanka | Samuel 12 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291409 | ||
| D. sp. | S18 | Sri Lanka | Samuel 18 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291410 | ||
| D. sp. | S22 | Sri Lanka | Samuel 22 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291411 | ||
| D. sp. | S25 | Sri Lanka | Samuel 25 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291412 | ||
| D. sp. | S26 | Sri Lanka | Samuel 26 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291413 | ||
| D. sp. | S28 | Sri Lanka | Samuel 28 | PDA | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291414 | ||
| D. sp. | W33 | Madagascar | Sieder 440 | W | KF291842 | KF291901 | KF291960 | KF291415 | KF291571 | KF291748 |
| D. sp. | W36 | Madagascar | Sieder et al. 258 | W | DQ923965 | EU980788 | DQ924072 | EU981203 | ||
| D. sp. | W77 | Madagascar | Sieder et al. 3079 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291416 | ||
| Euclea crispa (Thunb.) Gürke | Eb202 | Living coll. HBV (EB 4/2) | Wallnöfer 13949 | W | DQ923966 | EU980789 | DQ924073 | EU981204 | ||
| Euclea crispa | K21188 | Malawi | Chapman & Chapman 8085 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291435 | ||
| Euclea divinorum Hiern | Eb201 | Cult. HBV (EB 2/1, Salisburg 69) | Wallnöfer & Duangjai 13947 | W | DQ923967 | EU980790 | DQ924074 | EU981205 | ||
| Euclea natalensis A.DC. | K21186 | Zimbabwe | Timberlake & Cunliffe 4389 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291436 | ||
| Euclea natalensis | W08 | South Africa | Kurzweil E514 | W | DQ923968 | EU980791 | DQ924075 | EU981206 | ||
| Euclea pseudobenus E. Mey. ex A.DC | K21190 | Namibia | Ward 9205 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291437 | ||
| Euclea racemosa L. | K21183 | Somalia | Thulin 10739 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291438 | ||
| Euclea sp. | W58 | Tanzania | Kutalek 1-2001 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291439 | ||
| Euclea sp. | W59 | Tanzania | Mbeyela 2-2001 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291440 | ||
| Euclea undulata Thunb. | Eb200 | Cult. HBV (EB 5/2, 1973) | Wallnöfer 13897 | W | EU980874 | EU980792 | DQ924076 | EU981207 | KF291620 | KF291797 |
| Royena cordata E. Mey ex A.DC | K1144 | South Africa | Chase 1144 | K | DQ923975 | EU980799 | DQ924084 | EU981214 | ||
| Royena glabra L. | W05 | South Africa | Kurzweil 2097 | W | DQ923976 | EU980800 | DQ924085 | EU981215 | ||
| Royena lucida L. | Eb203 | South Africa | Wallnöfer & Duangjai 13943 | W | DQ923977 | EU980801 | DQ924086 | EU981216 | ||
| Royena lucida | W06 | South Africa | Kurzweil E513 | W | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291442 | ||
| Royena lycioides Desf. Ex A.DC | K977 | South Africa | Chase 977 | K | DQ923978 | EU980802 | DQ924087 | EU981217 | ||
| Royena sp. | K1145 | South Africa | Chase 1145 | K | KF291859 | KF291918 | KF291977 | KF291444 | ||
| Royena whyteana Hiern | Eb177 | Africa | Kiehn s.n. | WU | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291443 | KF291622 | KF291799 |
| Royena zombensis B.L. Burtt | K22558 | Tanzania | Abdallah & Vollesen 95/106 | K | Duangjai unpubl. | Duangjai unpubl. | Duangjai unpubl. | KF291445 | ||
| Lissocarpa benthamii Gürke | W61 | Venezuela | Berry et al. 7217 | PORT | DQ923969 | EU980793 | DQ924077 | EU981208 | ||
| Lissocarpa guianensis Gleason | W04 | Guyana | Arets s.n. | U | DQ923970 | EU980794 | DQ924078 | EU981209 | ||
| Lissocarpa stenocarpa Steyerm. | K20609 | Peru | Vásquez & Ortiz-Gentry 25233 | MO | DQ923971 | EU980795 | DQ924079 | EU981210 | ||
| Argania spinosa (L.) Skeels | K978 | Morocco | Chase 978 | K | DQ923981 | EU980805 | DQ924090 | KF291308 | ||
| Cleyera japonica Thunb. | K1690 | Japan | Chase 1690 | K | DQ923985 | EU980811 | DQ924094 | KF291309 | ||
| Halesia carolina L. | K910 | USA | Chase 910 | K | KF291621 | KF291798 | ||||
| Madhuca macrophylla (Hassk.) H.J. Lam | K1363 | Cult. Indonesia | Chase 1363 | K | DQ923982 | EU980806 | DQ924091 | KF291441 | ||
| Styrax benzoin Dryand. | K1371 | Indonesia | Chase 1371 | K | DQ923989 | EU980809 | DQ924098 | KF291446 | ||
| Styrax officinalis L. | K872 | Living coll. RGB Kew 1973-14474 | Chase 872 | K | KF291623 | KF291800 |
2.2. DNA extraction
For DNA extraction the sorbitol/high-salt CTAB method (Tel-Zur et al., 1999), modified for 2 ml micro-centrifuge tubes, was used. Tubes containing silica gel-dried material were frozen with liquid nitrogen (to keep material frozen during grinding to avoid enzymatic action) and then ground with glass-beads to a fine powder. Prior to extraction, ground material was washed three times with sorbitol buffer.
2.3. PCR and cycle sequencing
We sequenced four plastid regions: atpB, rbcL, trnK–matK (partial trnK intron and complete matK gene) and trnS–trnG, which collectively represent approximately 6.5 kb. Primers and PCR conditions are those of Duangjai et al. (2009). We added 136 accessions to the matrix of Duangjai et al. (2009).
Chloroplast-expressed glutamine synthetase (ncpGS) was amplified with primers designed for this study (GScpDio1F and GScpDioR; Table 4). Initial Diospyros sequences for primer design were obtained with the primers and PCR protocol of Yockteng and Nadot (2004). Primers were situated at the end of exon 7 (forward) and beginning of exon 11 (reverse), amplifying a fragment between 700 and 715 bp (Fig. 2). Primers used for PCR were also used for cycle sequencing (Tables 2 and 3).
Table 4.
Primers used in this study.
| Primer name | Fragment | Sequence (5’-3’) | References |
|---|---|---|---|
| GScp839F | ncpGS | CACCAATGGGGAGGTTATGC | Yockteng and Nadot (2004) |
| GScp1056R | ncpGS | CATCTTCCCTCATGCTCTTTGT | Yockteng and Nadot (2004) |
| GscpDio1F | ncpGS | CCAATGGGGAGGTTATGCCTGGACAG | This study |
| GScpDioR | ncpGS | CATCTTCCCTCATGCTCTTTGTACTG | This study |
| PHYA upstream | phyA | GACTTTGARCCNGTBAAGCCTTAYG | Mathews and Donoghue (1999) |
| PHYA downstream | phyA | GDATDGCRTCCATYTCRTAGTC | Mathews and Donoghue (1999) |
| PhyADioF | phyA | GTBAAGCCTTAYGAAGTCCCGATGA | This study |
| PhyADioFi | phyA | GTCAAYGAGGGGGATGRAGAGGGAG | This study |
| PhyADioR | phyA | GCRTCCATYTCRTAGTCCTTCCAAG | This study |
| PhyADioRi | phyA | CTGATTYTCCAAYTCTAACTCCTTGTTGAC | This study |
Fig. 2.
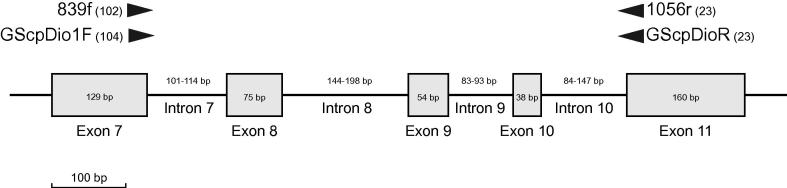
Schematic diagram of exon 7–exon 11 of ncpGS with primer positions and length of exons and introns. Numbers in parentheses give 5′ end of primers.
Table 2.
