Abstract
This study was undertaken to optimize skim milk and yeast extract concentration as a cultivation medium for optimal Bifidobacteria pseudocatenulatum G4 (G4) biomass and β-galactosidase production as well as lactose and free amino nitrogen (FAN) balance after cultivation period. Optimization process in this study involved four steps: screening for significant factors using 23 full factorial design, steepest ascent, optimization using FCCD-RSM, and verification. From screening steps, skim milk and yeast extract showed significant influence on the biomass production and, based on the steepest ascent step, middle points of skim milk (6% wt/vol) and yeast extract (1.89% wt/vol) were obtained. A polynomial regression model in FCCD-RSM revealed that both factors were found significant and the strongest influence was given by skim milk concentration. Optimum concentrations of skim milk and yeast extract for maximum biomass G4 and β-galactosidase production meanwhile low in lactose and FAN balance after cultivation period were 5.89% (wt/vol) and 2.31% (wt/vol), respectively. The validation experiments showed that the predicted and experimental values are not significantly different, indicating that the FCCD-RSM model developed is sufficient to describe the cultivation process of G4 using skim-milk-based medium with the addition of yeast extract.
1. Introduction
Probiotic can be defined as “living microorganisms which when administered in adequate amounts confer a health benefit on the host” [1]. The beneficial effects of probiotic include regulation in immune system, anticarcinogenic, preventing the infection by exogenous microorganisms, vitamin synthesis, and enhancing digestion and absorption of nutrients [2].
Bifidobacteria, the major species of intestinal flora in healthy human, have been widely used in probiotics formulation [3]. Bifidobacteria are anaerobic rods, Gram-positive, and nonspore forming bacteria [4]. Bifidobacterium pseudocatenulatum G4, isolated from breast-fed infants feces, has been identified and characterized as a potential probiotic with many beneficial health effects [5–7]. The antimicrobial properties, adherence ability, antagonistic activity, and ability to survive in gut environment for Bifidobacterium pseudocatenulatum G4 have also been reported [8].
Proliferation of bifidobacteria requires appropriate medium which supplies nurturing substances for growth. The use of commercial media to support bacterial growth is limited by its cost [9] and off-flavor generated in food products [10]. Milk is a highly nutritious growth medium for many microorganisms as it is rich in carbohydrates, fat, casein, protein, vitamins, and minerals [11]. However, some microorganisms are unable to grow well in this medium when specific enzymes required for lactose metabolization process are lacking [12]. β-Galactosidase is an enzyme required to break lactose to galactose and glucose. Most of the bifidobacteria preferred these carbon sources to promote their growth. The production of β-galactosidase can be enhanced using yeast extract as a nitrogen source [13]. Various concentrations of yeast extract (ranging from 0.3% to 1.0% wt/vol) were added into milk (ranging from 10% to 12% wt/vol) to enhance growth of probiotics [14, 15]. Various concentrations of glucose (ranging from 0.5% to 2.0% wt/vol) have also been used as a carbon source in the cultivation of probiotic like bifidobacteria [16].
In optimization of milk-based-medium for bifidobacteria growth, it is important to consider β-galactosidase production by the cells in order to facilitate lactose breakdown into more readily available sugar. In addition, the concomitant use of nitrogen sources is also important to avoid nutrients wastage. Most of the previous studies on the medium optimization for bifidobacteria focused mainly on the maximum biomass production [8, 17]. To the best of our knowledge, justification involving other responses of growth activities of bifidobacteria in milk medium in order to ensure that the medium has been used competently is not available in the literature.
The optimization of medium concentration or components by conventional approach involves large numbers of experiments which are time consuming and expensive and lack in representing the effects of individual factors. Thus, a statistical approach known as response surface methodology (RSM) has been applied as a tool for process improvement. The main advantage of RSM is the reduced number of experiments or trials required to evaluate multiple parameters and their interactions [18, 19]. The most commonly design approached is the central composite design. The design allows estimation of all the regression parameters required to fit a second-order model of given responses. Rotability character is the most preferred in any central composite design. This is because this characteristic provides constant variance of the estimated response corresponding to all new observation points that are at the same distance from the center point of the design (in terms of the coded variable).
Therefore, the main objective of this study was to optimize milk-based medium for the improvement of B. pseudocatenulatum G4 cultivation. In this optimization process, there were four responses; namely, (i) cells production, (ii) accumulation of lactose degrading enzyme (β-galactosidase), and (iii) residual concentration of lactose and free amino nitrogen remaining in the culture at the end of cultivation, were considered.
2. Materials and Methods
2.1. Bacterium and Inoculum Preparation
Probiotic bacterium, B. pseudocatenulatum G4 (G4), was used throughout this study and this bacterium was obtained from the Probiotic Laboratory, Faculty of Food Science and Technology, Universiti Putra Malaysia. G4 was previously isolated from breast-fed infant feces [5, 6] and was stored at −20°C in a mixture of glycerol and Trypticase Phytone Yeast (TPY) extract broth (Scharlau-Chemie, Barcelona, Spain), at a ratio of 80 : 20. TPY medium (Scharlau-Chemie, Barcelona, Spain) was used to maintain and to propagate the bacterium [4]. A single colony of the bacterium was transferred from TPY agar to TPY broth and incubated anaerobically at 37°C for 24 h. After two successive transfers in TPY broth at 37°C for 24 h under anaerobic condition using Anaerocult A gas packs (Merck, Darmstadt, Germany), the activated culture was then properly diluted and served as the standard inoculum for all cultivations.
