Abstract
Regulatory gene circuits with positive feedback loops control stem cell differentiation, but several mechanisms can contribute to positive feedback. Here, we dissect feedback mechanisms through which the transcription factor PU.1 controls lymphoid and myeloid differentiation. Quantitative live-cell imaging revealed that developing B-cells decrease PU.1 levels by reducing PU.1 transcription, whereas developing macrophages increase PU.1 levels by lengthening their cell cycles, which causes stable PU.1 accumulation. Exogenous PU.1 expression in progenitors increases endogenous PU.1 levels by inducing cell-cycle lengthening, implying positive feedback between a regulatory factor and the cell cycle. Mathematical modeling showed that this cell-cycle coupled feedback architecture effectively stabilizes a slow-dividing differentiated state. These results show that cell cycle duration functions as an integral part of a positive auto-regulatory circuit to control cell fate.
The transcription factor PU.1 is a central component of the regulatory gene network controlling lymphoid and myeloid development from haematopoietic progenitors (1–4). It is expressed at intermediate levels in progenitors, and its subsequent levels become a determinant of lymphoid and myeloid fate choices, with down-regulation of PU.1 required for B- and T- cell development and higher PU.1 levels favoring the development of macrophages or myeloid dendritic cells (5–8).
Differential regulation of PU.1 during lymphoid and myeloid development involves transcriptional positive feedback of PU.1 (9). PU.1 positively regulates its own transcription in myeloid cells and stem cells, but not in lymphoid cells (10–13), and forms additional positive feedback loops through mutual inhibition with other haematopoietic regulators (7, 14). Positive feedback can in principle generate multiple stable states with different levels of regulatory factors, possibly accounting for the observed differences in PU.1 levels. However, it is unclear how PU.1 is regulated during myeloid or lymphoid development, what feedback mechanisms are involved, and why particular feedback architectures may have been selected.
PU.1 promotes growth in several progenitor types (1, 15), but also coordinates cell-cycle arrest with differentiation in myeloid progenitors. Reduced PU.1 activity causes acute myeloid leukemia, where progenitors fail to initiate differentiation growth arrest (16–19); conversely, re-expression of PU.1 restores growth arrest (17, 20, 21). However, it is unclear whether PU.1’s effect on the cell cycle influences its ability to regulate its own levels and control differentiation.
Here, we analyzed PU.1 and cell cycle regulation in individual cells during early macrophage and B-cell development (Fig. 1A). We isolated fetal liver progenitors (FLPs, Lin-cKit+CD27+) from mice containing a bicistronic PU.1-GFP knock-in reporter (2), cultured them with cytokines supporting B-cell and macrophage differentiation, and analyzed PU.1-GFP levels over time by timelapse imaging or flow cytometry [Figs. 1, S1, S2; (22)]. Importantly, PU.1-GFP levels varied linearly with nuclear PU.1 protein levels in this culture system (Fig. S3). We found that progenitors initially expressed PU.1-GFP at uniform levels, but subsequently up-regulated or down-regulated PU.1-GFP over time (Fig. 1B–D, Fig. S4). Cells up-regulating PU.1-GFP expressed the macrophage markers CD11b and F4/80 but not the granulocyte marker Gr1, and were also large and adherent, reflecting differentiation into macrophages (Fig. 1B, 1C–top right; Fig. S4). In contrast, cells down-regulating PU.1-GFP expressed the B-cell marker CD19, and were also small and round, reflecting differentiation into B-cells (Fig. 1B, 1C – bottom right; Figs. S2, S4). Developing granulocytes and persisting progenitor-like cells maintained PU.1-GFP levels similar to starting progenitors (Fig. 1B, Fig. S4). Both macrophages and B cells preferentially developed from Fcγ receptor II/III (FcγR2/3)low FLPs, whereas FcγR2/3+ FLPs mostly differentiated into granulocytes (Fig. S5, and see below). These results validate the use of our system for analyzing PU.1 regulation during B-cell or macrophage differentiation.