PCR reactions.
| ncpGS | 1st phyA | 2nd phyA | |||
|---|---|---|---|---|---|
| 18 μl | 1.1xReddyMix (Thermo Scientific) | 18 μl | 1.1xReddyMix (Thermo Scientific) | 18 μl | 1.1xReddyMix (Thermo Scientific) |
| 0.4 μl | Primer GScpDio1F (20 pM) | 0.4 μl | Primer PHYA upstream (20 pM) | 0.4 μl | Primer PhyADioF (20 pM) |
| 0.4 μl | Primer GScpDioR (20 pM) | 0.4 μl | Primer PHYA down-stream (20 pM) | 0.4 μl | Primer PhyADioR (20 pM) |
| 0.7 μl | Water | 0.7 μl | Water | 0.3 μl | Water |
| 0.4 μl | BSA (20 mg/ml) | ||||
| 0.5 μl | DNA | 0.5 μl | DNA | 0.5 μl | PCR product |
BSA: bovine albumin serum (Thermo Scientific).
Table 3.
PCR conditions.
| ncpGS | 1st phyA | 2nd phyA | |||
|---|---|---|---|---|---|
| 95 °C for 2 min | 95 °C for 2 min | 95 °C for 2 min | |||
| 95 °C for 30 s | 95 °C for 30 s | 95 °C for 30 s | |||
| 58 °C for 30 s | 35 cycles | 52 °C for 30 s | 35 cycles | 60 °C for 30 s | 35 cycles |
| 72 °C for 2 min | 70 °C for 2 min | 72 °C for 1.5 min | |||
| 72 °C for 7 min | 70 °C for 7 min | 72 °C for 7 min | |||
Initial PCR products and sequences of PHYA were obtained with the locus-specific primers of Mathews and Donoghue (1999; PHYA upstream [2nd] and PHYA downstream [1st]). As these primers were not specific enough, we cloned the PCR products (see Section 2.4) to be able to design Diospyros-specific PHYA PCR and sequencing primers (PhyADioF, PhyADioR, PhyADioFi and PhyADioRi; Table 4; Fig. 3). However, as the new PCR primers designed for Diospyros did not amplify consistently, we used a two-step amplification protocol. In the first PCR, the universal PHYA primers were used, and then a second nested PCR was performed with the newly designed primers and the product from the first PCR as template. All primers are located in exon 1 of PHYA flanking a region of 1187 bp in length. PCR conditions and composition are provided in Tables 2 and 3. For cycle-sequencing, we used the two internal primers and the external reverse primer.
Fig. 3.
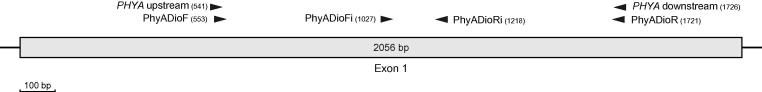
Schematic diagram of exon 1 of phyA with primer positions and length of exon. Numbers in parentheses give 5′ end of primers.
PCR products were cleaned with a mixture of exonuclease I and alkaline phosphatase (10 units exo I and one unit FastAP, both from Thermo Scientific) and incubated at 37 °C for 45 min followed by 15 min at 80 °C to inactivate enzymes. Cycle sequencing reactions were performed with 0.8 μl BigDye Terminator v3.1 (AB, Live Technologies), 1.0 μl primer (3.2 μM), 1.6 μl 5× sequencing buffer and 6.6 μl cleaned-up PCR product using 35 cycles of 96 °C for 10 s, 50 °C for 5 s and 60 °C for 3 min. Sequences were produced on a capillary sequencer (3730 DNA Analyzer, AB, Life Technologies) following the manufacturer’s protocols.
2.4. Cloning
Cloning was needed to produce PHYA from some accessions; these where than used for development of more specific primers. In addition, cloning of samples was necessary when we failed to obtain good sequences with the Diospyros-specific primers. PCR products were obtained using the universal PHYA primers, and after gel purification (Inivsorb Spin DNA Extraction Kit, Invitek), cleaned products were cloned using the pGEM-T Easy cloning system (Promega), following the manufacturer’s protocol. Cloned fragments were amplified using M13-f47 and M13-r48 primers and the following PCR conditions: initial denaturation 94 °C for 3 min, 35 cycles of denaturation 94 °C for 30 s, annealing 62 °C for 30 s and extension 72 °C for 2 min followed by a final extension at 72 °C for 7 min.
2.5. Sequence assembly, and editing, and phylogenetic analyses
Assembly and editing of sequences was done with the SeqMan Pro of the Lasergene v8.1 software package (DNASTAR); alignment was conducted with MUSCLE v3.8 (Edgar, 2004) and inspected visually using BioEdit v7.0.4 (Hall, 1999). Discrimination between the two copies of PHYA that were recovered from some species was done based on the alignment, and the ‘wrong’ (highly divergent) copy was excluded from further analyses. To test congruence between the data sets, ILD (incongruence length difference) test (Farris et al., 1994) implemented in PAUP∗ v4b10 (Swofford, 2003; termed the “partition homogeny test”) was carried out with 100 replicates. To speed up this analysis, the neo-endemic clade (where resolution is low due to lack of variability and therefore congruence is unlikely to be detected) was reduced to two accessions (D. sp. Pic N’ga BT318 and D. vieillardii BT025). Results of the ILD test indicated congruence of the four plastid data sets, and therefore the plastid data sets were combined; jModeltest indicated the same model could be used in all analyses without partitioning. Phylogenetic analyses were performed using PAUP∗ v4b10 (Swofford, 2003) for maximum parsimony (MP) and RaxML (Stamatakis, 2006) for maximum likelihood (ML) analyses. For both methods, bootstrap with 1000 replicates was performed to estimate clade support. For Bayesian inference, the program BEAST v1.7.4 (Drummond et al., 2012) was used. Parsimony and Bayesian analyses were run on the Bioportal computer cluster of the University Oslo (www.bioportal.uio.no), and likelihood analyses were run on CIPRS Science Gateway (http://www.phylo.org/portal2/; Miller et al., 2010). Estimation of evolutionary models and values was conducted with jModeltest v2.0.1 (Darriba et al., 2012; Guindon and Gascuel, 2003). For the Bayesian analyses the general time reversible nucleotide substitution model (GTR; Tavaré, 1986) with among site rate variation modelled with a gamma distribution (GTR + Γ) was used for ncpGS, whereas for plastid data the same model was used but with a proportion of invariable sites (GTR + Γ + I). For PHYA the Hasegawa–Kishino–Yano nucleotide substitution model (HKY; Hasegawa et al., 1985) was used with among site rate variation modelled with a gamma distribution and a proportion of invariable sites (HKY + Γ + I). Base frequencies (uniform), substitution rates between bases (gamma shape 10), alpha (gamma shape 10), kappa (gamma shape 10) and p-inv (uniform) were inferred by Modeltest from each data set. We used a relaxed uncorrelated log-normal clock model (Drummond et al., 2006). As speciation model, we used a Yule model (Gernhard, 2008; Yule, 1925). For further details see Supplementary material S1. Two independent Metropolis-coupled Markov chain Monte Carlo (MCMC) analyses each with 20 million generations were run sampling each 1000th generation. The initial 10% of trees obtained from each MCMC run were removed as burn in; the remaining trees of both runs were used to calculate a maximum clade credibility tree.
2.6. Dating the tree
To obtain an overarching dated tree, we used parts (atpB and rbcL sequences of Cornales and Ericales) of the data set of Bell et al. (2010) and combined it with our matrix. This matrix consisted of two plastid markers (atpB and rbcL), which were analysed as two partitions. Dating analyses were run in BEAST with an uncorrelated log-normal relaxed clock under the GTR + Γ + I model. The tree was calibrated with two fossils, Paleoenkianthus sayrevillensis (90 myr; Nixon and Crepet, 1993) as minimum age for Ericales and A. cryptostoma (34 myr; Basinger and Christophel, 1985) as minimum age for Diospyros clade II. Both groups (Ericales and Diospyros calde II) were defined as monophyletic, including the stem. Following tmrca (time of most recent common ancestor) settings used were: log normal prior distribution with a mean of 1.5, log standard deviation of 0.5 and an offset of 89 (Ericales) and 33 (Diospyros clade II). Priors for the molecular clock were: ucld.stdev: log normal, mean 0.9, log stdev 1, initial value 0.5, mean in real space; ucld.mean: CTMC rate reference (Ferreira and Suchard, 2008, initial value 1. Details of settings for BEAST analysis are provided in Section 2.5 (above) and Supplementary material S2. In addition to the plastid marker dating, we also conducted an analysis with our combined data set. We used the same settings as for the Bayesian analysis, but we added two calibration points: A. cryptostoma at 34 myr (Basinger and Christophel, 1985) as minimum age for Diospyros clade II and the split of Diospyros and its sister clade, Euclea plus Royena, 42 myr, which is the minimum age of that node based on dating exercises with the plastid markers. All settings for the molecular clock were the same as those for the plastid data set. The input file used for dating the combined analysis is provided in Supplementary material S3.