2.2. Medium Preparation
Cultivation was carried out in 250 mL SCHOTT DURAN bottle (Schott Duran, Mainz, Germany). The medium components consist of skim milk (NZMP medium heat skim milk powder, Auckland, New Zealand), yeast extract (Bio Springer, Maisons-Alfort Cedex, France), and glucose (Merck, Darmstadt, Germany). All components were prepared in separate bottle and sterilized at 121°C for 15 min. Once cooled at room temperature, the components were mixed prior to inoculation with 104 cfu/mL of G4. The bottle was then incubated at 37°C for 21 h in an anaerobic condition. The culture pH was not controlled in all experiments. During the cultivation, 10 mL of samples was taken at 3 h intervals for analysis.
2.3. Lactose Determination
The amount of lactose in the samples was determined using HPLC method as described by Hou et al. with some modifications [20]. One mL of sample was centrifuged at 13000 rpm for 10 min. The clear fraction was filtered through a 0.2 μm nylon membrane filter and injected onto HPLC system (Alliance 2690/5: Waters Corporation, Milford, CA), equipped with a 4.6 mm × 150 mm Agilent Zorbax Carbohydrate Analysis column (Agilent Technologies Inc., USA). The mobile phase used is 75% (v/v) acetonitrile (Fisher, HPLC grade). The flow rate was set at 1.4 mL min−1 and the analysis was carried out at 30°C using a refractive index detector (RI-1371, Waters Corporation, Milford, CA).
2.4. Free Amino Nitrogen Analyses
Free amino nitrogen (FAN) presence in the sample was determined using ninhydrin analysis described by Hwang and Ederer with some modifications [21]. A sample (1 mL) was centrifuged at 13000 rpm for 10 min. The clear fraction was collected and diluted into 50 mL distilled water and 2 mL of the diluted sample was then transferred into test tubes. Ninhydrin solution (0.35 g ninhydrin into 100 mL of a 1 : 1 mixture of acetone and butanol) was then added and the tubes were covered with a piece of parafilm to avoid the loss of solvent due to evaporation. The tubes were then heated in a boiling water bath (80°C–100°C) for 15 min before being transferred to a cold water bath. The dilution reagent (5 mL) was then added and mixed and the absorbance was read at 570 nm using spectrophotometer (UV-Vis 1601 spectrophotometer, Shimadzu, Japan). Distilled water was used as a blank and glycine (Merck, Darmstadt, Germany) was used as a standard.
2.5. β-Galactosidase Analyses
β-Galactosidase was assayed on nonpermeabilized and permeabilized cells. The culture was centrifuged at 11000 rpm for 10 min at 4°C. Subsequently, the resulting supernatant was discarded by aspiration. The cell pellet was washed twice with Z buffer (0.1 M phosphate—pH 7; 10 mM MgSO4·7H2O; 1 mM CaCl2) and concentrated by 10-fold. Permeabilization was accomplished by the addition of 0.5 mL of Triton X-100 (5% vol/vol in Z buffer) into 0.5 mL cell suspension with mixing using vortex mixer. The mixture was incubated at 37°C for 10 min and then centrifuged at 14000 rpm for 15 min and the resulting supernatant served as the enzyme source. The activity of β-galactosidase was then assayed essentially according to the method described by Nagy et al. (2001) [22]. The reaction mixture was composed of 0.5 mL of enzyme sample and 0.5 mL of 15 mM o-nitrophenyl β-d-galactopyranoside (Sigma Chemical Co., St. Louis, MO) in 0.03 M sodium phosphate buffer (pH 6.8). Once a faint yellow tint appeared, the reaction was terminated by adding 0.1 M sodium carbonate. The absorbance was measured using a spectrophotometer at 420 nm wavelength (UV-Vis 1601 spectrophotometer, Shimadzu, Japan). One unit of β-galactosidase activity was defined as the amount of enzyme catalyzing the formation of 1 μmol of o-nitrophenyl per min under the assay condition.
2.6. Microbiological Analyses
For viable cells enumeration, samples were serially diluted using 0.1% (wt/vol) sterile peptone water (Merck, Darmstadt, Germany) and plated in duplicate onto TPY agar. The plates were incubated in anaerobic jars containing Anaerocult A (Merck, Darmstadt, Germany) at 37°C for 48 h. All plates with 30 to 300 colonies were counted and the viability was expressed as log10 cfu/mL.
2.7. Experimental Design and Statistical Analyses
Design-Expert software (Stat-Ease Inc., Minneapolis, MN, USA, version 6.0.6) was used to design the experiment in this study. First, initial screening experiments were performed to evaluate the significant effects of skim milk, yeast extract, and glucose on biomass production using a 23 full factorial design. Each factor was coded at three levels between −1 and +1, with 5 times replication of center point (0), where the variables of factors were changed in the ranges. The factorial design resulting in 26 experimental runs (including duplicates and 5 middle points run). In the second step, a first-order empirical equation was applied to exclude insignificant factors and to generate the steepest ascent path to facilitate the maximum increase in response. The steepest ascent design was based on results of the equation from the screening step of which only factors with significant effects on response were used and fixed at their zero coded value. The insignificant factors were eliminated from the design. The path begins at the centre point of design and stretches out linearly in order to determine the suitable ranges within significant factors which tend to approach the optimal condition. The third step involved further optimization of significant factors using face-centered central composite design (FCCD). FCCD is an effective design that is used for sequential experimentation and provides reasonable amount of information for testing the goodness of fit and does not require large number of design points thereby reducing the overall cost associated with the experiment [23]. In this design, four responses, namely, biomass (y 1, log10 cfu/mL) and β-galactosidase production, (y 2, U/mL) as well as residual lactose (y 3, g/L) and FAN (y 4, mg/L) remaining in the culture were determined. There were three coded factor levels, −1, 0, and +1 (where −1 corresponded to the low level of each factor, 0 to the middle level, and +1 to the high level). The coded values were determined using
| (1) |
The center point was repeated five times in order to evaluate the curvature and the experiment replication facilitated the pure error estimation, so that the significant lack of fit of the models could be predicted. All the 26 experiments were carried out in duplicate for 21 h.