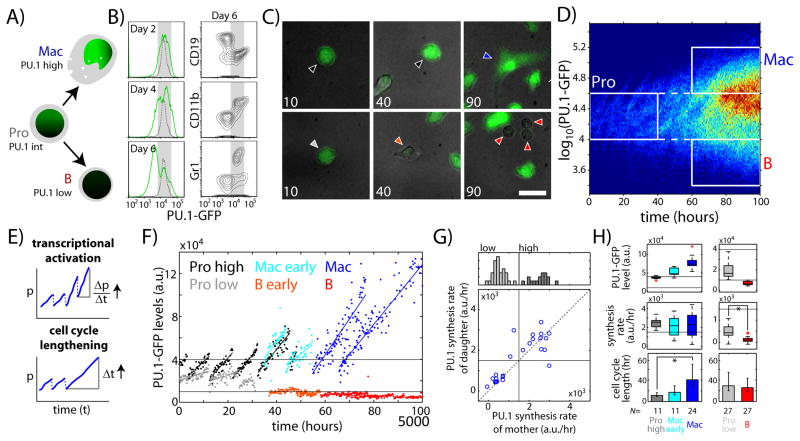
Fig. 1. Cell-cycle lengthening drives PU.1 up-regulation during macrophage development.
FLPs (Lin-cKit+CD27+) from E13.5 PU.1-GFP mice were cultured with B- and macrophage-supporting cytokines (SCF, IL-3, IL-7, Flt3L, M-CSF) and analyzed using timelapse imaging or flow cytometry. A) Schematic showing myeloid and lymphoid development from haematopoietic progenitor cells. B) Histograms (left) (left) show PU.1-GFP levels measured after the indicated number of days in culture. Dotted lines give initial PU.1-GFP levels. Flow cytometry plots (right) show CD19, CD11b and Gr-1 levels against PU.1-GFP after six days. C) Merged DIC (gray) and PU.1-GFP fluorescence (green) images of cultured FLPs, taken after the indicated number of hours. Cells with PU.1-GFP time traces shown in F) are marked with correspondingly colored arrowheads. Scale bar = 20 μm. D) Heat map showing PU.1-GFP levels over time for all imaged cells. Rectangles define progenitor (gray), macrophage (blue) and B-cell (red) populations. E) Alternative hypotheses for PU.1-GFP up-regulation in macrophages. The PU.1 synthesis rate for a single cell is given by (Δp/Δt) over the entire observed cell cycle. F) Representative single-cell PU.1-GFP time traces for different cell populations. Data are taken from lineages shown in Fig. S9. Horizontal lines give PU.1-GFP level thresholds for the defined cell populations. G) Histogram (top) showing distribution of PU.1 synthesis rates in progenitors. Scatterplot shows relationship between PU.1 synthesis rates in mother versus daughter cells. Horizontal and vertical lines indicate the threshold for progenitor sub-populations with higher and lower rates of PU.1 synthesis. H) Plots comparing mean PU.1-GFP levels (top), PU.1 synthesis rates (middle) and cell cycle lengths (bottom) in different cell populations. Red crosses indicate boxplot outliers. Bottom error bars represent 95% confidence intervals. Asterisks indicate significantly different means (p<10−7, one-tailed t-test). Data are representative of three independent experiments.
Changes in PU.1 levels during B-cell or macrophage differentiation may result from changes in either the rate of PU.1 synthesis or the rate of PU.1 removal (Fig. 1E), which would occur predominantly through dilution due to cell division (23, 24), as PU.1’s protein half-life is substantially longer than the progenitor cell-cycle length (Fig. S6). To determine how PU.1 levels were regulated, we measured PU.1 synthesis rates and cell cycle lengths for individual cells within defined progenitor (Pro), macrophage (Mac) and B-cell (B) populations (Fig. 1D, Fig. S7). PU.1 synthesis rates could be measured by the slopes of stable PU.1-GFP increase over time [(Δp/Δt for an observed cell cycle), Figs. 1E, S7; Fig. S8 shows GFP stability], independent of average PU.1-GFP levels. Although cell movement precluded comprehensive multigenerational tracking (Fig. S9), the movies allowed accurate measurements of average cell cycle lengths and PU.1 synthesis rates for different cell populations. Progenitors comprised two sub-populations with higher and lower rates of PU.1 synthesis (Fig. 1F, G). Switches between states with high and low PU.1 synthesis rates were infrequent across cell division (Fig. 1G), suggesting that these states are maintained stably in most cells. Macrophages had more PU.1-GFP and PU.1 protein than any of the progenitors, as expected. Surprisingly, however, their PU.1 synthesis rates were not higher than that of the progenitor sub-population with high PU.1 synthesis rates (Figs. 1F–H, S9). Instead, they had significantly longer cell-cycle lengths (Figs. 1F–H, S9), and descended from ancestors with shorter cell cycle lengths but similar PU.1 synthesis rates (Mac early, Fig. 1F–H). Thus, developing macrophages increase their PU.1 levels by lengthening their cell cycles, which allows PU.1 to accumulate to higher levels. In contrast, emerging B-cells had significantly lower PU.1 synthesis rates than progenitors but similar cell cycle lengths (Fig. 1F–H, Fig. S9). Thus, unlike macrophages, B cells decrease PU.1 levels by reducing PU.1 transcription.