2.7. Chromosome counts of Diospyros
Chromosome preparations were made using Feulgen staining following the protocol from Weiss-Schneeweiss et al. (2009). Root tips were collected from plant material growing in the Botanical Garden of the University of Vienna (HBV) and a private garden in New Caledonia. To arrest mitotic spindles, root tips were treated with 0.002 M 8-hydroxquinoline for 2 h at room temperature and 2 h at 4 °C (always in darkness because 8-hydroxquinoline is light sensitive). Pre-treated material was fixed for 12 h at room temperature in 3:1 ethanol:acetic acid and then stored at −20 °C until examined. Fixed root tips were washed in distilled water to remove fixative, hydrolysed in 5 N HCl for 30 min, washed again with distilled water and stained with Schiff’s reagent for approximately 2 h in the dark. Squash preparations were made under a coverslip in a drop of 45% acetic acid. Counts could only be made for few species because obtaining young, actively growing root-tips from New Caledonian Diospyros is difficult. Collecting root-tips from forest trees and shrubs is not possible because there are too many roots in the soil to determine which is from the plant of interest. An alternative method is to grow seedlings in the lab/greenhouse. Obtaining seeds from tropical plants is not easy because these species do not produce fruit at a specific time of the year and flowering is diffuse (only few flowers produced at a time), so one would have to visit the plants regularly for at least 1 year to collect seed material. The logistics of this in process in New Caledonia were difficult. In addition, we found germination of seeds and maintenance of Diospyros seedlings highly problematic. Fortunately, the material we were able to obtain is well distributed among the genome sizes obtained, so we can conclude more than would otherwise be possible.
2.8. Genome size estimations of Diospyros
Genome size was determined using flow cytometry performed on leaf material. Fresh tissue was used from plants growing in the HBV. In addition, recently collected silica-gel dried material from New Caledonia was used for several measurements because it was not possible to transport fresh leaf material from New Caledonia to the laboratory. Samples were chopped in Otto I buffer (Otto et al., 1981) together with leaves of the internal standard species, Solanum pseudocapsicum, 1C = 1.30 pg (Temsch et al., 2010) or Pisum sativum ´Kleine Rheinländeriń, 1C = 4.42 pg (Greilhuber and Ebert, 1994), according to the method of Galbraith et al. (1983). The isolate was filtered through a 30 μm nylon mesh, and RNA was digested with 15 mg/l RNase A for 30 min at 37 °C. Subsequently, DNA was stained in propidium iodide (50 mg/l) supplemented with Otto II buffer (Otto et al., 1981). Mean fluorescence intensity of a total of 15,000 particles was measured with a CyFlow cytometer (Partec, Münster, Germany) equipped with a green laser (Cobolt Samba, Cobolt AB, Stockholm, Sweden); the 1C-value was calculated according to the formula: (MFIobject/MFIStandard) × 1C-valueStandard, where MFI is the mean fluorescence intensity of the G1 nuclei population. Statistical significance of asymmetry between the results obtained from Diospyros species belonging to clade III and those from clades VII–XI was tested using SPSS 15.0 (SPSS, Chicago; IL, USA) and the non-parametric Mann–Whitney U-test because of non-homogeneity of variances between the two groups of variables (Levene’s test for equality of variances, p < 0.05).
3. Results
The data characteristics and statistics from the maximum parsimony analyses of all three individual and the combined data sets are provided in Table 5. Since the focus of this paper is the New Caledonian Diospyros species from clade III, only results pertaining to this group will be discussed in detail. The other species have been included to (i) investigate the utility of these markers for resolving phylogenetic relationships within Diospyros and (ii) further evaluate the hypothesis (proposed by Duangjai et al., 2009) that not all New Caledonian Diospyros resulted from a single colonisation event.
Table 5.
Data characteristics and statistics from the maximum parsimony analyses of all three individual and the combined data sets.
| Combined plastid markers | ncpGS | phyA | Combined data set | |
|---|---|---|---|---|
| Total no. of accessions | 294 | 177 | 177 | 129 |
| No. of outgroup accessions other than Ebenaceae | 4 | 2 | 2 | 1 |
| No. of outgroup accessions from Ebenaceae | 21 | 2 | 2 | 2 |
| No. of Diospyros accessions | 269 | 173 | 173 | 126 |
| No. of Diospyros species | 149 | 64 | 64 | 64 |
| No. of New Caledonian accessions | 98 | 134 | 134 | 86 |
| No. of New Caledonian species | 28 | 28 | 28 | 28 |
| No. of New Caledonian neoendemic accessions | 83 | 112 | 112 | 74 |
| No. of New Caledonian neoendemic species | 21 | 21 | 21 | 21 |
| Length of alignment | 6556 | 1039 | 1187 | 8542 |
| No. of variable characters | 1880 | 532 | 374 | 1845 |
| No. of parsimony informative characters | 1126 (17.2%) | 341 (32.8%) | 223 (18.8%) | 863 (10%) |
| No. of parsimony informative characters NCnc | 44 (0.7%) | 28 (2.7%) | 14 (1.2%) | 79 (0.9%) |
| Tree length of best parsimony tree (steps) | 3808 | 1171 | 689 | 3259 |
| Trees saved (parsimony analysis) | 210 | 4810 | 1870 | 930 |
| Consistency index | 0.603 | 0.663 | 0.685 | 0.692 |
| Retention index | 0.857 | 0.857 | 0.893 | 0.848 |
| Best fitting model | GTR + Γ + I | GTR + Γ | HKY + Γ + I |
3.1. Plastid markers
Parsimony analysis of the plastid data set produced 210 equally parsimonious trees, one of which (randomly selected) is shown to demonstrate comparative levels of divergence (Fig. 4). Clade names correspond to those of Duangjai et al. (2009). Resolution among the New Caledonian taxa of clade III is low, but monophyly of these taxa is strongly supported: bootstrap percentage MP (BMP) 88; bootstrap percentage ML (BML) 97; Bayesian posterior probability (BPP) 0.95. Furthermore, D. vieillardii (BMP 99, BML 98, BPP 1.00) and its position as sister (BMP 97, BML 96, BPP 1.00) to the rest of the clade are well supported. Within the NC clade III, only one group of three taxa (D. calciphila, D. inexplorata and D. sp. Pic N’ga) is supported in all three analyses (BMP 91, BML 92, BPP 1.00); this includes all accessions of each species forming unique clusters. There are a few more, weakly supported small groups in which individuals of one population fall together, but they are not consistent among the three analyses and fail to include all accessions of these species.
3.2. Low-copy nuclear markers
Nuclear markers contained proportionally more parsimony informative characters (ncpGS 2.7%, PHYA 1.2%) than the plastid markers (0.7%), but variation was still low. Some species form groups (Fig. 5), but they lack bootstrap and Bayesian posterior probability support. Among the three methods of analysis used for the ncpGS data set, Bayesian inference provides the best resolution (tree not shown), placing D. vieillardii (BBP 0.99) sister (BPP 1.00) to the rest of the NC clade. The relationship between D. veillonii and D. cherrieri (BPP 0.84) is weakly supported, but their position as subsequent sister of the rest of this clade is well supported (BPP 0.99). All individuals of D. umbrosa form a group with two individuals of D. trisulca (BBP 0.91). This set of accessions is subsequently sister (BBP 0.95) to the rest of the clade, within which there is no resolution. In the PHYA tree, there are only a few clades with strong support regardless of method of analysis. Clade III (BMP 100, BML 100, BPP 1.00) as monophyletic unit is confirmed, as well as the monophyly of NC clade III within it (BMP 77, BML 78, BPP 1.00). All included individuals of D. cherrieri fall together (BMP 84, BML 81, BPP 1.00) in the PHYA analyses. Only a single copy of ncpGS was recovered from all accessions investigated, as well as from most of the accessions of PHYA. Species from which two copies of PHYA were obtained when cloned are found in clades IX, X and XI (Fig. 4). The paralogous (divergent) copies of PHYA were easily detected and excluded from the phylogenetic analyses.
