Abstract
Pneumocystis pneumonia remains an important complication of immune suppression. The cell wall of Pneumocystis has been demonstrated to potently stimulate host inflammatory responses, with most studies focusing on β-glucan components of the Pneumocystis cell wall. In the current study, we have elaborated the potential role of chitins and chitinases in Pneumocystis pneumonia. We demonstrated differential host mammalian chitinase expression during Pneumocystis pneumonia. We further characterized a chitin synthase gene in Pneumocystis carinii termed Pcchs5, a gene with considerable homolog to the fungal chitin biosynthesis protein Chs5. We also observed the impact of chitinase digestion on Pneumocystis-induced host inflammatory responses by measuring TNFα release and mammalian chitinase expression by cultured lung epithelial and macrophage cells stimulated with Pneumocystis cell wall isolates in the presence and absence of exogenous chitinase digestion. These findings provide evidence supporting a chitin biosynthetic pathway in Pneumocystis organisms and that chitinases modulate inflammatory responses in lung cells. We further demonstrate lung expression of chitinase molecules during Pneumocystis pneumonia.
Keywords: Pneumocystis carinii, Cell wall, Chitin, Chs5, Chitinase, Chitotriosidase, Acidic mammalian chitinase
Introduction
Pneumocystis pneumonia is a clinically relevant respiratory complication of immunocompromised patients, such as those with HIV/AIDS [1, 2]. The incidence of Pneumocystis pneumonia is also rising in non-HIV infected patients receiving immunosuppressive treatment or anti-neoplastic chemotherapy [3]. Respiratory impairment leading to respiratory failure and death is caused by Pneumocystis jirovecii, a human-specific fungal species that resides in the alveoli of the lungs. P. carinii, the species that infects rats, represents an extremely useful animal model for the study of Pneumocystis pneumonia pathogenesis. Because Pneumocystis pneumonia continues to be a AIDS-defining opportunistic infection, ongoing studies to characterize the pathogenesis of Pneumocystis pneumonia are critically important [2]. In particular, studies indicate that lung inflammation contributes to respiratory impairment and injury during this infection [4].
The Pneumocystis cell wall armor plays a major role in the growth and survival of this opportunistic pathogen, as well as in the regulation of host defense responses. The cell walls of fungi are complex and dynamic structures that are composed of a thick network of branched glucan and chitin carbohydrates, and species-specific antigenic glycoproteins [5, 6]. Microscopy and biochemical studies demonstrate that the cell wall of the Pneumocystis species is composed of similar but unique carbohydrate structures and embedded proteins [7–12]. The cell wall biosynthesis mechanisms have been characterized in several fungal species [13–24], including β-glucan assembly pathways that have been defined in Pneumocystis [25–27]. Although most investigations have focused largely on β-glucans, studies suggesting the presence of cell wall chitin, an N-acetyl-D-glucosamine polymeric carbohydrate, in Pneumocystis have been reported previously. Immunolabeling studies have detected chitins in the cyst forms, intracystic bodies, and trophic forms of Pneumocystis from human lung biopsies and bronchoalveolar lavage specimens [28]. In addition, lectin and antiserum probes for N-acetyl-D-glucosamine revealed that several isolates of Pneumocystis express chitins at all stages of the life cycle [29].
The rigid carbohydrate-rich cell wall provides structural strength for this opportunistic pathogenic organism. However, glucans and chitins are also pathogen associated molecular patterns that elicit host responses. In Pneumocystis pneumonia, β-glucans have been directly attributed as potent stimulators of alveolar epithelial cell, alveolar macrophage, and dendritic cell inflammatory responses [4, 30–36]. Chitin-containing pathogens and other allergens have further been implicated as potent inducers of humoral Th2-specific pathways involving expression of mammalian chitinases [37]. These studies suggest that chitins also serve as important components of the cell wall. However, characterization of a chitin biosynthetic pathway in Pneumocystis and the potential role of host chitinases have not yet been studied.
In addition, the immunological roles of host mammalian chitinases have also recently been evaluated in various inflammatory disease models including asthma or allergen challenge with ovalbumin, Th2- and IL13-mediated inflammation, cigarette smoke challenge models, and pulmonary cryptococcosis [38, 39]. However, the activities of host chitinases have not yet been defined during active Pneumocystis pneumonia. Other recent studies support colonization of Pneumocystis in COPD and other chronic lung disease, which also may be impacted by the modulation of chitinases in the lower respiratory tract during these conditions [40].
In this light, we have sought to better understand chitin-mediated mechanisms in the pathogenesis of Pneumocystis pneumonia. In this study, we have evaluated host mammalian chitinase gene expression during Pneumocystis pneumonia. We have further identified a putative chitin synthase gene in Pneumocystis carinii by characterizing Pcchs5, a gene with considerable homolog to the S. cerevisiae and S. pombe Chs5 chitin biosynthesis proteins. We further illustrate the impact of chitinases in modifying Pneumocystis-induced host inflammatory responses by measuring TNFα release. We also evaluated mammalian chitinase expression by lung epithelial and macrophage cells that have been stimulated with Pneumocystis organisms and isolated cell walls in the presence and absence of exogenous chitinase digestion. These findings provide evidence for a chitin biosynthetic pathway in the Pneumocystis organism and that chitinases modulate host responses in lung cells.
Methods
Pneumocystis propagation and isolation
Pneumocystis carinii organisms were propagated in and isolated from female Long Evans rats (Harlan, Inc. Indianapolis, IN) lungs. Rats were immunosuppressed with dexamethasone (1.2 mg/L) added to their drinking water for 2 weeks prior to intratracheal inoculation of P. carinii. Infection was allowed to progress for 8–12 weeks. P. carinii were extracted for subsequent assays by resection of the lung followed by mincing and homogenization in normal saline. The homogenate was filtered through gauze to separate large cellular debris. The filtrate was then centrifuged at 6,600×g for 10 min at 4 °C. The pellets were resuspended in sterile water and incubated at room temperature for 5 min to allow lysis of remaining host cells. After centrifugation, the pellets were resuspended in 20 mL normal saline, passed through a 10-μm nitrocellulose filter and centrifuged to collect a purified population of P. carinii organisms. The P. carinii organisms were then either processed immediately for further analysis or the pellet flash frozen in a dry ice/methanol bath and stored at −80 °C.
Pneumocystis cell wall isolation
Pneumocystis carinii particulate cell wall carbohydrate fractions were isolated from approximately 30 rats [4, 36]. To accomplish this, the P. carinii were recovered by centrifugation, washed with double deionized water, autoclaved, and then allowed to cool overnight at 4 °C. The P. carinii pellets were then washed with water, resuspended in 50 mL water, and disrupted with 0.5 mm glass beads (Biospec Products Inc). The lysates were then transferred into clean centrifuge tubes. NaOH (1 M) was added to the lysates, stirred at room temperature for 16 h, and then centrifuged at ~13,700×g for 30 min (Beckman Coulter Avanti J-26 XP). The cell wall pellets were next resuspended in 1 M NaOH, incubated at 65 °C for 2 h, followed by several washes with water. The cell wall isolates were treated with chloroform and methanol (2:1), stirred at room temperature for 2 h and centrifuged. The pellets were washed once with each solution: 50 % methanol/50 % saline, 25 % methanol/75 % saline, 10 % methanol/90 % saline, 100 % saline. After the final wash, 10 μg/mL polymyxin B sulfate was added to the cell wall suspension and incubated at 4 °C overnight to bind any residual endotoxin, which may have been associated with the isolate. After several further washes with saline, the cell wall isolates were recovered, weighed and finally resuspended in the appropriate volume of Dulbecco’s phosphate-buffered saline (DPBS). Our cell wall fractionation procedures produce isolates that are in excess of 90 % carbohydrate [4, 36]. The presence of endotoxin in the final isolate wash was excluded using the Pyrosate assay (<0.25 EU/mL; Associates of Cape Cod) according to manufacturer’s instructions.