3. Results and Discussion
3.1. Initial Screening of Significant Medium Components and the Steepest Ascent
The effect of skim milk (x 1), yeast extract (x 2), and glucose (x 3) on biomass production (y 1) during the cultivation of G4 using 23 full factorial design is shown in Table 1. The maximum biomass (ranging from 4.495 to 7.638 log10 cfu/mL) was obtained at 21 h of cultivation. The analysis of variance (ANOVA) of the first-order model is shown in Table 2 while the regression analysis of the model is shown in Table 3. The model was significant and only 1.12% of the total variation was observed due to noise. The model was linear with insignificant (P > 0.05) curvature. The regression analysis of the variables showed that skim milk (P = 0.007) was the only significant factor while yeast extract and glucose were insignificant with P values higher than 0.05 (Table 3). Since yeast extract (x 2) was the sole nitrogen, it must be considered in the medium optimization. Improvement of growth of bifidobacteria could be achieved using yeast extract at concentration higher than 1.0% without the addition of glucose [17]. As a result, final-order equation (coded term) was generated based on the first-order model to determine the biomass production response (y 1) to the medium composition containing skim milk (x 1) and yeast extract (x 2) factors, which give
| (2) |
For every unit increase in x 1, an increase of 1.09 units in y 1 is predicted (2). On the other hand, for every unit increase in x 2, 0.30 units of increase in y 1is expected. Between these two factors, skim milk (x 1) gave more effect than yeast extract (x 2) on biomass production (y 1). This equation was further used as the fundamental scale in the subsequent step which was the steepest ascent. The path of the steepest ascent was determined based on the increase in 0.10% (wt/vol) of x 1 and the movement was generated along the path until no improvement occurred. Five design units were developed based on 0.10/0.10 = 1. Hence, the movement of x 2 was 0.28 design units (0.3/1.09 = 0.28). As for natural factor, x 1 movement was based on coded factor × lower point of x 1 (1 × 2 = 2). Thus, movement of x 2 natural factor involved 0.28 × 0.5 = 0.14. The path coordination of the steepest ascent was generated and is shown in Table 4. The highest biomass production can be observed in the third step of the steepest ascent coordinates with the value of 7.68 ± 0.17 log10 cfu/mL from the combination of skim milk (6% (wt/vol)) and yeast extract (1.89% (wt/vol)). After the third step of coordinates, declining in biomass production was observed. Consequently, this combination was selected as the middle point for further optimization.
Table 1.
The 23 full factorial design and responses for screening experiments.
| Run | Factors | Response | ||
|---|---|---|---|---|
| Skim milk (% wt/vol) | Yeast extract (% wt/vol) | Glucose (% wt/vol) | Maximum biomass (log10 CFU per milliliter) | |
| 1 | 2 (−1) | 0.5 (−1) | 0.5 (−1) | 4.495 |
| 2 | 2 (−1) | 0.5 (−1) | 1.5 (+1) | 4.789 |
| 3 | 2 (−1) | 3 (+1) | 0.5 (−1) | 5.335 |
| 4 | 2 (−1) | 3 (+1) | 1.5 (+1) | 5.800 |
| 5 | 4 (0) | 1.75 (0) | 1 (0) | 6.417 |
| 6 | 4 (0) | 1.75 (0) | 1 (0) | 6.671 |
| 7 | 4 (0) | 1.75 (0) | 1 (0) | 6.075 |
| 8 | 4 (0) | 1.75 (0) | 1 (0) | 6.824 |
| 9 | 4 (0) | 1.75 (0) | 1 (0) | 6.874 |
| 10 | 6 (+1) | 0.5 (−1) | 0.5 (−1) | 7.158 |
| 11 | 6 (+1) | 0.5 (−1) | 1.5 (+1) | 7.173 |
| 12 | 6 (+1) | 3 (+1) | 0.5 (−1) | 7.204 |
| 13 | 6 (+1) | 3 (+1) | 1.5 (+1) | 7.638 |
Table 2.
ANOVA results of the first-order model for 23 full factorial design.
| Source of variation or factors | DFa | Sum of squares | Mean square | F or t value | P value |
|---|---|---|---|---|---|
| Modelb | 7 | 10.75 | 1.54 | 14.09 | 0.0112 |
| Curvature | 7 | 0.43 | 0.43 | 0.071 | 0.1182 |
| Pure error | 4 | 0.44 | 0.11 | 3.94 | |
| Correlation total | 12 | 11.61 |
aDF: degree of freedom.
b R 2: 96.10%.
Significant at α = 0.05.
Table 3.
Regression analysis of the 23 full factorial design with maximum biomass (log10 cfu/mL) as the response.
| Variable | F Value | P value |
|---|---|---|
| x 1 a | 87.94 | 0.0007 |
| x 2 | 6.40 | 0.0647 |
| x 3 | 1.67 | 0.2658 |
| x 1 x 2 | 2.06 | 0.2246 |
| x 1 x 3 | 0.11 | 0.7572 |
| x 2 x 3 | 0.40 | 0.5623 |
| x 1 x 2 x 3 | 0.07 | 0.8034 |
| Model | 14.09 | 0.0112 |
| Curvature | 3.94 | 0.1182 |
| C.V = 2.78% | R 2 = 0.96 | Adjusted R 2 = 0.90 |
a x 1 represents skim milk concentration in % (wt/vol).
x 2 represents yeast extract concentration % (wt/vol).
x 3 represents glucose concentration % (wt/vol).