Fig. 5.
Maximum parsimony trees inferred from the nuclear data sets, branch length scaled to same value on both trees. Bold branches have more than 70% support in all three analysis. New Caledonian taxa are coloured, red represents clade III NC. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Combined data set
The ILD test found the trees of the plastid and low-copy nuclear markers to be congruent with p-values of 0.01, which indicates that combined analysis was appropriate. In trees inferred from the combined data set (Fig. 6), species of clade III were highly supported (BMP 100, BML 100, BPP 1.00); they include the species of NC clade III, Indian Ocean islands, Thailand and Hawai‘i. Diospyros vera is sister to D. sandwicensis (BMP 100, BML 100, BPP 1.00) and then the NC clade III. NC clade III is moderately to well supported (BMP 83, BML 96, BPP 0.96). The position of D. vieillardii (BMP 100, BML 99, BPP 1.00) as sister to the rest of the clade is strongly supported (BMP 92, BML 98, BPP 1.00). All accessions of each of the two species, D. umbrosa (BMP < 70, BML 75, BPP 1.00) and D. flavocarpa (BMP < 70, BML < 70, BPP 0.99), form unique groups, which together are sister (BMP 100, BML 100, BPP 1.00) to the rest of the group. A sister relationship between D. cherrieri (BMP 96, BML 99, BPP 1.00) and D. veillonii (BMP 78, BML 86, BPP 1.00) is supported (BMP 75, BML 88, BPP 1.00). A clade comprising D. calciphila, D. inexplorata (both on coralline substrates) and D. sp. Pic N’ga (ultramafic substrate) is well supported (BMP 97, BML 99, BPP 1.00).
Fig. 6.
Baysian maximum clade credibility tree inferred from the combined data set. Bold branches have more than 70% support in all three analysis, nodes with at least one support value ⩾70% are indicated with blue dots (BPP/BMP/BML). New Caledonian taxa are coloured, red represents clade III NC. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Dating analysis
We performed two dating analyses. The first one was based on a joint matrix of our plastid sequences together with the data set of Bell et al. (2010), which included many families across the whole Ericales with Cornales as outgroup. This dating analysis was used to get age estimates for the crown node of Ebenaceae, the two subfamilies Ebenoideae and Lissocarpoideae, the split of the three genera of Ebenoideae (Diospyros versus Euclea/Royena) and the main clades of Diospyros. The second dating analysis was based on our combined data set, which was used to infer ages of clades and species within Diospyros. The dating analysis of the over-arching matrix of plastid markers (Fig. S4, Supplementary material) indicates that the two subfamilies of Ebenaceae, Lissocarpoideae and Ebenoideae, diverged around 54 mya (42–65; 95% highest posterior density interval). The split of Diospyros from its sister genera, Euclea plus Royena occurred around 42 mya (35–50). The following conclusions are based on the dating analysis of the combined data set (Fig. 7). The Australian clade of Diospyros (clade II, Fig. 4), including five species from New Caledonia, separated from the rest of the genus around 34 mya (33–36), the New Caledonian and Australian members of this clade diverged around 20 mya (11–29). Divergence among the New Caledonian members began only about 6 mya (3–10). The two large main groups (clades V–XI and clade III, Fig. 4) diverged about 32 mya (25–35). The last common ancestor of D. fasciculosa and D. olen existed around 15 mya (11–19). Diospyros olen is around 5 myr (3–9) old and D. fasciculosa about 6 myr (3–10). Lineages of clade III started to diversify about 19 mya (13–21). Lineages forming NC clade III arrived in New Caledonia around 9 mya (6–13). Diospyros vieillardii is around 7 myr (5–10) old. The clade comprising D. cherrieri and D. veillonii is around 5 myr (3–8) old, and the two species separated around 3 mya (1–5). The clade including D. flavocarpa, D. umbrosa and one accession of D. trisulca is 5 myr (3–7) old. Diospyros flavocarpa is around 4 myr (3–6) old. The relationship between D. umbrosa and D. trisulca is not highly supported, but suggests an age of around 3 myr (2–5) for D. umbrosa. The group comprising D. calciphila, D. inexplorata and D. sp. Pic N’ga appears to be around 2 myr (1–3) old and started to diversify around 0.9 mya (0.5–2). Resolution between other species is too limited to say anything about their ages.
Fig. 7.
Chronogram based on the combined data set. Ages are given (in million years) for nodes with more than 0.85 BPP. Nodes which were calibrated are marked with a black dot. Yellow bars represent the 95% highest posterior density interval. New Caledonian taxa are coloured, red represents clade III NC.
3.5. Chromosome counts and genome size
Chromosome counts made for Diospyros fasciculosa, D. inconstans, D. macrocarpa, D. minimifolia, D. pentamera, D. pustulata, D. texana, D. veillonii and D. yatesiana indicate that they are all diploid, 2n = 30. The counts from the underlined species are here reported for the first time in literature. The other counts confirm results of White (1992).
Measurements of genome size showed differences among the New Caledonian species of Diospyros. Diospyros olen has with 1C = 0.86 pg, the smallest genome of the New Caledonian Diospyros species examined, followed by D. fasciculosa with 1C = 1.13 pg (both clade XI). The investigated species from the NC clade III have larger genomes (mean value 1C = 1.90 pg) than the two mentioned above (Table 6). We were not able to examine New Caledonian species from clade II. Finally, across whole genus Diospyros there is a significant difference (Mann Whitney U test, p < 0.001) in genome size between clade III on the one hand and clades VI–XI on the other (Fig. 8). However, D. pentamera of clade II has a comparatively large genome (1C = 1.97 pg).
Table 6.
Genome size of Diospyros and other genera from Ebenoideae. S.D.: standard deviation, N: number of measurements (replicates), S.p.: Solanum pseudocapsicum, P.s.: Pisum sativum ‘Kleine Rheinländerin’.