Detection of a Pneumocystis chitin synthase gene
A partial Pneumocystis chitin synthase gene sequence was identified from the Pneumocystis carinii Genome Project database (http://pgp.cchmc.org/). This sequence was used to generate a 616-bp radiolabeled probe using RadPrime DNA labeling system (Invitrogen, Inc.) and [32P]-ATP followed by filtration through Sepharose mini QuickSpin columns (Roche, Inc). The primer sequences employed for probe generations were: forward primer 5′-CTATCAGGCCTTGCATTGAATACGGAG-3′ and reverse primer 5′-CATCTTCCTCGCTAATTGGTCTTGGAG-3′. For Southern blot analysis, P. carinii from three rat lungs were purified as described. Genomic DNA was extracted using MasterPure Yeast DNA Purification Kit (Epicentre Bio-technologies, Inc.) according to the manufacturer’s protocol. P. carinii genomic DNA (10 μg) was digested with 50 units of restriction enzymes, HinDIII, XbaI, or EcoRI, (Promega, Inc.) for 4 h at 37 °C. Genomic DNA (10 μg) from normal rat lung tissue was also digested under the same conditions as controls. The digested genomic DNA was electrophoresed through a 1 % agarose gel in TAE buffer and transferred onto a Nytran Supercharge nylon membrane using downward capillary flow (TurboBlotter, Whatman) of 20× saline sodium citrate buffer overnight at room temperature, followed by UV crosslinking. Radiolabeled probes were hybridized to the membranes using ExpressHyb Hybridization solution (Clontech, Inc.) at a concentration of 1.0 × 106 cpm/mL. Prehybridization, hybridization, and washing were done according to manufacturer’s protocol. The membranes were exposed to X-ray film with two intensifying screens at −70 °C for 48 h.
Identifying the complete coding sequence of Pcchs5, a putative Pneumocystis chitin synthase
A partial Pneumocystis chitin synthase gene sequence was used to derive primers for rapid amplification of cDNA ends (RACE). The full-length coding region was generated using the RACE strategy in a manner parallel to our previous studies [26]. The final cDNA product was cloned into pCR-BluntII-TOPO® (Invitrogen) and fully sequenced (GenBank accession # GU722292). The final genomic DNA product was also cloned and sequenced (GenBank accession # GU722293). The open reading frame was identified as a chitin biosynthesis protein, Pcchs5, analyzed by sequence alignment in AlignX (Life Tech) and BLASTX [41], and identification of conserved domains [42, 43].
Complementation of S. cerevisiae function with Pcchs5
To further verify the function of PcChs5, we performed complementation studies in Chs5-deficient yeast, using a Calcofluor White M2R sensitivity assay. It has been demonstrated previously in S. cerevisiae that chs5 gene deletion causes increased resistance to Calcofluor and upon restoration of the chs5 gene by complementation Calcofluor sensitivity is restored [44]. Accordingly, we next assessed the growth of the S. cerevisiae Δchs5 strain transformed with the PcChs5 DNA cloned in the pYES2.1 TOPO® TA expression vector (Invitrogen, Inc.). To compare expression of the yeast chs5Δ harboring the PcChs5 gene, the control yeast expression vector pYES2.1/V5-His/lacZ was transformed into the parental wild-type or chs5Δ strains. After the overnight growth of the various strains at 30 °C in minimal medium with 2 % galactose, yeast cells were diluted to ~6 × 106 cells/mL. Yeast cells were then serially diluted and plated on minimal media plates containing 2 % galactose alone, or in the presence of 100 μg/ mL of Calcofluor White M2R (Sigma Aldrich, Inc.) prepared as described [45]. Finally, the yeasts were cultured for 2 additional days at 30 °C, and the growth was assessed.
Macrophage inflammatory response to Pneumocystis cell wall isolates in the presence and absence of exogenous chitinase digestion
Macrophage RAW 264.7 cells (ATTC No. TIB-71) were cultured and maintained in Dulbecco’s Modified Eagle Medium (DMEM) with 10 % heat-inactivated fetal bovine serum (FBS) and 1 % penicillin/streptomycin. Cells were plated into 96-well plates at a density of 1.0 × 106 cells/ mL and allowed to adhere by incubation at 37 °C for 4–6 h. P. carinii cell wall isolates were meanwhile resuspended in DMEM (100 μg/mL) and incubated with cell wall hydrolases, endo-1,3-1,4-β-D-glucanohydrolase (Lichenase, Megazyme, Inc., Catalog No. E-LICHN) and β-N-acetylglucosaminidase (Chitinase, Catalog No. P0732L, New England Biolabs) at 37 °C for 4 h. The hydrolase reactions were then heat inactivated at 65 °C for 20 min. P. carinii cell wall isolates were digested with either 1 U or 10 U of enzyme (or D-PBS control) and were added to the RAW cells and incubated at 37 °C for 18 h. Tumor necrosis factor (TNFα) secreted into the cell culture media was measured by ELISA (eBioscience, Inc.).
Mammalian chitinase expression in Pneumocystis-infected rats
Lungs from healthy control uninfected rats and Pneumocystis-infected rats were extracted and placed in RNA stabilization reagent (RNAlater®). Lung tissue samples were homogenized, lysed, and the total RNA extracted and purified using RNeasy Plus (Qiagen). The cDNAs from all samples were generated by reverse transcription of mRNA (from 5 μg total RNA) using SuperScript III First Strand Synthesis System and Oligo(dT) primer (Invitrogen). The mRNA transcripts of two host mammalian chitinases, a chitotriosidase (Chit1) and an acidic mammalian chitinase (AMCase), were measured by quantitative real-time PCR using Platinum SYBR Green (Invitrogen, Inc.), detected and analyzed by the CFX96 Real-Time PCR Detection System (Bio-Rad, Inc.). PCR amplifications were performed on an iCycler Apparatus (BIORAD, Inc.) with a basal sensitivity of 10 pg of nucleic acid per sample. The primer sequences for each mammalian chitinase gene are as follows: Chit1 forward primer 5′-GAAGGGACTGGGA GGGGCCATGG-3′ and reverse primer 5′-ACAGTCGCC CCCCTCCACAGCTG-3′; AMCase forward primer 5′-CTTTGGTGGTGCCATGATCTGGGCC-3′ and reverse primer 5′-GTCATCTGCCACAGGGTAGAGGCCATCTG-3′. The PCR quantitation method was semi-quantitative, meaning that the relative fold expression was normalized to mammalian Histone H3 transcript expression for loading normalization of rat cDNA in each of the samples studied. Data points represent the change in transcript levels relative to Histone H3 in normal lungs. At least four animals were tested for each condition.
In vitro analysis of Pneumocystis-induced host lung cell chitinase mRNA expression
Rat lung epithelial RLE-6TN cells (ATCC No. CRL-2300) were cultured and maintained in DMEM/F-12 medium with 10 % FBS and 1 % penicillin/streptomycin. RAW cells were cultured as described above. RLE cells were plated at a density of 75,000 cells/well and RAW cells at a density of 150,000 cells/well in 48-well plates and incubated at 37 °C overnight. Pneumocystis cell wall isolates were isolated as described. Isolated P. carinii cell walls were suspended in mammalian cell culture media at concentrations of 50, 100, and 200 μg/mL for 18 h. The media were then aspirated and the cell monolayers frozen at −80 °C overnight. Cells were then processed for Chit1 and AMCase mRNA quantification as described above. Data points represent the relative expression levels compared with D-PBS controls.
Statistical analysis
All assays were performed on at least three occasions with the data representing the mean ± the standard error of the mean. To evaluate differences between means, initially multi-group analyses were performed using one-way analysis of variance (ANOVA), and Student’s two group T tests for paired measurements if the measured variable was normally distributed. For measured variables that were non-Gaussian in distribution, the corresponding non-parametric tests were employed, namely the Kruskal–Wallis test for multi-group analyses, and the Wilcoxon test for paired two group analyses. P <0.05 was defined as a statistically significant difference. Statistical analyses were performed using the GraphPad Prism version 5.0b software program.