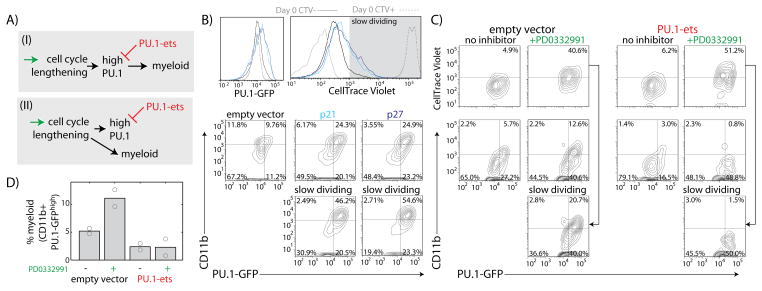
Increased PU.1 levels caused by cell-cycle lengthening may be functionally important for macrophage differentiation, or may simply reflect a consequence of differentiation growth arrest (Fig. 2A). To distinguish between these two possibilities, we tested whether artificial cell cycle lengthening promotes myeloid differentiation in a PU.1-dependent manner. We induced cell-cycle lengthening in FLPs by two different methods–either by retroviral transduction of cyclin-dependent kinase (CDK)-inhibitors p21Cip1 (Cdkn1a) or p27Kip1 (Cdkn1b) (Fig. 2B, Fig. S10), or by treatment with PD0332991, a CDK4/6 inhibitor (25) (Fig. 2C, D). Induced cell-cycle lengthening in progenitors increased PU.1-GFP and PU.1 levels, and increased the percentage of myeloid cells, with these increases being most dramatic in the slowest-dividing cells (Fig 2B, C). This differentiation depended on PU.1 activity, because in cells transduced with a competitive inhibitor of PU.1 (PU.1-ets, Fig. S11), PD0332991 treatment still increased PU.1-GFP, but no longer increased the fraction of CD11b-expressing cells as in empty vector (EV)-transduced cells (Fig. 2C, D). These results suggest that PU.1 accumulation as a result of cell-cycle lengthening is functionally important for macrophage differentiation.
Figure 2. PU.1 accumulation due to cell-cycle lengthening is important for myeloid differentiation.
A) Two hypotheses for the function of high PU.1 levels in differentiating macrophages. B) FLPs were transduced with empty vector (EV), p21, or p27, cultured for four days and analyzed by flow cytometry. Histograms (top) show CellTrace Violet and PU.1-GFP levels for different transduced populations. Gray shaded area indicates slow-dividing cell gate. Flow plots (bottom) show CD11b versus PU.1-GFP levels for different transduced cell populations. C) FLPs transduced with EV or PU.1 antagonist (PU.1-ets) were cultured for 3 days with or without 2.1 μM CDK4/6 inhibitor PD0332991, and analyzed by flow cytometry. Flow plots show CellTrace Violet (top) or CD11b (bottom) versus PU.1-GFP for the different conditions. D) Effects of PD0332991 and PU.1-ets transduction on the percentage of myeloid cells. Bars represent means of two independent experiments, and circles give individual measurements.
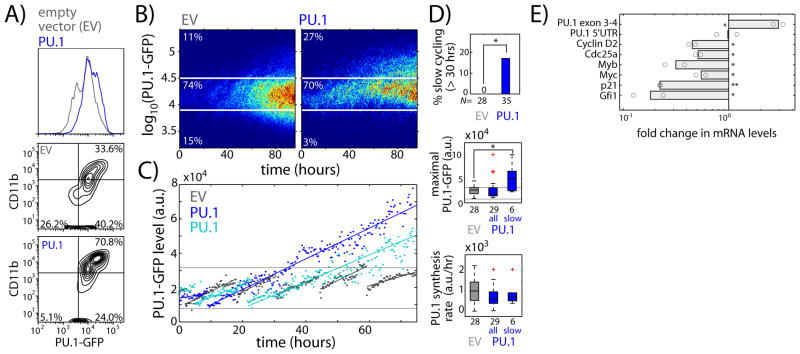
To examine how positive transcriptional feedback regulates PU.1’s own expression (10–13), we tested how PU.1 and dominant negative PU.1 transduction affected transcription of the PU.1-GFP reporter. Forced expression of PU.1-ets in FLPs reduced PU.1-GFP levels (Fig. S12), implying that a threshold level of PU.1 activity is important for maximal PU.1 expression. Conversely, flow cytometry and imaging showed that exogenous PU.1 up-regulated PU.1-GFP and CD11b, while inhibiting PU.1-GFP down-regulation and CD19 up-regulation (Fig. 3A–B, Fig. S13). However, imaging analysis (cf. Fig. 1E) showed that exogenous PU.1 expression did not increase endogenous PU.1 synthesis rates; instead, it induced cell cycle lengthening in a sub-population of progenitors, which in turn led to the significant increase in PU.1-GFP levels (Fig. 3C–D, Fig. S14). This cell-cycle lengthening occurred preferentially in FcγR2/3low FLPs (Fig. S5C), which accounted for most of the macrophage potential in the FLP population. Thus, high PU.1 levels promote cell-cycle lengthening in cells capable of generating macrophages, which in turn allows high PU.1 levels to be stably maintained. Taken together, our results provide evidence for a regulatory circuit architecture involving positive feedback on a transcription factor through the cell cycle.