| Name | Acc. nr | 1C-value | S.D. | N | Standard | Material |
|---|---|---|---|---|---|---|
| D. calciphila | BT313 | 1.99 | 1 | S.p. | Dry | |
| D. calciphila | BT316 | 1.97 | 1 | P.s. | Silicagel | |
| D. cherrieri | BT262 | 1.65 | 0.0092 | 5 | S.p. | Dry |
| D. cherrieri | BT293 | 1.57 | 0.0117 | 2 | S.p. | Silicagel |
| D. discolor | EBE100026 | 0.92 | 0.0020 | 3 | S.p. | Fresh |
| D. erudita | BT260 | 2.17 | 1 | S.p. | Dry | |
| D. erudita | BT261 | 2.13 | 0.0367 | 3 | S.p. | Dry |
| D. erudita | BT280 | 1.88 | 0.0253 | 3 | S.p. | Silicagel |
| D. fasciculosa | BT012 | 1.19 | 0.0031 | 3 | P.s. | Silicagel |
| D. fasciculosa | BT106 | 1.13 | 0.0064 | 3 | P.s. | Silicagel |
| D. fasciculosa | BT144 | 1.22 | 0.0032 | 3 | P.s. | Silicagel |
| D. fasciculosa | BT167 | 1.02 | 0.0318 | 4 | P.s. | Silicagel |
| D. fasciculosa | BT212 | 1.09 | 0.0227 | 2 | P.s. | Silicagel |
| D. fasciculosa | BT335 | 1.14 | 0.0300 | 3 | P.s. | Silicagel |
| D. glans | BT019 | 2.03 | 1 | S.p. | Silicagel | |
| D. glans | BT093 | 2.02 | 0.0153 | 3 | S.p. | Dry |
| D. impolita | BT101 | 1.79 | 1 | P.s. | Silicagel | |
| D. impolita | BT105 | 1.90 | 0.0132 | 3 | P.s. | Silicagel |
| D. inconstans | 1.13 | 0.0019 | 3 | S.p. | Fresh | |
| D. inexplorata | BT304 | 1.94 | 0.0693 | 3 | P.s. | Silicagel |
| D. kaki | Sharon | 2.29 | 0.0121 | 3 | S.p. | Dry |
| D. lotus | EBE | 0.87 | 0.0075 | 8 | S.p. | Fresh |
| D. lotus | EBE03002 | 0.86 | 0.0012 | 3 | S.p. | Fresh |
| D. mespiliformis | EBE000001 | 1.24 | 0.0029 | 3 | P.s. | Fresh |
| D. mespiliformis | EBE100027 | 1.27 | 0.0035 | 3 | P.s. | Fresh |
| D. minimifolia | BT230 | 1.57 | 0.0445 | 3 | P.s. | Silicagel |
| D. olen | BT001 | 0.82 | 0.0062 | 3 | S.p. | Silicagel |
| D. olen | BT036 | 0.87 | 0.0475 | 3 | S.p. | Silicagel |
| D. olen | BT096 | 0.86 | 0.0042 | 3 | S.p. | Dry |
| D. olen | BT186 | 0.90 | 0.0041 | 3 | S.p. | Dry |
| D. pancheri | BT077 | 2.28 | 0.0129 | 3 | S.p. | Dry |
| D. parviflora | BT085 | 2.16 | 0.0493 | 3 | P.s. | Dry |
| D. pentamera | EBE030020 | 1.97 | 0.0020 | 3 | S.p. | Fresh |
| D. perplexa | BT002 | 2.27 | 1 | S.p. | Silicagel | |
| D. pustulata | BT137 | 1.54 | 0.0490 | 2 | P.s. | Silicagel |
| D. revolutissima | BT222 | 2.05 | 0.0148 | 4 | P.s. | Dry |
| D. texana | EBE020015 | 0.89 | 0.8849 | 3 | S.p. | Fresh |
| D. texana | EBE100029 | 0.89 | 0.0019 | 3 | S.p. | Fresh |
| D. tridentata | BT205 | 2.21 | 0.0246 | 2 | S.p. | Dry |
| D. tridentata | BT206 | 2.09 | 1 | P.s. | Dry | |
| D. umbrosa | BT171 | 1.61 | 1 | P.s. | Silicagel | |
| D. umbrosa | BT247 | 1.51 | 0.0894 | 3 | P.s. | Silicagel |
| D. vieillardii | BT100 | 1.55 | 1 | P.s. | Dry | |
| D. vieillardii | BT216 | 1.57 | 0.0238 | 3 | P.s. | Dry |
| D. yatesiana | 0.60 | 0.0010 | 5 | S.p. | Fresh | |
| E. divinorum | EBE000002 | 1.98 | 0.0014 | 3 | S.p. | Fresh |
| E. undulata | EBE100002 | 0.74 | 0.0220 | 3 | S.p. | Fresh |
| R. whyteana | EBE030021 | 0.79 | 0.0031 | 3 | S.p. | Fresh |
| R. whyteana | EBE030022 | 0.78 | 0.0007 | 3 | S.p. | Fresh |
Fig. 8.
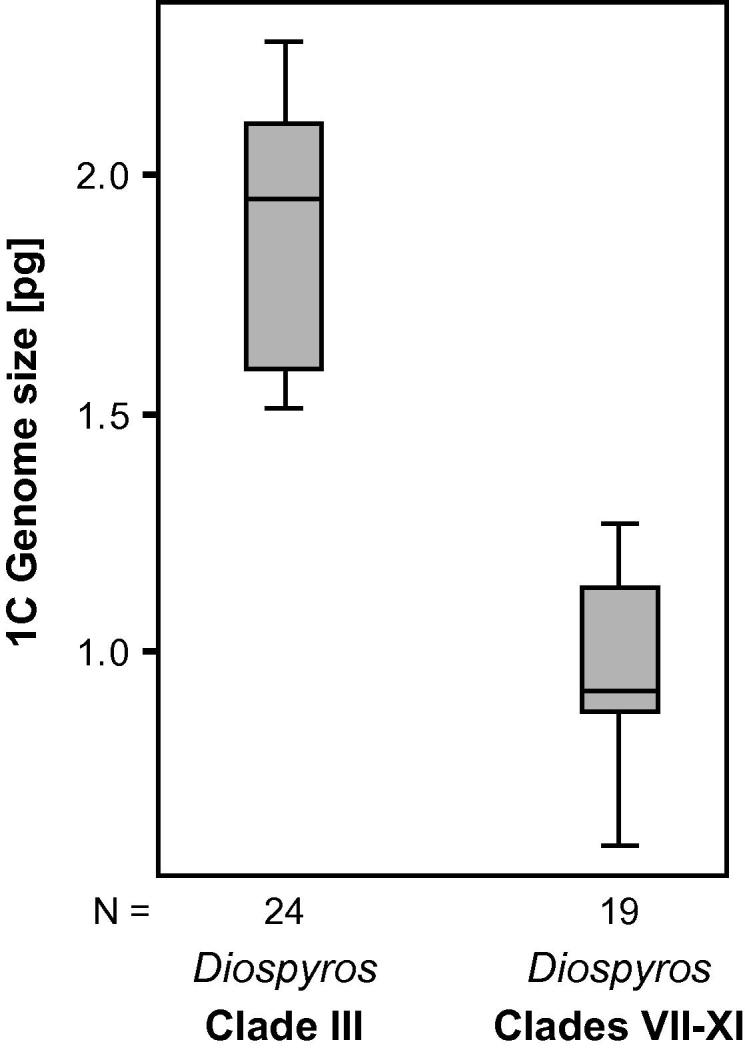
Boxplot of genome size differences between taxa from clade III and those from clades VII–XI.
4. Discussion
Previous phylogenetic studies of Diospyros based on plastid markers demonstrated low levels of sequence divergence among New Caledonian species belonging to clade III (Duangjai et al., 2009), and inclusion of additional species in our investigation did not improve resolution in this group. Low-copy nuclear markers have been shown to be highly informative and useful for resolving phylogenetic relationships at lower taxonomic levels in some taxa (e.g. Passiflora: Yockteng and Nadot, 2004; Paeonia: Tank and Sang, 2001). The low-copy markers ncpGS and PHYA used here, however, did not improve resolution in this clade of 21 closely related species, thus preventing detection of hybrids and elucidation of geographical patterns (Fig. 5). There are also examples where low-copy nuclear markers were not able to fully resolve phylogenetic relationships between closely related species, especially on islands (e.g. Pillon et al., 2009a, 2013; Green et al., 2011). Nonetheless, the analysis based on combined plastid and nuclear data provides some resolution of relationships within the NC clade III. Of the 21 entities included in the analyses, seven species and one unidentified taxon formed well defined and inclusive clusters (Fig. 6). The remaining 14 species failed to form groups including all individuals of a particular species, but in many cases it was simply that some accessions were part of a polytomy and did not cluster consistently with any group.
In light of our results, members of the NC clade III appear little diverged but still form a strongly supported clade, which our dating analyses indicate are the result of recent rapid radiation. Only a few studies have examined the adaptive basis and processes involved in speciation in New Caledonia (e.g. Pillon et al., 2009b; Murienne et al., 2009). Rapid radiation has been observed in isolated areas such as islands (e.g. Givnish et al., 2009; Knope et al., 2012), high mountains (e.g. Hughes and Eastwood, 2006) and valleys (e.g. Givnish et al., 2007, 2011; Richardson et al., 2001). Island floras often show high levels of endemism and closely related species groups that result from a single colonisation event followed by rapid speciation, some of which have been hypothesised to represent adaptive radiations (e.g. Hawaiian silverswords, Baldwin and Sanderson, 1998; Hawaiian Bidens, Knope et al., 2012; Araucaria in New Caledonia, Gaudeul et al., 2012). The low levels of variation and resolution detected in the NC clade III prevent us from examining factors that may be promoting speciation on New Caledonia.