Results
Identification of a putative chitin synthase gene in Pneumocystis carinii
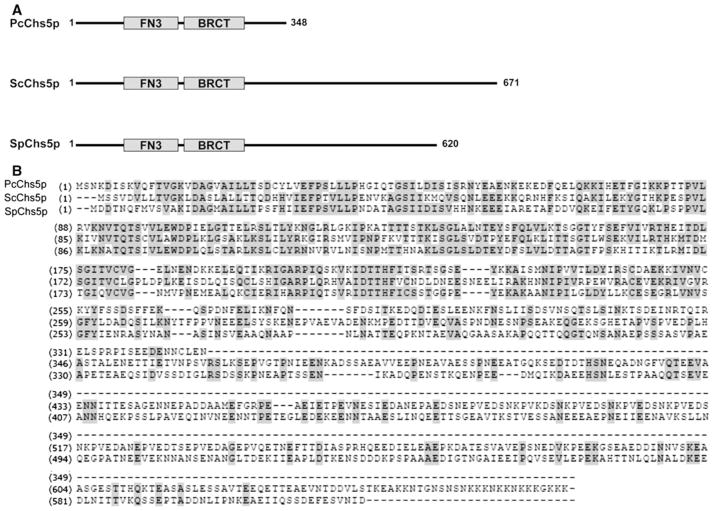
Structural and biochemical studies of Pneumocystis organisms have shown evidence of chitin-containing compounds within the cell wall, but the mechanisms of chitin generation in P. carinii have not yet been determined [7, 28, 29]. Accordingly, we further sought evidence of a chitin biosynthetic pathway in Pneumocystis carinii. An in silico search revealed a 729-bp partial chitin synthase gene sequence (Pcchs5) within the available partial coverage of the Pneumocystis Genome Project database (http://pgp.cchmc.org/). To complete the cloning of this sequence, we designed gene-specific primers for rapid amplification and sequencing of cDNA ends (RACE), revealing a 1047-bp open reading frame (GenBank # GU722292). Subsequent amplification of genomic DNA revealed a 1096-bp region containing one intron (GenBank # GU722293). The amino acid sequence was predicted to contain two motifs, a Breast Cancer Suppressor Protein carboxy-terminal domain (BRCT) and a Fibronectin type 3 domain (FN3), conserved domains present in both the Saccharomyces cerevisiae and Schizosaccharomyces pombe Chs5 (Fig. 1). The percent identity for the predicted PcChs5 protein across the whole length of the protein was found to be 44 % similarity in comparison with the S. cerevisiae orthologue. This level of amino acid similarity compared with the S. cerevisiae Chs5 increased 65 % when comparisons were extended to include like-charged amino acids. In comparison of the predicted whole length PcChs5 protein with S. pombe Chs5, the basal similarity was 46 %, increasing to 69 % when similarly charged amino acids were counted as conserved.
Fig. 1.
Conserved domains of the putative Pneumocystis chitin synthase Chs5. a Conserved domains of Chs5 in P. carinii (PcChs5), S. cerevisiae (ScChs5), and S. pombe (SpChs5) have a Breast Cancer Suppressor Protein carboxy-terminal motif (BRCT) and a Fibronectin type 3 motif (FN3). b Amino acid sequence of PcChs5 reveals 42 % identity to ScChs5 and 48 % identity to SpChs5 within conserved regions
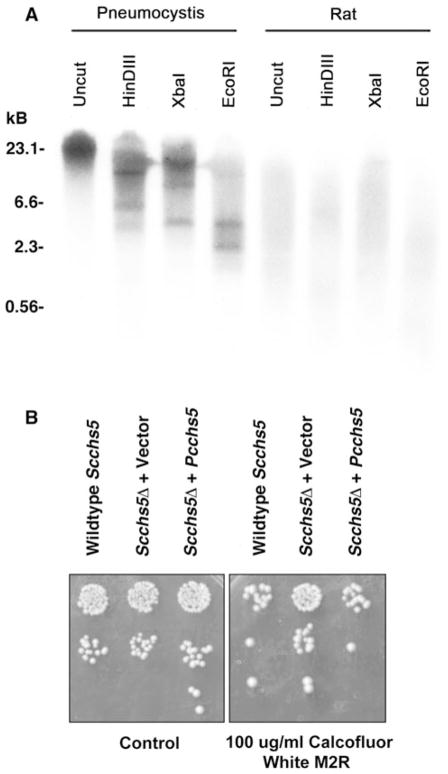
To confirm that the available sequence was indeed of P. carinii origin, rather than an amplification artifact, we further evaluated whether the Pcchs5 sequence was present in Pneumocystis genomic DNA by Southern blot analysis. Pneumocystis carinii organisms were isolated from rat lungs and genomic DNA extracted. DNA was digested with various restriction enzymes (HinDIII, XbaI, EcoRI), separated by agarose gel electrophoresis, and transferred to nitrocellulose membrane. A Pcchs5-specific radioactive nucleotide probe was generated followed by hybridization of the probe to the transferred genomic DNA. The probe corresponded to amino acids 137-341 of the PcChi5 protein as depicted in Fig. 1. The hybridization demonstrated bands in lanes representing digested P. carinii genomic DNA, but no hybridization occurred with digested rat genomic DNA (Fig. 2a). The observations of greater than one band within any given digestion condition, such as with EcoRI, would indicate a restriction endonuclease recognition sequence somewhere within the gene, including within any intron regions. Alternatively, the probe may also cross react and recognize other chitin synthases present within the Pc genome that have not yet been described. Thus, Pneumocystis carinii appears to contain a genetic coding sequence with significant homology to other fungal chitin synthase genes.
Fig. 2.
A functional chitin synthase gene (Pcchs5) is present within the Pneumocystis genome. a Southern blot analysis illustrates the presence of chitin synthase in Pneumocystis. Genomic DNA was digested with restriction enzymes HinDIII, XbaI and EcoRI and labeled with a 616-bp Pcchs5 gene-specific radioactive probe. Rat genomic DNA was used as negative control. b PcChs5 complements Scchs5Δ-deficient yeast. Deficiency of Chs5 function in yeast results in relative resistance to Calcofluor white, an agent that binds chitin in the fungal cell wall, compared to the wild-type parental yeast. Transformation of Scchs5Δ-deficient yeast with empty vector displays such resistance during culture in the presence of Calcofluor. In contrast, transformation of Scchs5Δ-deficient yeast with Pcchs5 restores the sensitivity to Calcofluor parallel to the parental wild-type yeast
Finally, to further verify the potential function of PcChs5, we proceeded to complement PcCh5 function in chs5-deficient yeast (Fig. 2b). It has been previously demonstrated that S. cerevisiae deficient in the chs5 gene exhibit resistance to Calcofluor. However, upon restoration of the chs5 gene function by complementation Calcofluor sensitivity is restored [44]. Under these conditions, complementation of chs5Δ yeast with either wild-type ScChs5 or PcChs5 provided comparable level of Calcoflour sensitivity. Hence, under these conditions Pneumocystis carinii PcChs5 provided identical function to wild-type S. cerevisiae ScChs5.
Chitinase digestion reduces Pneumocystis cell wall carbohydrate induced host inflammatory reactivity
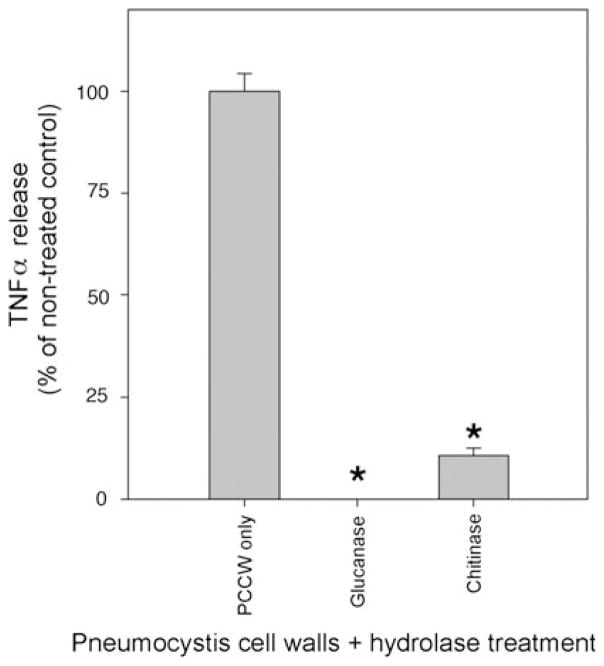
Host immune responses to Pneumocystis cell wall carbohydrates have been well documented [31, 33–36, 46, 47]. However, we further postulated that chitinases might act to reduce innate host inflammatory responses. Accordingly, we next examined the role of chitinase digestion of P. carinii cell wall carbohydrate isolates on macrophage cytokine responses. To address this, cultured macrophage cells (RAW) were exposed to purified P. carinii cell wall carbohydrates isolates in the presence and absence of digestion with chitinase, and macrophage TNFα release responses were measured (Fig. 3). As we previously observed, treatment of P. carinii cell walls isolates with an endo-1,3-1,4-β-D-glucanohydrolase (glucanase) greatly reduced TNFα production compared with non-treated controls, confirming our prior studies that P. carinii cell wall β-glucan stimulates host inflammatory responses [36]. Similarly, however, when the Pneumocystis cell wall isolates were treated with chitinase, the TNFα responses of macrophages to the P. carinii cell walls isolates were also significantly reduced by up to 90 % (P ≤ 0.001 compared with control without digestion). Thus, the overall reduction of TNFα release following chitinase digestion indicates that chitin components of the Pneumocystis cell wall may participate in host inflammatory responses to the P. carinii cell wall.