Table 4.
The steepest ascent coordination path for all chosen factors at coded and natural levels.
| Step | Coded factora | Natural factorb (% wt/vol) | Maximum biomass (log10 cfu/mL)e | ||
|---|---|---|---|---|---|
| ε 1 | ε 2 | x 1 | x 2 | ||
| Basec | 0 | 0 | 4 | 1.75 | 5.77 ± 0.24 |
| Δd | 1 | 0.28 | 2 | 0.14 | 4.05 ± 0.09 |
| Base + Δ | 1 | 0.28 | 6 | 1.89 | 7.68 ± 0.17 |
| Base + 2Δ | 2 | 0.56 | 8 | 2.03 | 7.66 ± 0.68 |
| Base + 3Δ | 3 | 0.84 | 10 | 2.17 | 7.65 ± 0.26 |
| Base + 4Δ | 4 | 1.12 | 12 | 2.31 | 7.63 ± 0.66 |
| Base + 5Δ | 5 | 1.40 | 14 | 2.45 | 7.61 ± 0.04 |
aCoded factor ε1: skim milk, ε2: yeast extract.
bNatural factor x1: skim milk, x2: yeast extract.
cBased: 0 for coded factor and middle point for natural factor.
dMovement units (coded factor ε1: 0.1/0.1, ε2: 0.3/1.09) (natural factor: coded factor x lower point).
eMaximum biomass of B. pseudocatenulatum G4 (log10 cfu/mL) at 21 h of cultivation period. Values are mean of triplicates.
3.2. Optimization of Medium Component
The experimental responses for the optimization of skim milk and yeast extract are shown in Table 5. Skim milk concentration (x 1) was in the range of 4% (wt/vol) to 8% (wt/vol) with 6% (wt/vol) as a center point whereas, for yeast extract (x 2), the range of 1% (wt/vol) to 2.8% (wt/vol) with 1.9% (wt/vol) as a center point was fixed further optimization. Center points with a coded (0,0) were repeated five times. The importance of medium components for cultivation process can be considered by their effect on biomass production (y 1) and β-galactosidase production (y 2) as well as the residual lactose (y 3) and free amino nitrogen (y 4) remaining in the culture. The responses (y 1, y 2, y 3, and y 4) were fitted with quadratic polynomial model and subsequently produced the response surface as expressed in (3). Consider
| (3) |
where x 1 and x 2 represent coded independent factors of skim milk and yeast extract, respectively. Meanwhile, β 0, β 1, β 2, β 11, and β 22 are coefficients and ε i is the random error.
Table 5.
Experimental design and results using face-centered full factorial design (FCCD).
| Run | Skim milk, x 1 | Yeast extract, x 2 | Responsesa | |||||
|---|---|---|---|---|---|---|---|---|
| Coded value | Actual value | Coded value | Actual value | Max. biomass, y 1 (log10 cfu/mL) | β-Galactosidase production y 2 (U/mL) | Lactose balance y 3 (g/L) | FAN balance y 4 (mg/L) | |
| 1 | −1 | 4.0 | −1 | 1.0 | 5.15 | 0.19 | 4.59 | 198.24 |
| 2 | −1 | 4.0 | 0 | 1.9 | 5.18 | 3.03 | 4.51 | 209.58 |
| 3 | −1 | 4.0 | +1 | 2.8 | 5.22 | 3.13 | 4.40 | 213.24 |
| 4 | 0 | 6.0 | −1 | 1.0 | 7.09 | 7.06 | 10.83 | 229.15 |
| 5 | 0 | 6.0 | 0 | 1.9 | 7.15 | 8.37 | 11.53 | 237.06 |
| 6 | 0 | 6.0 | 0 | 1.9 | 7.11 | 9.40 | 11.58 | 231.04 |
| 7 | 0 | 6.0 | 0 | 1.9 | 7.12 | 9.32 | 11.81 | 234.65 |
| 8 | 0 | 6.0 | 0 | 1.9 | 7.14 | 8.35 | 11.91 | 238.98 |
| 9 | 0 | 6.0 | 0 | 1.9 | 7.16 | 9.38 | 11.84 | 233.14 |
| 10 | 0 | 6.0 | +1 | 2.8 | 7.13 | 8.41 | 12.90 | 359.76 |
| 11 | +1 | 8.0 | −1 | 1.0 | 7.26 | 7.02 | 22.41 | 287.53 |
| 12 | +1 | 8.0 | 0 | 1.9 | 7.23 | 6.27 | 19.10 | 290.59 |
| 13 | +1 | 8.0 | +1 | 2.8 | 7.11 | 6.18 | 25.25 | 318.70 |
aAll factorial and axial points are means of duplicate.