As all lineages of New Caledonian Diospyros seem to have arrived relatively recent on this island, the terms paleo-endemics and neo-endemics used by Duangjai et al. (2009) were not used here. The common ancestor of clade III diverged about 19 mya (Fig. 7), and the earlier diverging species occur mainly in Africa and on islands of the western Indian Ocean (e.g. Madagascar). Our results in combination with the DIVA analysis from Duangjai et al. (2009) indicate that, from there, this group spread eastwards via Southeast Asia, where it arrived around 15 mya, and then reached the Hawaiian Archipelago and New Caledonia around 9–10 mya. This time of colonisation is consistent with that found for other plant groups (reviewed in Pillon, 2012) and animals (e.g. Nattier et al., 2011). The close relationship of New Caledonian and Hawaiian endemic Diospyros shows that migration around the Pacific Ocean has taken place, but to make more definite conclusions about the direction of dispersal, data from species present on other islands between New Caledonia and Hawai‘i are needed. In contrast to long-held hypotheses that many taxa are Gondwanan relicts (e.g. Lowry II, 1998; Swenson et al., 2001), our results suggest that all groups of New Caledonian Diospyros are much younger than 37 myr (when New Caledonia re-emerged) and arrived, like many others, via long-distance dispersal (e.g. Bartish et al., 2011; Espeland and Murienne, 2011; Murienne, 2009).
The closely related species of the NC clade III are distinguishable from one another by morphological characters (e.g. leaf, flower, fruit and calyx characters), and many of them are found in different habitats (e.g. humid/dry, different substrate types, different elevations, etc.). Leaf morphology shows adaptation to the environment in which a species occurs (e.g. species found in dry habitats have sclerophyllous leaves; for details of species descriptions see White, 1992, 1993). In most plant groups, closely related species rarely occur in sympatry, but not in New Caledonia where this seems to be a common pattern in several groups (J. Munzinger pers. obs.), including Diospyros. However, Diospyros has been reported to be one of the few genera outside New Caledonia (e.g. Madagascar) with several co-occurring species (pers. comm. P.S. Ashton). The habitats occupied by the New Caledonian Diospyros species belonging to clade III can be roughly divided into seven groups (Table 7). D. vieillardii, a common species found all over Grande Terre and the islands north of the main island in maquis vegetation, occurs on a variety of substrates, including ultramafic. Diospyros umbrosa/D. flavocarpa are sister to the remainder of the clade excluding D. vieillardii (Fig. 6). D. umbrosa occurs only on ultramafic substrates in comparatively humid forests mainly consisting of Nothofagus and Araucaria. D. flavocarpa is found on schist in middle elevation forests in northeastern Grande Terre. Diospyros cherrieri (a local endemic in dry forests on basalts at the western coast of Grande Terre, Fig. 1I) and D. veillonii (a local endemic in dry coastal forests on black clay on the western side of Grande Terre, Fig. 1F) are together sister to the rest of the clade (minus those mentioned above). The clade comprising D. calciphila, D. inexplorata (littoral forests on coralline substrates) and D. sp. from Pic N’ga (maquis on ultramafic substrate on Ile des Pins) is well supported. Relationships among all other members of the clade could not be resolved with the markers used, although most of them are morphologically and ecologically well defined. This phenomenon (morphological and ecological distinctiveness, but no resolution) is found, for example, in D. labillardieri (lanceolate leaves, hanging branches; river edges in middle elevation forests on schist, Fig 1D), D. pancheri (obcordate pubescent leaves, hanging branches, humid forests at low elevation on ultramafic soils, Fig. 1E) and many others. Due to the poor resolution of the phylogenetic trees, possible grouping of New Caledonian Diospyros species according to their ecological niches remains untested.
Table 7.
Main habitats of New Caledonian neoendemic Diospyros species.
| Habitat | Species | |
|---|---|---|
| Maquis on ultramafic substrates | D. erudita, D. pancheri, D. parviflora, D. tireliae, D. vieillardii | |
| Dry forests on non-ultramafic substrates | D. cherrieri, D. perplexa, D. yahouensis | |
| Humid forests at low elevations | Ultramafic substrates | D. pancheri, D. parvilfora, D. umbrosa |
| Calcareous rocks | D. tridentata | |
| Humid mountain forests on schist | D. flavocarpa, D. labillardierei, D. trisulca | |
| Dry coastal forests | Black clays | D. veillonii, D. minimifolia, D. pustulata (the latter two can also occur on calcareous substrates) |
| Schist | D. impolita | |
| Ultramafic substrates | D. revolutissima | |
| Various substrates | D. pancheri | |
| Coastal forests on coralline substrates | D. calciphila, D. impolita | |
| Humid forests on the east coast | D. glans | |
A greater than threefold variation within the genome size of Diospyros is observed, although the chromosome counts performed here and elsewhere indicates that they are diploid with 2n = 30, and we hypothesize that the most recent common ancestor of Diospyros had a large genome because species belonging to earlier diverging clades (e.g. E. divinorum and D. pentamera) have large genomes. Developing firmer ideas about evolution of genome size in Diospyros would require many more measurements of species from throughout the phylogenetic tree, especially species from islands in the Indian and Pacific Ocean, which will be key to assessing evolution of genome size in NC clade III. The limited data available today suggest that polyploidy seems to be rare among wild Diospyros species. The diversification of species of the NC clade III remains an overall poorly understood subject, despite our extensive efforts to find variation relevant to addressing these questions. It seems that we can eliminate polyploidy as one feature of their evolution, but the question of the involvement of hybridization cannot be eliminated without the use of more variable markers. To address patterns of speciation and factors promoting divergence, we will have to turn more markers used in population genetic studies, such as AFLPs, microsatellites or fingerprinting methods based on next generation sequencing methods.
Acknowledgments
This work was funded by a grant from the Austrian Science Fund (FWF, Project-Number: P 22159-B16) awarded to R. Samuel. The authors thank the team of the Department of Systematic and Evolutionary Botany as well as the team of the Botanic Department of IRD Noumea for support with this study. Special thanks go to V. Klenja for the lab work. Thanks to the following persons for their help with lab work, field work and ideas to improve our manuscript: J.-P. Butin, C. Chambrey, G. Dagostini, E. Grasserbauer, V. Hequet, G. Kohl, D. & I. Létocart, F. Maghuly, W. Nigote, O. Paun, G. Schneeweiss, J. Schönenberger, H. Vandrot, Fam. Villegente, B. Wallnöfer and H. Weiss-Schneeweiss.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ympev.2013.07.002.
Appendix A. Supplementary material
Supplementary Figure 1.
Dated phylogeny of Ericales based on the a joint matrix the data set of Bell et al. (2010) together with our plastid sequences. Taxa from families other than Ebenaceae are collapsed to family level, taxa other than Diospyros are collapsed to generic level. Multiple accessions of a species are collapsed to species level. The NC clade III part of the tree is mostly collapsed due to lack of support of respective nodes. Nodes which were calibrated with fossils are marked with a black dot. Yellow bars represent the 95% highest posterior density interval. New Caledonian taxa are coloured, red represents clade III NC.
BEAST input file for the Bayesian analysis of the combined data set. For Bayesian analyses of the individual data sets we used the same settings as in the combined analysis for the respective data set.
BEAST input file for dating analysis of the plastid marker data set from Bell et al. (2010) merged with our data.
BEAST input file for dating analysis of the combined data set.
References
- Alba R., Kelmsnson P.M., Cordonnier-Pratt M.-M., Pratt L.H. The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Molecular Biology and Evolution. 2000;17:362–373. doi: 10.1093/oxfordjournals.molbev.a026316. [DOI] [PubMed] [Google Scholar]
- APG III An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Baldwin B.G., Sanderson M.J. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proceedings of the National Academy of Sciences USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartish I.V., Antonelli A., Richardson J.E., Swenson U. Vicariance or long-distance dispersal: historical biogeography of the pantropical subfamily Chrysophylloideae (Sapotaceae) Journal of Biogeography. 2011;38:177–190. [Google Scholar]
- Basinger J.F., Christophel D.C. Fossil flowers and leaves of the Ebenaceae from the Eocene of southern Australia. Canadian Journal of Botany. 1985;63:1825–1843. [Google Scholar]
- Bell C.D., Soltis D.E., Soltis P.S. The age and diversification of the angiosperms re-revisited. American Journal of Botany. 2010;97:1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- Bennett M.D., Leitch I.J. Genome size evolution in plants. In: Gregory T.R., editor. The Evolution of the Genome. Elsevier; San Diego: 2005. pp. 90–151. [Google Scholar]
- Bennett, M.D., Leitch, I.J., 2010. Angiosperm DNA C-Values Database (Release 7.0, December 2010). <http://www.kew.org/cvalues/>.