Fig. 3.
Pneumocystis cell wall isolates induce host TNFα response that is reduced by chitinase (and glucanase) digestion. Macrophages (RAW) were challenged with isolated Pneumocystis carinii cell wall isolates (PCCW) that were either digested with β-glucanase (10 U) and chitinase (1 U) or left undigested (control) and TNFα release into the culture media assessed by ELISA. (*denotes P ≤ 0.001 compared with undigested control)
Pneumocystis infection induces mammalian chitinase gene expression in host lungs
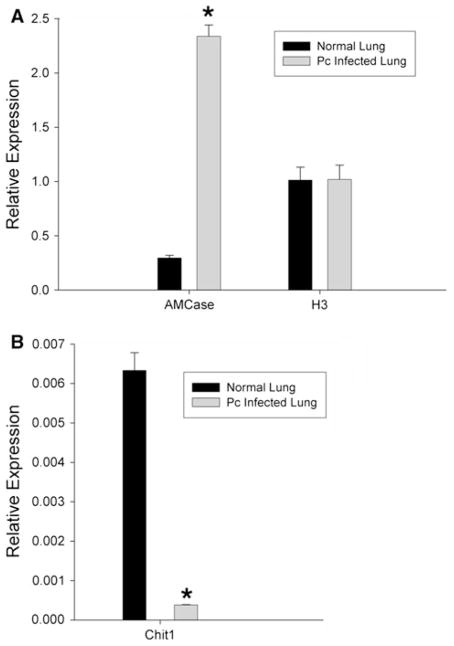
Mammalian chitinases have been recently implicated in host defense mechanisms against chitin-containing pathogens and allergens [38, 39]. We therefore investigated the effects of Pneumocystis infection on the expression of mammalian chitinases in rat lungs. Lungs from uninfected controls and Pneumocystis-infected rats were extracted and processed for RNA purification and generation of cDNA. The mRNA levels of the chitinase, chitotriosidase (Chit1), and an acidic mammalian chitinase (AMCase) were quantified by semi-quantitative PCR as described under methods, using mammalian histone H3 to normalize for loading (Fig. 4). The values reported are the fold changes relative to the Histone H3 housekeeping gene expressed constitutively in lungs. There was an eightfold increase in AMCase expression (Fig. 4a) from normal lungs (relative quantity 0.2962) compared with P. carinii-infected lungs (relative quantity 2.3380) (P <0.0001).
Fig. 4.
Pneumocystis infection induces mammalian chitinase expression in the lungs. Pneumocystis pneumonia was induced in rats as described under methods. After 8 weeks of infection, the animals were killed and whole lung fixed in RNAlater® and mammalian host chitinase expression determined by semi-quantitative PCR as described in the methods, using primers recognizing a acidic mammalian chitinase (AMCase) and b chitotriosidase (Chit1). For comparison, identical aged uninfected rats were used as controls. To exclude the possibility that the immune suppression was altering gene expression in the lung, we similarly quantified rat H3 histone (H3) expression as an additional control. (*denotes P <0.001 compared with uninfected control rat lung)
As an added control, we also evaluated the expression of AMCase in dexamethasone-treated rats, which also received low-level TMP suppression (2 mg/mL in their drinking water) to suppress latent Pneumocystis reactivation, in order to be certain that the dexamethasone itself did not induce the expression of AMCase. Indeed, in the presence of dexamethasone alone, but with no Pneumocystis infection, there was actually a 51.7 ± 0.1 % reduction in the basal level of AMCase expression, further indicating that the noted induction of AMCase during Pneumocystis pneumonia was due to infection with the organisms rather than effects of the dexamethasone alone.
Furthermore, the magnitude of Chit1 expression levels was also observed to much less than AMCase overall. However, Chit1 expression levels did demonstrate a 17-fold decrease in expression during Pneumocystis pneumonia (Fig. 4b) compared with normal lungs (relative quantity 0.0063) compared with P. carinii-infected lungs (relative quantity 0.00038) (P = 0.0002). Taken together, these data indicate that mammalian chitinase expression profiles are differentially modulated in mammalian lungs during Pneumocystis pneumonia.
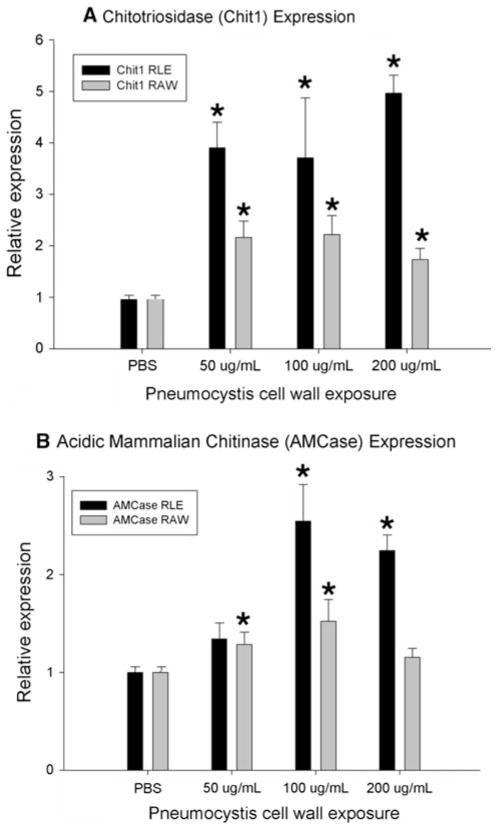
Mammalian chitinases in host epithelial and macrophage cells are induced by Pneumocystis cell wall carbohydrates
Studies have shown that expression of mammalian chitinases in lung epithelial cells and macrophages are important mediators of both anti-infective responses and allergen-induced inflammation [39, 48, 49]. To further investigate the role of mammalian chitinase expression seen in the Pneumocystis infection animal model, Chit1 and AMCase expressions were quantified in lung epithelial and macrophage cells that were exposed to Pneumocystis carbohydrate cell wall isolates (Fig. 5). Purified Pneumocystis carbohydrate cell wall isolates were suspended in cell culture media (50, 100, and 200 μg/mL) then incubated with rat lung epithelial (RLE) and murine macrophage (RAW) cells. The relative expression levels of Chit1 compared with histone H3 increased up to fivefold in RLE cells (P = 0.031) and up to 2.5-fold in RAW cells (P = 0.008), while the relative expression levels of AM-Case increased up to 2.5-fold in RLE cells (P = 0.036) and up to 1.5-fold in RAW cells (P = 0.008), when compared with cells that were treated with PBS only. These results suggest that Pneumocystis cell wall carbohydrates isolates are potent inducers of mammalian chitinases in lung epithelial and macrophage following challenge with Pneumocystis.
Fig. 5.