3.2.1. Optimization of Biomass Production (y 1)
By applying regression analysis on the experimental data, biomass production can be described by the second-order equation
| (4) |
Skim milk (x 1, x 1 2) was shown to be highly significant (P < 0.001) whereas yeast extract (x 2, x 2 2) presented less influence towards biomass production. As a result, the quadratic model was further reduced with insignificant model term excluded. However, the addition of yeast extract (x 2), as nitrogen source, in the milk medium must be considered to enhance growth of bifidobacteria [13]. Therefore, yeast extract is included in (4) to (5). Consider
| (5) |
Table 6 shows ANOVA result of quadratic model. Biomass production is significant as indicated by P value (P < 0.001). The R 2 implies the sample variation of 99.93% for biomass production (y 1) and this indicates that only about 0.07% of total variation is not explained by the model, indicating good agreement between the experimental and predicted values for biomass production. The lack of fit measures the failure of the model to represent data in the experimental domain at points, which are not included in the regression [24]. The value of lack of fit for regression is insignificant (P > 0.05), suggesting that the model fitted well to the data in the experimental region. Moreover, second-order terms were found sufficient and higher order terms were not necessary.
Table 6.
ANOVA and regression analysis for the response of biomass production (y 1): optimization of skim milk (% wt/vol) and yeast extract (% wt/vol).
| Source of variation | Sum of squire or coefficient estimate | DFa | MS or Std error | F value | P value |
|---|---|---|---|---|---|
| Modelb | 8.95 | 4 | 2.24 | 2893.35 | <0.001 |
| Residual | 6.186E − 003 | 8 | 7.732E − 004 | ||
| Lack of fit | 4.466E − 003 | 4 | 1.116E − 003 | 2.60 | 0.1890 |
| Pure error | 1.720E − 003 | 4 | 4.300E − 004 | ||
| Total | 8.95 | 12 | |||
| Factorc | |||||
| Intercept | 7.14 | 1 | |||
| x 1 | 1.01 | 1 | 0.011 | 7889.68 | <0.001d |
| x 2 | −6.667E − 003 | 1 | 0.011 | 0.34 | 0.5732 |
| x 1 2 | −0.94 | 1 | 0.015 | 3667.73 | <0.001d |
| x 1 x 2 | −0.055 | 1 | 0.014 | 15.65 | 0.0042d |
aDF: degree of freedom.
b R 2 = 99.93%.
c x 1, skim milk (% wt/vol), x 2, yeast extract (% wt/vol).
dSignificant at α = 0.05.
The response surface model shown in Figure 1 indicated that skim milk concentration (% wt/vol) was the most important factor in the medium. Lactose in milk was able to supply readily utilizable carbon for growth of G4. Increased milk concentration, with small addition of yeast extract as growth enhancer, significantly increased biomass production. Lactose in milk induced β-galactosidase biosynthesis in Bifidobacterium strain, which break down lactose into glucose and galactose more efficiently during the cultivation process [13]. The response surface obtained is a stationary edge system, whereby the maximum value is a plane platform rather than point form. Thus, there is flexibility in choosing the appropriate optimum points [25].
Figure 1.
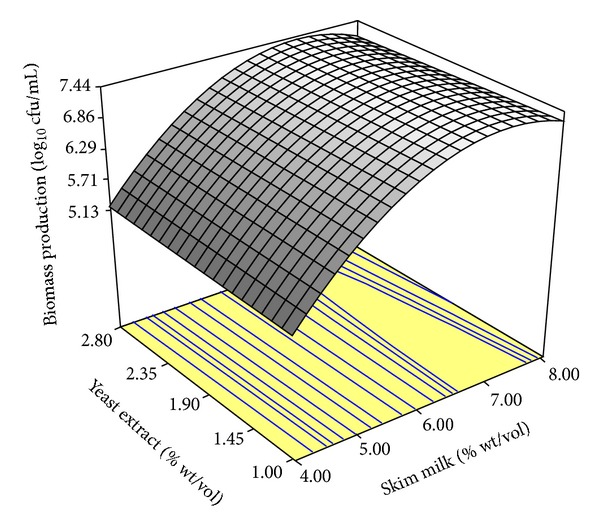
Response surface methodology for biomass production (log10 cfu/mL) from the adjusted quadratic mathematical model.
Based on the optimum point acquired from response surface methodology, the combination of 7.17% (wt/vol) skim milk, x 1, and 1.02% (wt/vol) yeast extract, x 2, was predicted to produce 7.44 log10 cfu/mL biomass. This prediction was verified by a validation experiment. A maximum biomass of 7.35 log10 cfu/mL was obtained from replication experiments. Even though the experimental value was lower than the predicted value, no statistical difference (P > 0.05) was observed. In order to observe other growth activities of G4 in optimized medium, analysis of β-galactosidase production as well as determination of lactose and free amino nitrogen residual in the culture at 21 h of cultivation was performed. As shown in Table 7, the optimized medium (7.17% (wt/vol) skim milk and 1.02% (wt/vol) yeast extract) was able to support G4 growth up to 7 log10 cfu/mL with substantially high β-galactosidase production at 21 h of cultivation. However, high amount of lactose and free amino nitrogen residual was still observed at the end of cultivation. To avoid wastage of nutrients remaining in the medium, further optimization step was carried out by considering residual nutrients remaining in the culture at the end of cultivation. This step was conducted and presented in the following section.
Table 7.
Comparison experiments using optimized skim milk and yeast extract based on biomass production, β-galactosidase production, lactose, and free amino nitrogen residual at 21 h of cultivation period.
| Skim milk (% wt/vol)* | Biomass production (log10 cfu/mL) | β-Galactosidase production (U/mL) | Lactose (g/L) | Free amino nitrogen (mg/L) |
|---|---|---|---|---|
| 0 | 1.03 ± 0.11a∗∗ | 0.35 ± 0.21a | 0.00 ± 0.00a | 141.57 ± 18.90a |
| 4 | 7.19 ± 0.07b | 6.36 ± 0.16b | 7.25 ± 2.02b | 153.80 ± 15.46a |
| 7.17 (optimized concentrations) | 7.35 ± 0.06b | 6.34 ± 0.08b | 15.92 ± 3.23c | 198.73 ± 13.67a |
| 10 | 7.78 ± 0.55b | 5.54 ± 1.02b | 21.39 ± 3.29c | 251.57 ± 18.71b |
| 12 | 7.52 ± 0.63b | 5.23 ± 0.56b | 25.98 ± 3.19c | 282.27 ± 11.13b |
| TPY | 7.79 ± 0.45b | 0.05 ± 0.11a | 0.00 ± 0.00a | 374.20 ± 16.22c |
*Cultivation was conducted in different concentrations of skim milk with 1.02% yeast extract supplementation.