- Bennett J.R., Mathews S. Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A1. American Journal of Botany. 2006;93:1039–1051. doi: 10.3732/ajb.93.7.1039. [DOI] [PubMed] [Google Scholar]
- Bremer K., Friis E.M., Bremer B. Molecular phylogenetic dating of asterid flowering plants shows early Cretaceous diversification. Systematic Biology. 2004;53:496–503. doi: 10.1080/10635150490445913. [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. JModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Ho S.Y.W., Phillips M.J., Rambaut A. Relaxed phylogenetics and dating with confidence. Molecular Biology and Evolution. 2006;29:1969–1973. doi: 10.1371/journal.pbio.0040088. (PLoS Biology 4, 699–710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duangjai S., Wallnöfer B., Samuel R., Munzinger J., Chase M.W. Generic delimitation and relationships in Ebenaceae sensu lato: evidence from six plastid DNA regions. American Journal of Botany. 2006;93:1808–1827. doi: 10.3732/ajb.93.12.1808. [DOI] [PubMed] [Google Scholar]
- Duangjai S., Samuel R., Munzinger J., Forest F., Wallnöfer B., Barfuss M.H.J., Fischer G., Chase M.W. A multi-locus plastid phylogenetic analysis of the pantropical genus Diospyros (Ebenaceae), with an emphasis on the radiation and biogeographic origins of the New Caledonian endemic species. Molecular Phylogenetics and Evolution. 2009;52:602–620. doi: 10.1016/j.ympev.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emshwiller E., Doyle J.J. Chloroplast-expressed glutamine synthetase (ncpGS): potential utility for phylogenetic studies with an example from Oxalis (Oxalidaceae) Molecular Phylogenetics and Evolution. 1999;12:310–319. doi: 10.1006/mpev.1999.0613. [DOI] [PubMed] [Google Scholar]
- Espeland M., Murienne J. Diversity dynamics in New Caledonia: towards the end of the museum model? BMC Evolutionary Biology. 2011;11:254. doi: 10.1186/1471-2148-11-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris J.S., Källersjö M., Kluge A.G., Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. [Google Scholar]
- Ferreira M.A.R., Suchard M.A. Bayesian analysis of elapsed times in continuous-time Markov chains. The Canadian Journal of Statistics. 2008;36:355–368. [Google Scholar]
- Forest F. Calibrating the Tree of Life: fossils, molecules and evolutionary timescales. Annals of Botany. 2009;104:789–794. doi: 10.1093/aob/mcp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Maddox J.M., Ayres N.M., Sharma D.P., Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Gaudeul M., Rouhan G., Gardner M.F., Hollingsworth P.M. AFLP markers provide insights into the evolutionary relationships and diversification of New Caledonian Araucaria species (Araucariaceae) American Journal of Botany. 2012;99:68–81. doi: 10.3732/ajb.1100321. [DOI] [PubMed] [Google Scholar]
- Geeraerts A., Raeymaekers J.A.M., Vinckier S., Pletsers A., Smets E., Huysmans S. Systematic palynology in Ebenaceae with focus on Ebenoideae: morphological diversity and character evolution. Review of Palaeobotany and Palynology. 2009;153:336–353. [Google Scholar]
- Gernhard T. The conditioned reconstructed process. Journal of Theoretical Biology. 2008;253:769–788. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Givnish T.J., Millam K.C., Berry P.E., Sytsma K.J. Phylogeny, adaptive radiation, and historical biogeography of Bromeliaceae inferred form ndhF sequence data. Aliso. 2007;23:3–26. [Google Scholar]
- Givnish T.J., Millam K.C., Mast A.R., Paterson T.B., Theim T.J., Hipp A.L., Henss J.M., Smith J.F., Wood K.R., Sytsma K.J. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae) Proceedings of the Royal Society B. 2009;276 doi: 10.1098/rspb.2008.1204. 407–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish T.J., Barfuss M.H.J., Van Ee B., Riina R., Schulte K., Horres R., Gonsiska P.A., Jabaily R.S., Crayn D.M., Smith J.A.C., Winter K., Brown G.K., Evans T.M., Holst B.K., Luther H., Till W., Zizka G., Berry P.E., Sytsma K.J. Phylogeny, adaptive radiation, and historical biogeography in Bromeliacae: insight from an eight-locus plastid phylogeny. American Journal of Botany. 2011;98:872–895. doi: 10.3732/ajb.1000059. [DOI] [PubMed] [Google Scholar]
- Grandcolas P., Murienne J., Robillard T., Desutter-Grandcolas L., Jourdan H., Guilbert E., Deharveng L. New Caledonia: a very old Darwinian island? Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 2008;363:3309–3317. doi: 10.1098/rstb.2008.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A.F., Ramsey T.S., Ramsey J. Phylogeny and biogeography of ivies (Hedera spp., Araliaceae), a polyploid complex of woody vines. Systematic Botnay. 2011;36:1114–1127. [Google Scholar]
- Greilhuber J., Ebert I. Genome size variation in Pisum sativum. Genome. 1994;37:646–655. doi: 10.1139/g94-092. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guo Y.-P., Wang S.-Z., Vogl C., Ehrendorfer F. Nuclear and plastid haplotypes suggest rapid diploid and polyploid speciation in the N Hemisphere Achillea millefolium complex (Asteraceae) BMC Evolutionary Biology. 2012;12:2. doi: 10.1186/1471-2148-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hasegawa M., Kishino H., Yano T.-A. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hughes C., Eastwood R. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. PNAS. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knope M.L., Morden C.W., Funk V.A., Fukami T. Area and the rapid radiation of Hawaiian Bidens (Asteraceae) Journal of Biogeography. 2012;39:1206–1216. [Google Scholar]
- Ladiges P.Y., Cantrill D. New Caledonia–Australian connections: biogeographic patterns and geology. Australian Systematic Botany. 2007;20:383–389. [Google Scholar]
- Leitch I.J. Genome size through the ages. Heredity. 2007;99:121–122. doi: 10.1038/sj.hdy.6800981. [DOI] [PubMed] [Google Scholar]
- Lowry P.P., II . Diversity, endemism and extinction in the flora of New Caledonia: a review. In: Peng C.F., Lowry P.P. II, editors. Rare, Threatened, and Endangered Floras of Asia and the Pacific Rim. Institute of Botany; Taipei, Taiwan: 1998. pp. 181–206. [Google Scholar]
- Magallón S. Using fossils to break long branches in molecular dating: a comparison of relaxed clocks applied to the origin of angiosperms. Systematic Biology. 2010;59:384–399. doi: 10.1093/sysbio/syq027. [DOI] [PubMed] [Google Scholar]
- Mathews S., Donoghue M.J. The root of angiosperm phylogeny inferred from duplicate phytochrome genes. Science. 1999;286:947–950. doi: 10.1126/science.286.5441.947. [DOI] [PubMed] [Google Scholar]
- Mathews S., Clements M.D., Beilstein M.A. A duplicate gene rooting of seed plants and the phylogenetic position of flowering plants. Philosophical Transaction of the Royal Society B Biological Sciences. 2010;365:383–395. doi: 10.1098/rstb.2009.0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin S. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany. 2001;49:271–300. [Google Scholar]
- Millán-Martínez M. Fossil record and age of the Asteridae. Botanical Reviews. 2010;76:83–135. [Google Scholar]
- Miller, M.A., Pfeiffer, W., Schwarz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, Louisiana, 14 November, 2010, pp. 1–8.