Mammalian chitinases are induced by Pneumocystis cell wall carbohydrate isolates. Lung epithelial (RLE) and macrophages (RAW) were challenged with Pneumocystis cell wall carbohydrate isolates. Relative increases in mRNA expression levels of Chit1 and AMCase mammalian chitinases compared with histone H3 were measured by PCR. (*denotes P <0.05 compared with PBS control)
We further sought to confirm whether surface components present on whole Pneumocystis organisms would also induce mammalian chitinase expression in lung cells. Indeed, whole Pc induced significant release of both AMCase and Chit1 from RLE and RAW cells, though at somewhat lower levels than observed using the purified cell wall carbohydrate preparations. For instance, using rat lung epithelial cells, the relative expression of AMCase expression was 1.4-fold increased by freshly isolated whole P. carinii organisms (10 Pc per host cell) and Chit1 expression was increased 1.6-fold compared with control PBS-treated cells (P <0.001 for both comparisons). Similarly, RAW macrophages stimulated with P. carinii increased AMCase expression by 1.6-fold and Chit1 expression by 1.3-fold compared with control (P <0.01 for both comparisons). Thus, whole P. carinii organisms are also capable of inducing mammalian chitinase expression in lung epithelial cells and macrophages. We finally attempted to determine whether digestion of the P. carinii organisms and cell wall isolates with either glucanase or chitinase (using 1 U and 10 U/mL each). Interestingly, the levels of either AMCase or Chit 1 expression were not significantly altered by either chitinase or glucanase digestions and in some experimental runs trended toward slightly increased expression (data not shown). We postulate that the stimulatory signals inducing mammalian chitinase gene expression are complex and may involve multiple Pneumocystis surface components. Digestion of one component may have acted simply to unmask other moieties also capable of stimulating mammalian chitinase gene expression.
Discussion
Pneumocystis pneumonia is a serious respiratory infection that continues to affect immunocompromised patient populations [1, 2, 50]. Despite expanded use of prophylaxis strategies, this opportunistic disease will not likely be eradicated and will continue to be an important factor in respiratory compromise in patients with suppressed immunity [51–53]. Although an ecological niche for Pneumocystis organisms has not been identified outside of the mammalian host, studies of the organism’s pathobiology within the lung, including evidence of person-to-person transmission, colonization, and proliferation, indicate the requirement for generation and remodeling of a protective cell wall [54].
The cell wall is a major regulator of organism growth and survival. This carbohydrate-rich layer consists of a network of branched glucan and chitin polymers as characterized in numerous fungal species. In Pneumocystis, we have previously characterized several cell wall associated genes that are involved in the biosynthesis, stability, degradation, and remodeling of the cell wall β-glucan structures. These genes include gsc1 that encodes a protein mediating β-glucan deposition [25], the environmentally responsive kinases cbk1 and phr1 involved in regulation of cell wall integrity, and the eng2 endo-β-glucanase gene that mediates cell wall degradation [26, 27, 55–57]. Our prior studies have focused mainly on the β-glucan components of the organism, illustrating its apparent importance in cell wall morphology. However, chitin is another component of the fungal cell wall, and investigations of this carbohydrate polymer in Pneumocystis are also of considerable importance.
The current studies are the first to identify a complete putative chitin synthase gene (Pcchs5) within the Pneumocystis genome and to illustrate conserved domains with homology to chitin biosynthesis proteins and similar function to genes that have been characterized in S. cerevisiae and S. pombe. In further support of a chitin-remodeling pathway, our previous studies have demonstrated that the Pneumocystis Cbk1 cell wall biosynthesis kinase further regulates chitinase activity when heterologously expressed in yeast [26]. The characterization of these genes that regulate chitin generation and degradation should provide tools to enhance our understanding of Pneumocystis cell wall morphology, its role in organism viability and proliferation, and its impact on host inflammatory responses.
How the Pneumocystis PcCh5 relates to chitin synthases in other Ascomycetes is difficult to discern at this time, as other members of this phylum contain between two and up to 20 chitin synthase genes. These genes are further subdivided into up to seven various classes [58]. This diversity of chitin synthase genes may reflect in part redundancy of function or a variety of specialized activities under various culture or environmental conditions. It remains highly likely that Pc also contain other various chitin synthase genes and cognate proteins.
Fungal cell wall proteins and carbohydrates are potent elicitors of host inflammatory and immunological responses. For instance, surface glycoprotein components of Pneumocystis have been widely studied because of their high specificity and the potential to stimulate humoral responses and complement activation [8, 59–62]. The great variety of the major surface glycoprotein isoforms and differences in antigenicity suggest a potential mechanism for evasion of host immune responses during Pneumocystis infection [63–66]. Additional studies indicate that cell wall β-glucans and chitins participate in important host inflammatory responses. Pneumocystis β-glucans have been directly attributed as potent stimulators of alveolar epithelial, macrophage and dendritic cell inflammatory responses [4, 67]. Relevant mechanisms such as dectin-1 receptor binding [34, 68], NF-κb activation [31, 33], lactosylceramide-mediated mechanisms [32], and fas–fas ligand binding [30] lead to the generation of TNFα, IL-8, and IFN-γ-inducible protein 10, and chemokine production following β-glucan challenge of lung cells [35, 36].
The relatively large quantities of β-glucan carbohydrates contained within the Pneumocystis cyst wall preparations provided our initial rationale for characterizing β-glucan-induced host immune responses. However, the current studies clearly indicate that Pneumocystis cell wall chitin components may also participate in modulating host responses to the fungal cell wall. Our data demonstrate that digestion of Pneumocystis cell wall with chitinases reduces the net inflammatory activation of lung cells following P. carinii cell wall carbohydrate challenge. It is possible that the chitin moieties themselves in the Pneumocystis cell wall may directly possess pro-inflammatory activity. However, at the present time, this is not directly testable because a method to isolate purified Pneumocystis chitins independent of β-glucans has not yet been developed. It also remains possible that digestion of chitin moieties in our Pneumocystis cell wall carbohydrate isolates disrupts embedding β-glucan molecules and hence releases some of these β-glucan chains, thereby reducing host cell inflammatory cell responses. Until specific chitin receptor blocking reagents or methods to isolate this less prevalent cell wall component are formulated, this cannot be fully resolved. Nonetheless, the data do strongly support that Pneumocystis cell wall chitin digestion does in fact modulate host inflammatory responses to the organisms. Thus, the host generation of chitinase molecules may serve to blunt exuberant host inflammatory responses during this infection.
Our data further indicate that Pneumocystis cell wall components induce the expression of the mammalian chitinases, Chit1 and AMCase, in specific lung cell types. Recently, a family of mammalian chitinases and chitinase-like proteins, known as the 18-glycosyl hydrolase family, have been implicated both in host defenses against chitin-containing pathogens and in allergic inflammation of the lung [37–39, 69]. Two of these enzymes, chitotriosidase (Chit1) and acidic mammalian chitinase (AMCase), have been demonstrated to possess true chitin hydrolase activity [70–72]. Mammals and other higher organisms lack endogenous cell wall glucan and chitin, and therefore, these hydrolytic enzymes are viewed as a form of innate immune defense to limit fungal growth in the host. However, in addition, recent studies have demonstrated that AMCase has a role in Th2-specific IL-13-mediated inflammation in inflammatory and allergic airways diseases such as asthma [39]. While host immune responses during fungal infections have been largely focused on Th1 cell-mediated immunity, the current studies of Chit1 and AM-Case expression may also suggest that Th2 pathways are, in part, activated during Pneumocystis lung infection.
In the whole-animal infection studies, we observed an increase in the AMCase and decrease in the Chit1 mRNA levels of the Pc-infected lungs indicative of a differential regulation of the two chitinases. However, in the lung cell culture studies using RAW macrophages and RLE lung epithelial cells, we noted increased expression of both the AMCase and Chit1 chitinases. The reasons for this disparity are not yet fully understood. However, we postulate that in the whole-animal infection model, the inflammatory milieu may well alter overall chitinase expression. It is also possible that other cell types beyond macrophages and epithelial cells may also participate in the expression of chitinases in vivo during Pneumocystis pneumonia. Such conditions are, unfortunately, impossible to fully investigate in vitro at this time.
It should also be noted that previous studies by Icenhour et al. have shown that >98% of all rats from commercial vendor can be demonstrated by PCR to have low-level colonization when obtained from the vendor [73]. Furthermore, this colonization occurs within hours of birth [74]. To our knowledge, all commercial rats that are available generally have some low level of Pc colonization. Thus, the basal levels of chitinases expressed in rats reflect this native low-level colonization. Unfortunately, it is impossible to have a ready source of non-latently colonized rats for comparison.