**Values in the same column with different letters were significantly different (P < 0.05).
3.2.2. β-Galactosidase Production (y 2), Lactose (y 3), and Free Amino Nitrogen (y 4) Presence as Responses
Further optimization of G4 medium cultivation was carried out by including β-galactosidase production, lactose, and free amino nitrogen balance at 21 h of cultivation as responses. The responses (y 2, y 3, and y 4) were fitted with the second-order polynomial equations
| (6) |
The statistical significance of the model was evaluated by the P value of the analysis of variance (ANOVA). ANOVA statistics for the three responses, y 2 (β-galactosidase), y 3 (lactose residual), and y 4 (free amino nitrogen residual), at 21 h of the cultivation period are shown in Table 8. Quadratic models for y 2 and y 4 as well as linear model for y 3 are highly significant, as shown by low probability value (P < 0.001). The models fitted well to the experimental design as lack of fit for all three responses is insignificant (P > 0.05). Moreover, the coefficient of determinations (R 2) is close to 1 (y 2: 0.97, y 3: 0.99, and y 4: 0.98), indicating high degree of correlation between observed and predicted values.
Table 8.
ANOVA analysis for responses y 2 (β-galactosidase: U/mL), y 3 (lactose: g/L), and y 4 (FAN: mg/L).
| Source | Sum squares | DF | Mean square | F value | P value |
|---|---|---|---|---|---|
| y 2 | |||||
| Model | 96.50 | 5 | 19.30 | 52.91 | <0.0001 |
| Residual | 2.55 | 7 | 0.36 | ||
| Lack of fit | 1.33 | 3 | 0.44 | 1.46 | 0.3521 |
| Pure error | 1.22 | 4 | 0.30 | ||
| R 2 = 0.9742 | |||||
| y 3 | |||||
| Model | 151.33 | 2 | 75.66 | 615.28 | <0.0001 |
| Residual | 1.23 | 10 | 0.12 | ||
| Lack of fit | 1.05 | 6 | 0.17 | 3.83 | 0.1075 |
| Pure error | 0.18 | 4 | 0.046 | ||
| R 2 = 0.9919 | |||||
| y 4 | |||||
| Model | 14609.65 | 5 | 2921.93 | 123.58 | 0.0001 |
| Residual | 165.51 | 7 | 23.64 | ||
| Lack of fit | 126.17 | 3 | 42.06 | 4.28 | 0.0971 |
| Pure error | 39.34 | 4 | 9.84 | ||
| R 2 = 0.9888 |
The response surface plot of β-galactosidase production (y 2) is shown in Figure 2(a). High production of β-galactosidase was observed at the middle range of skim milk (6.00 to 7.00% (wt/vol)). On the other hand, reduced production of β-galactosidase was detected at high and low point of skim milk concentration. The use of high skim milk concentration (8.00% (wt/vol)) might contribute to the presence of high lactose concentration and this could suppress β-galactosidase production. Medium containing high lactose concentration (>5% [wt/vo]), may attributed to increase in the concentration of internally glucose. It has been reported that the increased of internal glucose would suppressed the biosynthesis of β–galactosidase by tested organisms [26]. The presence of low lactose concentration in the medium using low skim milk concentration (4.00% (wt/vol)) may not induce β-galactosidase production.
Figure 2.
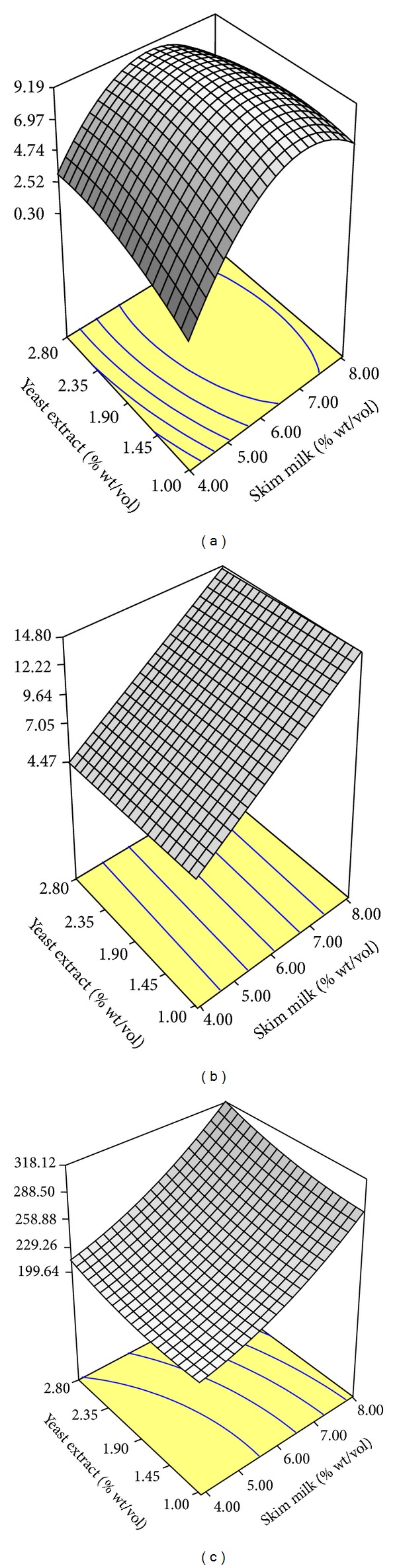
Response surface plot of the effect of skim milk and yeast extract concentration (% wt/vol) for the responses of (a) β-galactosidase production (U/mL), (b) lactose residual (g/L), and (c) free amino nitrogen residual (mg/L) at 21 h of cultivation period.