- Mittermeier R.A., Gil P.R., Hoffmann M., Pilgrim J., Brooks T., Mittermeier C.G., Lamoreux J., da Fonseca G.A.B. CEMEX; Mexico City: 2004. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions. [Google Scholar]
- Moody M.L., Rieseberg L.H. Sorting through the chaff, nDNA gene trees for phylogenetic inference and hybrid identification of annual sunflowers (Helianthus sect. Helianthus) Molecular Phylogenetics and Evolution. 2012;64:145–155. doi: 10.1016/j.ympev.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Morat P., Jaffré T., Tronchet F., Munzinger J., Pillon Y., Veillon J.-M., Chalopin M. Le référentiel taxonomique Florical et les caractéristiques de la flore vasculaire indigène de la Nouvelle-Calédonie. Adansonia. 2012;34:177–219. [Google Scholar]
- Murienne J. Testing biodiversity hypothesis in New Caledonia using phylogenetics. Journal of Biogeography. 2009;36:1433–1434. [Google Scholar]
- Murienne J., Grandcolas P., Piulachs M., Bellés X., D’Haese C., Legendre F., Pellens R., Guilbert E. Evolution on a shaky piece of Gondwana: is local endemism recent in New Caledonia? Cladistics. 2005;21:2–7. doi: 10.1111/j.1096-0031.2004.00042.x. [DOI] [PubMed] [Google Scholar]
- Murienne J., Guilbert E., Grandcolas P. Species’ diversity in the New Caledonian endemic genera Cephalidiosus and Nobarnus (Insecta: Heteroptera: Tingidae), an approach using phylogeny and species’ distribution modelling. Biological Journal of the Linnean Society. 2009;97:177–184. [Google Scholar]
- Myers N., Mittermeier R.A., Mittermeier C.G., da Fonseca G.A.B., Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nattier R., Robillard T., Desutter-Grandcolas L., Couloux A., Grandcolas P. Older than New Caledonia emergence? A molecular phylogenetic study of the eneopterine crickets (Orthoptera: Grylloidea) Journal of Biogeography. 2011;38:2195–2209. [Google Scholar]
- Nie Z.-L., Wen J., Azuma H., Qui Y.-L., Sun H., Meng Y., Sun W.-B., Zimmer E.A. Phylogenetic and biogeographic complexity of Magnoliaceae in the Northern Hemisphere inferred from three nuclear data sets. Molecular Phylogenetics and Evolution. 2008;48:1027–1040. doi: 10.1016/j.ympev.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Nixon K.C., Crepet W.L. Late cretaceous fossil flowers with ericalean affinity. American Journal of Botany. 1993;80:616–623. [Google Scholar]
- Otto F., Oldiges H., Göhde W., Jain V.K. Flow cytometric measurement of nuclear DNA content variations as a potential in vivo mutagenicity test. Cytometry. 1981;2:189–191. doi: 10.1002/cyto.990020311. [DOI] [PubMed] [Google Scholar]
- Parisod C., Alix K., Just J., Petit M., Sarilar V., Mhiri C., Ainouche M., Chalhoub B., Grandbastien M.A. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytologist. 2009;186:37–45. doi: 10.1111/j.1469-8137.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- Pelletier B. Geology of the New Caledonia region and its implications for the study of the New Caledonian biodiversity. In: Payri C., Richer de Forges B., editors. Compendium of marine species from New Caledonia, Documents Scientifiques et Techniques II4. Institut de Recherche pour le Développement Nouméa; 2006. pp. 17–30. [Google Scholar]
- Pellicer J., Fay M.F., Leitch I.J. The largest eukaryotic genome of them all? Botanical Journal of the Linnean Society. 2010;164:10–15. [Google Scholar]
- Petrov D.A. Evolution of genome size: new approaches to an old problem. Trends in Genetics. 2001;17:23–28. doi: 10.1016/s0168-9525(00)02157-0. [DOI] [PubMed] [Google Scholar]
- Pillon Y. Time and tempo of diversification in the flora of New Caledonia. Botanical Journal of the Linnean Society. 2012;170:288–298. [Google Scholar]
- Pillon Y., Hopkins H.C., Munzinger J., Amir H., Chase M.W. Cryptic species, gene recombination and hybridization in the genus Spiraeanthemum (Cunoniaceae) from New Caledonia. Botanical Journal of the Linnean Society. 2009;161:137–152. [Google Scholar]
- Pillon Y., Munzinger J., Amir H., Hopkins H.C., Chase M.W. Reticulate evolution on a mosaic of soils: diversification of the New Caledonian endemic genus Codia (Cunoniaceae) Molecular Ecology. 2009;18:2263–2275. doi: 10.1111/j.1365-294X.2009.04178.x. [DOI] [PubMed] [Google Scholar]
- Pillon Y., Munzinger J., Amir H., Lebrun M. Ultramafic soils and species sorting in the flora of New Caledonia. Journal of Ecology. 2010;98:1108–1116. [Google Scholar]
- Pillon Y., Johansen J., Sakishima T., Chamala S., Barbazuk W.B., Roalson E.H., Price D.K., Stacy E.A. Potential use of low-copy nuclear genes in DNA barcoding: a comparison with plastid genes in two Hawaiian plant radiations. BMC Evolutionary Biology. 2013;13:35. doi: 10.1186/1471-2148-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie M.D., Doyle J.A. Dating clades with fossils and molecules: the case of Annoaceae. Botanical Journal of the Linnean Society. 2012;169:84–116. [Google Scholar]
- Richardson J.E., Pennington R.T., Pennington T.D., Hollingsworth P.M. Rapid diversification of a species-rich genus of Neotropical rain forest trees. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- Salard M., Avias J. Contribution à la connaissance de la flore fossile de la Nouvelle-Calédonie. Palaeontographica, Abteilung B, Paläophytologie. 1968;124:1–44. [Google Scholar]
- Silvestro D., Schnitzler J., Zizka G. A Bayesian framework to estimate diversification rates and their variation through time and space. BMC Evolutionary Biology. 2011;11:311–325. doi: 10.1186/1471-2148-11-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swenson U., Hill R.S., McLoughlin S. Biogeography of Nothofagus supports the sequence of Gondwana break-up. Taxon. 2001;50:1025–1041. [Google Scholar]
- Swofford, D.L., 2003. PAUP∗. Phylogenetic Analysis Using Parsimony (∗ and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.
- Tamura M., Tao R., Yonemori K., Utsunomiya N., Sugiura A. Ploidy level and genome size of several Diospyros species. Journal of the Japanese Horticultural Society. 1998;67:306–312. [Google Scholar]
- Tank D.C., Sang T. Phylogenetic utility of the glycerol-3-phosphate acyltransferase gene: evolution and implications in Paeonia (Paeoniaceae) Molecular Phylogenetics and Evolution. 2001;19:421–429. doi: 10.1006/mpev.2001.0931. [DOI] [PubMed] [Google Scholar]
- Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences. 1986;17:57–86. [Google Scholar]
- Tel-Zur N., Abbo S., Myslabodsky D., Mizrahi Y. Modified CTAB procedure for DNA isolation from ephiphytic cacti of the genera Hylocereus and Selenicereus (Cactaceae) Plant Molecular Biology Reporter. 1999;17:249–254. [Google Scholar]
- Temsch E.M., Greilhuber J., Krisai R. Genome size in liverworts. Preslia. 2010;82:63–80. [Google Scholar]
- Weiss-Schneeweiss H., Villaseñor J.L., Stuessy T.F. Chromosome numbers, karyotypes, and evolution in Melampodium (Asteraceae) International Journal of Plant Science. 2009;170:1168–1182. [Google Scholar]
- White F. Twenty-two new and little known species of Diospyros (Ebenaceae) from New Caledonia with comments on section Maba. Bulletin du Muséum National d’Histoire naturelle 4ème série – Section B. Adansonia. 1992;2:179–222. [Google Scholar]
- White F. Muséum National d’Histoire Naturelle; Paris: 1993. Flore de la Nouvelle-Calédonie et Dépendances. 19. Ébénacées. [Google Scholar]
- Yockteng R., Nadot S. Phylogenetic relationships among Passiflora species based on the glutamine synthetase nuclear gene expressed in chloroplast (ncpGS) Molecular Phylogenetics and Evolution. 2004;31:379–396. doi: 10.1016/S1055-7903(03)00277-X. [DOI] [PubMed] [Google Scholar]
- Yule G.U. A mathematical theory of evolution, based on the conclusions of Dr. J.C. Willis, FRS. Philosophical Transactions of the Royal Society of London Series B. 1925;213:21–87. [Google Scholar]
- Zimmer E.A., Wen J. Using nuclear gene data for plant phylogenetics: progress and prospects. Molecular Phylogenetics and Evolution. 2012;65:774–785. doi: 10.1016/j.ympev.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl H., Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson G., Vogel H.J., editors. Evolving Genes and Proteins. Academic Press; New York: 1965. pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BEAST input file for the Bayesian analysis of the combined data set. For Bayesian analyses of the individual data sets we used the same settings as in the combined analysis for the respective data set.
BEAST input file for dating analysis of the plastid marker data set from Bell et al. (2010) merged with our data.
BEAST input file for dating analysis of the combined data set.