Despite the availability of various anti-Pneumocystis agents including trimethoprim-sulfamethoxazole, pentamidine, clindamycin-primaquine, and atovaquone, recent concerns support the need to develop additional agents [75–77]. While these agents are effective in reducing pathogen burden, drug-related toxicities limit use in some patients. Further, reports of mutations in Pneumocystis dihydropteroate synthase, dihydrofolate reductase, and cytochrome B targets of these drugs suggest the emergence of drug-resistant strains [78, 79]. In that light, the fungal cell wall is an attractive target for the future development of effective non-toxic antifungals that are minimally affected by fungal resistance. Echinocandins, compounds that inhibit the glucan synthesis pathway, have been studied in animal models of P. carinii pneumonia, but there are not yet definitive data that these agents are effective in human Pneumocystis pneumonia [80–83]. Because chitin is another major component of the fungal cell wall, understanding the Pneumocystis chitin biosynthesis and remodeling mechanisms may further provide an addition novel therapeutic target for this infection.
Acknowledgments
This work was funded by the Mayo Foundation and NIH grants R01-HL62150 and R01-HL55934 to AHL. LRV was supported by institutional training grant T32-HL07897.
Contributor Information
Leah R. Villegas, Department of Internal Medicine, Thoracic Diseases Research Unit, Division of Pulmonary and Critical Care, Mayo Clinic, 8-24 Stabile Building, Rochester, MN 55905, USA
Theodore J. Kottom, Department of Internal Medicine, Thoracic Diseases Research Unit, Division of Pulmonary and Critical Care, Mayo Clinic, 8-24 Stabile Building, Rochester, MN 55905, USA
Andrew H. Limper, Email: limper.andrew@mayo.edu, Department of Internal Medicine, Thoracic Diseases Research Unit, Division of Pulmonary and Critical Care, Mayo Clinic, 8-24 Stabile Building, Rochester, MN 55905, USA. Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, MN 55905, USA
References
- 1.Morris A, Lundgren JD, Masur H, Walzer PD, Hanson DL, Frederick T, Huang L, Beard CB, Kaplan JE. Current epidemiology of Pneumocystis pneumonia. Emerg Infect Dis. 2004;10(10):1713–1720. doi: 10.3201/eid1010.030985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan JE, Hanson D, Dworkin MS, Frederick T, Bertolli J, Lindegren ML, Holmberg S, Jones JL. Epidemiology of human immunodeficiency virus-associated opportunistic infections in the United States in the era of highly active antiretroviral therapy. Clin Infect Dis. 2000;30(Suppl 1):S5–S14. doi: 10.1086/313843. [DOI] [PubMed] [Google Scholar]
- 3.Catherinot E, Lanternier F, Bougnoux ME, Lecuit M, Couderc LJ, Lortholary O. Pneumocystis jirovecii Pneumonia. Infect Dis Clin North Am. 2010;24(1):107–138. doi: 10.1016/j.idc.2009.10.010. 10.1016/j.idc. 2009.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Limper AH, Lebron F, Evans SE, Hahn RY. Pneumocystis carinii: cell wall beta-glucan-mediated pulmonary inflammation. J Eukaryot Microbiol. 2003;50(Suppl):646. doi: 10.1111/j.1550-7408.2003.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 5.Latge JP. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. 2007;66(2):279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- 6.Kollar R, Petrakova E, Ashwell G, Robbins PW, Cabib E. Architecture of the yeast cell wall. The linkage between chitin and beta(1–>3)-glucan. J Biol Chem. 1995;270(3):1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- 7.Roth A, Wecke J, Karsten V, Janitschke K. Light and electron microscopy study of carbohydrate antigens found in the electronlucent layer of Pneumocystis carinii cysts. Parasitol Res. 1997;83(2):177–184. doi: 10.1007/s004360050229. [DOI] [PubMed] [Google Scholar]
- 8.De Stefano JA, Myers JD, Du Pont D, Foy JM, Theus SA, Walzer PD. Cell wall antigens of Pneumocystis carinii trophozoites and cysts: purification and carbohydrate analysis of these glycoproteins. The Journal of eukaryotic microbiology. 1998;45(3):334–343. doi: 10.1111/j.1550-7408.1998.tb04545.x. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto Y, Matsuda S, Tegoshi T. Yeast glucan in the cyst wall of Pneumocystis carinii. J Protozool. 1989;36(1):21S–22S. doi: 10.1111/j.1550-7408.1989.tb02674.x. [DOI] [PubMed] [Google Scholar]
- 10.De Stefano JA, Cushion MT, Puvanesarajah V, Walzer PD. Analysis of Pneumocystis carinii cyst wall. II. Sugar composition. J Protozool. 1990;37(5):436–441. doi: 10.1111/j.1550-7408.1990.tb01168.x. [DOI] [PubMed] [Google Scholar]
- 11.De Stefano JA, Cushion MT, Sleight RG, Walzer PD. Analysis of Pneumocystis carinii cyst wall. I. Evidence for an outer surface membrane. J Protozool. 1990;37(5):428–435. doi: 10.1111/j.1550-7408.1990.tb01167.x. [DOI] [PubMed] [Google Scholar]
- 12.Linke MJ, Cushion MT, Walzer PD. Properties of the major antigens of rat and human Pneumocystis carinii. Infect Immun. 1989;57(5):1547–1555. doi: 10.1128/iai.57.5.1547-1555.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabib E, Sburlati A, Bowers B, Silverman SJ. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1989;108(5):1665–1672. doi: 10.1083/jcb.108.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas CM. Fungal beta(1,3)-D-glucan synthesis. Med Mycol. 2001;39(Suppl 1):55–66. doi: 10.1080/mmy.39.1.55.66. [DOI] [PubMed] [Google Scholar]
- 15.Hartland RP, Fontaine T, Debeaupuis JP, Simenel C, Delepierre M, Latge JP. A novel beta-(1-3)-glucanosyltransferase from the cell wall of Aspergillus fumigatus. J Biol Chem. 1996;271(43):26843–26849. doi: 10.1074/jbc.271.43.26843. [DOI] [PubMed] [Google Scholar]
- 16.Sorais F, Barreto L, Leal JA, Bernabe M, San-Blas G, Nino-Vega GA. Cell wall glucan synthases and GTPases in Paracoccidioides brasiliensis. Med Mycol. 2010;48(1):35–47. doi: 10.3109/13693780802713356. [DOI] [PubMed] [Google Scholar]
- 17.de Medina-Redondo M, Arnaiz-Pita Y, Fontaine T, Del Rey F, Latge JP, Vazquez de Aldana CR. The beta-1,3-glucano-syltransferase gas4p is essential for ascospore wall maturation and spore viability in Schizosaccharomyces pombe. Mol Microbiol. 2008;68(5):1283–1299. doi: 10.1111/j.1365-2958.2008.06233.x. [DOI] [PubMed] [Google Scholar]
- 18.Cabib E, Blanco N, Grau C, Rodriguez-Pena JM, Arroyo J. Crh1p and Crh2p are required for the cross-linking of chitin to beta(1-6)glucan in the Saccharomyces cerevisiae cell wall. Mol Microbiol. 2007;63(3):921–935. doi: 10.1111/j.1365-2958.2006.05565.x. [DOI] [PubMed] [Google Scholar]
- 19.Sestak S, Hagen I, Tanner W, Strahl S. Scw10p, a cell-wall glucanase/transglucosidase important for cell-wall stability in Saccharomyces cerevisiae. Microbiology. 2004;150(Pt 10):3197–3208. doi: 10.1099/mic.0.27293-0. [DOI] [PubMed] [Google Scholar]
- 20.Pereira M, Felipe MS, Brigido MM, Soares CM, Azevedo MO. Molecular cloning and characterization of a glucan synthase gene from the human pathogenic fungus Paracoccidioides brasiliensis. Yeast. 2000;16(5):451–462. doi: 10.1002/(SICI)1097-0061(20000330)16:5<451::AID-YEA540>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JR, Douglas CM, Li W, Jue CK, Pramanik B, Yuan X, Rude TH, Toffaletti DL, Perfect JR, Kurtz M. A glucan synthase FKS1 homolog in cryptococcus neoformans is single copy and encodes an essential function. J Bacteriol. 1999;181(2):444–453. doi: 10.1128/jb.181.2.444-453.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mio T, Adachi-Shimizu M, Tachibana Y, Tabuchi H, Inoue SB, Yabe T, Yamada-Okabe T, Arisawa M, Watanabe T, Yamada-Okabe H. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis. J Bacteriol. 1997;179(13):4096–4105. doi: 10.1128/jb.179.13.4096-4105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly R, Register E, Hsu MJ, Kurtz M, Nielsen J. Isolation of a gene involved in 1,3-beta-glucan synthesis in Aspergillus nidulans and purification of the corresponding protein. J Bacteriol. 1996;178(15):4381–4391. doi: 10.1128/jb.178.15.4381-4391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shematek EM, Braatz JA, Cabib E. Biosynthesis of the yeast cell wall. I. Preparation and properties of beta-(1 leads to 3)glucan synthetase. J Biol Chem. 1980;255(3):888–894. [PubMed] [Google Scholar]