A linear response surface plot of lactose residual (y 3) is shown in Figure 2(b). At high skim milk concentration (8.00% (wt/vol)) residual lactose concentration at 21 h of cultivation reached a maximum value with minor changes in yeast extract (1% to 2.8% (wt/vol)). In contrast, very low residual glucose concentration was detected at low skim milk concentration (4.00% (wt/vol)). This response indicated that skim milk concentration (4% to 8% (wt/vol)) was closely associated with residual lactose concentration. On the other hand, yeast extract ranging from 1% to 2.8% (wt/vol)) had less significant effect on the response. However, the presence of yeast extract was required to support β-galactosidase production (Figure 2(a)). Minimum residual lactose concentration at 21 h of cultivation was considered in this optimization method to avoid wastage of carbon source. From this point of view, low skim milk concentration is preferred to be used as cultivation medium for G4.
Figure 2(c) shows response surface plot representing free amino nitrogen residual concentration in the culture at 21 h of cultivation. The presence of nitrogen in medium was attributed to yeast extract and skim milk as well as dead cells and was believed to sustain bacterial growth. Similar to lactose, minimum residual concentration of nitrogen remaining in the culture after 21 h was considered in the optimization to avoid wastage of nitrogen source. The response surface generated based on the second-order coefficient shows that skim milk and yeast extract presented a quadratic effect (Figure 2(c)). The use of skim milk at high concentration with increasing yeast extract concentration (ranging from 1% to 2.8% (wt/vol)) significantly influenced the residual concentration of free amino nitrogen in the culture. On the other hand, reduced residual concentration of free amino nitrogen in the culture was observed at low skim milk concentration (4.00% (wt/vol)) with decreasing yeast extract concentration (ranging from 2.8% to 1% (wt/vol)). This result indicated that interactions between skim milk and yeast extract might have a stronger influence on residual concentration of free amino nitrogen remaining in the culture. Consequently, the use of low skim milk and yeast extract concentration significantly improved the nitrogen uptake efficiency with low residual concentration remaining in the culture at the end of cultivation.
3.3. Validation of Optimized Medium
The optimum concentrations for skim milk and yeast extract were 5.89% (wt/vol) and 2.31% (wt/vol), respectively. The responses criteria for the optimization process were maximum for biomass and β-galactosidase production. On the other hand, the minimum values were set for residual concentration of lactose and free amino nitrogen. An experiment was performed under the predicted optimal conditions in order to validate the optimized medium. The experimental values fitted well to the predicted results with no significant difference (P > 0.05). Therefore, this result was encountered in the process of validation of response surface methodology optimization.
4. Conclusion
Results from this study have demonstrated that the optimization of milk-based medium using response surface methodology (RSM) greatly improved G4 cultivation performance, in terms of final cell concentration and β-galactosidase as well as residual concentration of lactose and amino nitrogen remaining in the culture. The quadratic model (biomass, β-galactosidase production, and free amino nitrogen residual) and linear model (lactose residual) were found sufficient for the optimization of medium for G4. The optimal medium that consists of 5.89% (wt/vol) skim milk and 2.31% (wt/vol) yeast extract gave the final biomass count of 107 cfu/mL, β-galactosidase activity of 8.74 U/mL, residual lactose concentration of 9.15 g/L, and residual free amino nitrogen concentration of 226.07 mg/mL. In addition, the use of this optimal medium gave comparable biomass count and β-galactosidase activity as those obtained in cultivation using high skim milk concentrations (10% to 13% (wt/vol)). However, residual lactose and free amino nitrogen concentration remaining in the culture at the end of cultivation were reduced by about two times lower as compared to those observed in cultivation using high skim milk concentration.
Acknowledgments
This study was funded by the Ministry of Higher Education, Malaysia, under Project no. 600-RMI/FRGS 5/13 15/2013. The first author is also grateful to Universiti Teknologi MARA (UiTM), Malaysia, and Universiti Putra Malaysia (UPM) for the support throughout the conduct of this study.
Conflict of Interests
The authors declare no conflict of interests.
References
- 1.FAO/WHO. Health and nutritional properties of probiotics in food including powder milk with live lactic: report of a joint FAO/WHO expert consultation on acid bacteria expert consultation on evaluation of health and nutritional properties of probitics in food including powder milk with live lactic acid bacteria. 2001
- 2.Ohashi Y, Ushida K. Health-beneficial effects of probiotics: its mode of action. Animal Science Journal. 2009;80(4):361–371. doi: 10.1111/j.1740-0929.2009.00645.x. [DOI] [PubMed] [Google Scholar]
- 3.Mitsouka T. Taxonomy and ecology of bifidobacteria. Bifidobacteria Microflora. 1984;(3):11–28. [Google Scholar]
- 4.Scardovi V. Genus Bifidobacterium . In: Mair NNS, editor. Bergey’s Manual of Systemic Bacteriology. New York, NY, USA: William and Wilkins; 1986. pp. 1418–1434. [Google Scholar]
- 5.Shuhaimi M, Ali AM, Saleh NM, Yazid AM. Differentiation of Bifidobacterium isolates from faeces of infant by RAPD. Bioscience Microflora. 2001;20:89–94. [Google Scholar]
- 6.Shuhaimi M, Ali AM, Saleh NM, Yazid AM. Utilisation of Enterobacterial Repetitive Intergenic Consensus (ERIC) sequence-based PCR to fingerprint the genomes of Bifidobacterium isolates and other probiotic bacteria. Biotechnology Letters. 2001;23(9):731–736. [Google Scholar]
- 7.Shuhaimi M, Ali AM, Norihan MS, Yazid AM. Classification of Bifidoacteriumisolates from infant faeces using PCR-based and 16S rdnapartial sequence analysis methods. Bioscience Microflora. 2002;21:155–161. [Google Scholar]
- 8.Wong S, Wendy YK, Kabeir BM, Shuhaimi M, Rosfarizan M, Yazid AM. Survival of Bifidobacterium pseudocatenulatum strain isolated from breast-fed infants to simulated gastric pH environment. Malaysian Applied Biology. 2006;35:57–62. [Google Scholar]
- 9.Rodrigues LR, Teixeira JA, Oliveira R. Low-cost fermentative medium for biosurfactant production by probiotic bacteria. Biochemical Engineering Journal. 2006;32(3):135–142. [Google Scholar]
- 10.Mark J. Successful probiotic bacteria. In: Shortt C, O’Brien J, editors. Handbook of Functional Dairy Products. Boca Raton, Fla, USA: CRC Press; 2004. pp. 36–61. [Google Scholar]
- 11.Marshall VM, Tamime AY. Starter cultures employed in the manufacture of biofermented milks. International Journal of Dairy Technology. 1997;50(1):35–41. [Google Scholar]
- 12.Østlie HM, Helland MH, Narvhus JA. Growth and metabolism of selected strains of probiotic bacteria in milk. International Journal of Food Microbiology. 2003;87(1-2):17–27. doi: 10.1016/s0168-1605(03)00044-8. [DOI] [PubMed] [Google Scholar]
- 13.Hsu CA, Yu RC, Chou CC. Production of β-galactosidase by Bifidobacteria as influenced by various culture conditions. International Journal of Food Microbiology. 2005;104(2):197–206. doi: 10.1016/j.ijfoodmicro.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Avonts L, Uytven EV, Vuyst LD. Cell growth and bacteriocin production of probiotic Lactobacillus strains in different media. International Dairy Journal. 2004;14(11):947–955. [Google Scholar]
- 15.Shihata A, Shah NP. Proteolytic profiles of yogurt and probiotic bacteria. International Dairy Journal. 2000;10(5-6):401–408. [Google Scholar]
- 16.Dave RI, Shah NP. Evaluation of media for selective enumeration of Streptococcus thermophilus, Lactobacillus delbrueckii ssp. bulgaricus, Lactobacillus acidophilus, and Bifidobacteria. Journal of Dairy Science. 1996;79(9):1529–1536. doi: 10.3168/jds.S0022-0302(96)76513-X. [DOI] [PubMed] [Google Scholar]
- 17.Stephanie W, Kabier BM, Shuhaimi M, Rosfarizan M, Yazid AM. Growth optimization of a probiotic candidate, Bifidobacterium pseudocatenulatum G4, in milk medium using response surface methodology. Biotechnology and Bioprocess Engineering. 2007;12(2):106–113. [Google Scholar]
- 18.Chen M-J, Chen K-N, Lin C-W. Optimization on response surface models for the optimal manufacturing conditions of dairy tofu. Journal of Food Engineering. 2005;68(4):471–480. [Google Scholar]
- 19.Karacan F, Ozden U, Karacan S. Optimization of manufacturing conditions for activated carbon from Turkish lignite by chemical activation using response surface methodology. Applied Thermal Engineering. 2007;27(7):1212–1218. [Google Scholar]
- 20.Hou J-W, Yu R-C, Chou C-C. Changes in some components of soymilk during fermentation with bifidobacteria. Food Research International. 2000;33(5):393–397. [Google Scholar]
- 21.Hwang MN, Ederer GM. Rapid hippurate hydrolysis method for presumptive identification of group B streptococci . Journal of Clinical Microbiology. 1975;1(1):114–115. doi: 10.1128/jcm.1.1.114-115.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy Z, Kiss T, Szentirmai A, Biro S. Carbon source regulation of β-galactosidase biosynthesis in Penicillium chrysogenum . Journal of Basic Microbiology. 2001;41:351–362. doi: 10.1002/1521-4028(200112)41:6<351::AID-JOBM351>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 23.Körbahti BK, Aktaş N, Tanyolaç A. Optimization of electrochemical treatment of industrial paint wastewater with response surface methodology. Journal of Hazardous Materials. 2007;148(1-2):83–90. doi: 10.1016/j.jhazmat.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Gao H, Gu W-Y. Optimization of polysaccharide and ergosterol production from Agaricus brasiliensis by fermentation process. Biochemical Engineering Journal. 2007;33(3):202–210. [Google Scholar]
- 25.Myers RH, Montgomery DC. Response Surface Methodology: Process and Product Optimization Using Designed Experiments. 2nd edition. New York, NY, USA: John Wiley & Sons; 2002. [Google Scholar]
- 26.Demirhan E, Apar DK, Özbek B. Product inhibition of whey lactose hydrolysis. Chemical Engineering Communications. 2008;195(3):293–304. [Google Scholar]