- 25.Kottom TJ, Limper AH. Cell wall assembly by Pneumocystis carinii. Evidence for a unique gsc-1 subunit mediating beta -1,3-glucan deposition. J Biol Chem. 2000;275(51):40628–40634. doi: 10.1074/jbc.M002103200. [DOI] [PubMed] [Google Scholar]
- 26.Kottom TJ, Limper AH. Pneumocystis carinii cell wall biosynthesis kinase gene CBK1 is an environmentally responsive gene that complements cell wall defects of cbk-deficient yeast. Infect Immun. 2004;72(8):4628–4636. doi: 10.1128/IAI.72.8.4628-4636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vohra PK, Kottom TJ, Limper AH, Thomas CF., Jr Pneumocystis carinii BCK1 complements the Saccharomyces cerevisiae cell wall integrity pathway. J Eukaryot Microbiol. 2003;50(Suppl):676–677. doi: 10.1111/j.1550-7408.2003.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 28.Walker AN, Garner RE, Horst MN. Immunocytochemical detection of chitin in Pneumocystis carinii. Infect Immun. 1990;58(2):412–415. doi: 10.1128/iai.58.2.412-415.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garner RE, Walker AN, Horst MN. Morphologic and biochemical studies of chitin expression in Pneumocystis carinii. J Protozool. 1991;38(6):12S–14S. [PubMed] [Google Scholar]
- 30.Carmona EM, Vassallo R, Vuk-Pavlovic Z, Standing JE, Kottom TJ, Limper AH. Pneumocystis cell wall beta-glucans induce dendritic cell costimulatory molecule expression and inflammatory activation through a Fas–Fas ligand mechanism. J Immunol. 2006;177(1):459–467. doi: 10.4049/jimmunol.177.1.459. [DOI] [PubMed] [Google Scholar]
- 31.Evans SE, Hahn PY, McCann F, Kottom TJ, Pavlovic ZV, Limper AH. Pneumocystis cell wall beta-glucans stimulate alveolar epithelial cell chemokine generation through nuclear factor-kappaB-dependent mechanisms. Am J Respir Cell Mol Biol. 2005;32(6):490–497. doi: 10.1165/rcmb.2004-0300OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahn PY, Evans SE, Kottom TJ, Standing JE, Pagano RE, Limper AH. Pneumocystis carinii cell wall beta-glucan induces release of macrophage inflammatory protein-2 from alveolar epithelial cells via a lactosylceramide-mediated mechanism. J Biol Chem. 2003;278(3):2043–2050. doi: 10.1074/jbc.M209715200. [DOI] [PubMed] [Google Scholar]
- 33.Lebron F, Vassallo R, Puri V, Limper AH. Pneumocystis carinii cell wall beta-glucans initiate macrophage inflammatory responses through NF-kappaB activation. J Biol Chem. 2003;278(27):25001–25008. doi: 10.1074/jbc.M301426200. [DOI] [PubMed] [Google Scholar]
- 34.Steele C, Marrero L, Swain S, Harmsen AG, Zheng M, Brown GD, Gordon S, Shellito JE, Kolls JK. Alveolar macrophage-mediated killing of Pneumocystis carinii f. sp. muris involves molecular recognition by the Dectin-1 beta-glucan receptor. J Exp Med. 2003;198(11):1677–1688. doi: 10.1084/jem.20030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassallo R, Standing J, Limper AH. Beta-glucan from Pneumocystis carinii stimulates TNF alpha release from alveolar macrophages. J Eukaryot Microbiol. 1999;46(5):145S. [PubMed] [Google Scholar]
- 36.Vassallo R, Standing JE, Limper AH. Isolated Pneumocystis carinii cell wall glucan provokes lower respiratory tract inflammatory responses. J Immunol. 2000;164(7):3755–3763. doi: 10.4049/jimmunol.164.7.3755. [DOI] [PubMed] [Google Scholar]
- 37.Elias JA, Homer RJ, Hamid Q, Lee CG. Chitinases and chitinase-like proteins in T(H)2 inflammation and asthma. J Allergy Clin Immunol. 2005;116(3):497–500. doi: 10.1016/j.jaci.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 38.Vicencio AG, Narain S, Du Z, Zeng WY, Ritch J, Casadevall A, Goldman DL. Pulmonary cryptococcosis induces chitinase in the rat. Respir Res. 2008;9:40. doi: 10.1186/1465-9921-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304(5677):1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 40.Morris A, Sciurba FC, Lebedeva IP, Githaiga A, Elliott WM, Hogg JC, Huang L, Norris KA. Association of chronic obstructive pulmonary disease severity and Pneumocystis colonization. Am J Respir Crit Care Med. 2004;170(4):408–413. doi: 10.1164/rccm.200401-094OC. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:W433–W437. doi: 10.1093/nar/gkm352. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos B, Duran A, Valdivieso MH. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17(5):2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ram AF, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protoc. 2006;1(5):2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- 46.Saijo S, Fujikado N, Furuta T, Chung SH, Kotaki H, Seki K, Sudo K, Akira S, Adachi Y, Ohno N, Kinjo T, Nakamura K, Kawakami K, Iwakura Y. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat Immunol. 2007;8(1):39–46. doi: 10.1038/ni1425. [DOI] [PubMed] [Google Scholar]
- 47.Hahn PY, Limper AH. The role of inflammation in respiratory impairment during Pneumocystis carinii pneumonia. Semin Respir Infect. 2003;18(1):40–47. doi: 10.1053/srin.2003.50004. [DOI] [PubMed] [Google Scholar]
- 48.Hartl D, He CH, Koller B, Da Silva CA, Kobayashi Y, Lee CG, Flavell RA, Elias JA. Acidic mammalian chitinase regulates epithelial cell apoptosis via a chitinolytic-independent mechanism. J Immunol. 2009;182(8):5098–5106. doi: 10.4049/jimmunol.0803446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Homer RJ, Zhu Z, Cohn L, Lee CG, White WI, Chen S, Elias JA. Differential expression of chitinases identify subsets of murine airway epithelial cells in allergic inflammation. Am J Physiol. 2006;291(3):L502–L511. doi: 10.1152/ajplung.00364.2005. [DOI] [PubMed] [Google Scholar]
- 50.Monnet X, Vidal-Petiot E, Osman D, Hamzaoui O, Durrbach A, Goujard C, Miceli C, Bouree P, Richard C. Critical care management and outcome of severe Pneumocystis pneumonia in patients with and without HIV infection. Crit Care. 2008;12(1):R28. doi: 10.1186/cc6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vargas SL, Hughes WT, Santolaya ME, Ulloa AV, Ponce CA, Cabrera CE, Cumsille F, Gigliotti F. Search for primary infection by Pneumocystis carinii in a cohort of normal, healthy infants. Clin Infect Dis. 2001;32(6):855–861. doi: 10.1086/319340. [DOI] [PubMed] [Google Scholar]
- 52.Wakefield AE. Detection of DNA sequences identical to Pneumocystis carinii in samples of ambient air. J Eukaryot Microbiol. 1994;41(5):116S. [PubMed] [Google Scholar]
- 53.Casanova-Cardiel L, Leibowitz MJ. Presence of Pneumocystis carinii DNA in pond water. J Eukaryot Microbiol. 1997;44(6):28S. doi: 10.1111/j.1550-7408.1997.tb05752.x. [DOI] [PubMed] [Google Scholar]
- 54.Thomas CF, Jr, Limper AH. Current insights into the biology and pathogenesis of Pneumocystis pneumonia. Nat Rev Microbiol. 2007;5(4):298–308. doi: 10.1038/nrmicro1621. [DOI] [PubMed] [Google Scholar]
- 55.Thomas CF, Jr, Vohra PK, Park JG, Puri V, Limper AH, Kottom TJ. Pneumocystis carinii BCK1 functions in a mitogen-activated protein kinase cascade regulating fungal cell-wall assembly. FEBS Lett. 2003;548(1–3):59–68. doi: 10.1016/s0014-5793(03)00730-0. [DOI] [PubMed] [Google Scholar]
- 56.Kottom TJ, Thomas CF, Jr, Limper AH. Characterization of Pneumocystis carinii PHR1, a pH-regulated gene important for cell wall Integrity. J Bacteriol. 2001;183(23):6740–6745. doi: 10.1128/JB.183.23.6740-6745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villegas LR, Kottom TJ, Limper AH. Characterization of PCEng2 a {beta}-1,3-Endoglucanase Homologue in Pneumocystis carinii with Activity in Cell Wall Regulation. Am J Respir Cell Mol Biol. 2009 doi: 10.1165/rcmb.2009-0131OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merzendorfer H. The cellular basis of chitin synthesis in fungi and insects: common principles and differences. Eur J Cell Biol. 2011;90(9):759–769. doi: 10.1016/j.ejcb.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 59.Lundgren B, Lipschik GY, Kovacs JA. Purification and characterization of a major human Pneumocystis carinii surface antigen. J Clin Investig. 1991;87(1):163–170. doi: 10.1172/JCI114966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keely SP, Stringer JR. Complexity of the MSG gene family of Pneumocystis carinii. BMC genomics. 2009;10:367. doi: 10.1186/1471-2164-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walzer PD, Djawe K, Levin L, Daly KR, Koch J, Kingsley L, Witt M, Golub ET, Bream JH, Taiwo B, Morris A. Long-term serologic responses to the Pneumocystis jirovecii major surface glycoprotein in HIV-positive individuals with and without P. jirovecii infection. J Infect Dis. 2009;199(9):1335–1344. doi: 10.1086/597803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Limper AH. Alveolar macrophage and glycoprotein responses to Pneumocystis carinii. Semin Respir Infect. 1998;13(4):339–347. [PubMed] [Google Scholar]
- 63.Tanabe K, Takasaki S, Watanabe J, Kobata A, Egawa K, Nakamura Y. Glycoproteins composed of major surface immunodeterminants of Pneumocystis carinii. Infect Immun. 1989;57(5):1363–1368. doi: 10.1128/iai.57.5.1363-1368.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Angus CW, Tu A, Vogel P, Qin M, Kovacs JA. Expression of variants of the major surface glycoprotein of Pneumocystis carinii. J Exp Med. 1996;183(3):1229–1234. doi: 10.1084/jem.183.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lasbury ME, Angus CW, Klivington D, Durant PJ, Bartlett MS, Smith JW, Lee CH. Recombinant major surface glycoprotein of Pneumocystis carinii elicits a specific immune response but is not protective in immuno suppressed rats. J Eukaryot Microbiol. 1999;46(5):136S–137S. [PubMed] [Google Scholar]
- 66.Kutty G, Maldarelli F, Achaz G, Kovacs JA. Variation in the major surface glycoprotein genes in Pneumocystis jirovecii. J Infect Dis. 2008;198(5):741–749. doi: 10.1086/590433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vassallo R, Thomas CF, Jr, Vuk-Pavlovic Z, Limper AH. Alveolar macrophage interactions with Pneumocystis carinii. J Lab Clin Med. 1999;133(6):535–540. doi: 10.1016/s0022-2143(99)90182-8. [DOI] [PubMed] [Google Scholar]
- 68.Rapaka RR, Goetzman ES, Zheng M, Vockley J, McKinley L, Kolls JK, Steele C. Enhanced defense against Pneumocystis carinii mediated by a novel dectin-1 receptor Fc fusion protein. J Immunol. 2007;178(6):3702–3712. doi: 10.4049/jimmunol.178.6.3702. [DOI] [PubMed] [Google Scholar]
- 69.Lee CG, Da Silva CA, Lee JY, Hartl D, Elias JA. Chitin regulation of immune responses: an old molecule with new roles. Curr Opin Immunol. 2008;20(6):684–689. doi: 10.1016/j.coi.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Renkema GH, Boot RG, Muijsers AO, Donker-Koopman WE, Aerts JM. Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins. J Biol Chem. 1995;270(5):2198–2202. doi: 10.1074/jbc.270.5.2198. [DOI] [PubMed] [Google Scholar]
- 71.Boot RG, Blommaart EF, Swart E, Ghauharalivan der Vlugt K, Bijl N, Moe C, Place A, Aerts JM. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J Biol Chem. 2001;276(9):6770–6778. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 72.Zheng T, Rabach M, Chen NY, Rabach L, Hu X, Elias JA, Zhu Z. Molecular cloning and functional characterization of mouse chitotriosidase. Gene. 2005;357(1):37–46. doi: 10.1016/j.gene.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Icenhour CR, Rebholz SL, Collins MS, Cushion MT. Widespread occurrence of Pneumocystis carinii in commercial rat colonies detected using targeted PCR and oral swabs. J Clin Microbiol. 2001;39(10):3437–3441. doi: 10.1128/JCM.39.10.3437-3441.2001. 10.1128/JCM.39.10.3437-3441. 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Icenhour CR, Rebholz SL, Collins MS, Cushion MT. Early acquisition of Pneumocystis carinii in neonatal rats as evidenced by PCR and oral swabs. Eukaryot Cell. 2002;1(3):414–419. doi: 10.1128/EC.1.3.414-419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson T, Khan IA, Avery MA, Grant J, Meshnick SR. Quantitative structure-activity relationship studies of a series of sulfa drugs as inhibitors of Pneumocystis carinii dihydropteroate synthetase. Antimicrob Agents Chemother. 1998;42(6):1454–1458. doi: 10.1128/aac.42.6.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaneshiro ES, Sul D, Hazra B. Effects of atovaquone and diospyrin-based drugs on ubiquinone biosynthesis in Pneumocystis carinii organisms. Antimicrob Agents Chemother. 2000;44(1):14–18. doi: 10.1128/aac.44.1.14-18.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Francesconi I, Wilson WD, Tanious FA, Hall JE, Bender BC, Tidwell RR, McCurdy D, Boykin DW. 2,4-Diphenyl furan diamidines as novel anti-Pneumocystis carinii pneumonia agents. J Med Chem. 1999;42(12):2260–2265. doi: 10.1021/jm990071c. [DOI] [PubMed] [Google Scholar]
- 78.Nahimana A, Rabodonirina M, Helweg-Larsen J, Meneau I, Francioli P, Bille J, Hauser PM. Sulfa resistance and di-hydropteroate synthase mutants in recurrent Pneumocystis carinii pneumonia. Emerg Infect Dis. 2003;9(7):864–867. doi: 10.3201/eid0907.020753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kessl JJ, Hill P, Lange BB, Meshnick SR, Meunier B, Trumpower BL. Molecular basis for atovaquone resistance in Pneumocystis jirovecii modeled in the cytochrome bc(1) complex of Saccharomyces cerevisiae. J Biol Chem. 2004;279(4):2817–2824. doi: 10.1074/jbc.M309984200. [DOI] [PubMed] [Google Scholar]
- 80.Schmatz DM, Romancheck MA, Pittarelli LA, Schwartz RE, Fromtling RA, Nollstadt KH, Vanmiddlesworth FL, Wilson KE, Turner MJ. Treatment of Pneumocystis carinii pneumonia with 1,3-beta-glucan synthesis inhibitors. Proc Natl Acad Sci USA. 1990;87(15):5950–5954. doi: 10.1073/pnas.87.15.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurtz MB, Abruzzo G, Flattery A, Bartizal K, Marrinan JA, Li W, Milligan J, Nollstadt K, Douglas CM. Characterization of echinocandin-resistant mutants of Candida albicans: genetic, biochemical, and virulence studies. Infect Immun. 1996;64(8):3244–3251. doi: 10.1128/iai.64.8.3244-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Douglas CM, Marrinan JA, Li W, Kurtz MB. A Saccharomyces cerevisiae mutant with echinocandin-resistant 1,3-beta-D-glucan synthase. J Bacteriol. 1994;176(18):5686–5696. doi: 10.1128/jb.176.18.5686-5696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walker LA, Munro CA, de Bruijn I, Lenardon MD, McKinnon A, Gow NA. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4(4):e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]