Fig. 3. PU.1 up-regulates its own expression during macrophage development by inducing cell-cycle lengthening.
FLPs transduced with EV or PU.1 retroviral constructs were sorted and cultured with multi-lineage supporting cytokines (SCF, IL-3, IL-7, Flt3L). A) Histogram showing PU.1-GFP levels (top), and flow plots showing CD11b versus PU.1-GFP levels after four days of culture (bottom). B) Heat maps comparing time evolution of PU.1-GFP levels for imaged EV- or PU.1-transduced cells. C) Representative PU.1-GFP time traces for EV or PU.1-transduced cells, taken from lineage trees shown in Fig. S14. D) Box-plots comparing EV- and PU.1-transduced progenitors, showing percentage of slow dividing cells (top), along with maximal PU.1-GFP levels (middle), and PU.1 synthesis rate (bottom) for both the entire PU.1-transduced progenitor population and slow-dividing progenitors alone. Red crosses indicate outliers. Asterisks indicate significantly different means (% slow dividing, p < 0.05, χ2 test, d.f. = 1; maximal PU.1-GFP, one-tailed t-test, p<0.005). Data are representative of two independent experiments. E) EV or PU.1-transduced FcγR2/3low FLPs were cultured for 2 days, harvested for RNA and analyzed using qRT-PCR. Bar chart shows mRNA level fold change for the indicated genes in PU.1-transduced as compared to EV-transduced cells. Bars represent the means of two independent experiments, and circles represent individual measurements (*-p < 0.1; **-p < 0.01; two-tailed t-test).
Insight into cell-cycle lengthening mechanisms emerged from analysis of regulatory gene expression in PU.1-transduced progenitors (Fig. 3E). Consistent with PU.1 auto-regulation through cell-cycle lengthening rather than transcriptional acceleration, PU.1 transduction did not affect endogenous PU.1 mRNA levels, but significantly reduced the levels of cell-cycle promoting factors Cyclin D2 (Ccnd2) and Cdc25a. Consistent with other studies (26–28), exogenous PU.1 also reduced the levels of Myb and Myc, growth-promoting proto-oncogenes that are down-regulated during normal differentiation. Exogenous PU.1 also reduced levels of p21 and Gfi1, which can mediate quiescence, although these are up-regulated by PU.1 in stem cells (13). Thus, the mechanisms underlying PU.1-mediated cell cycle arrest during macrophage differentiation appear distinct from those operating in stem cell quiescence.
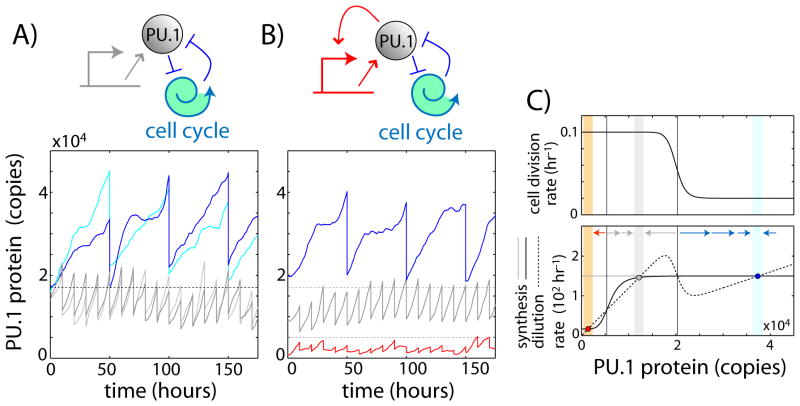
How can positive feedback between PU.1 and the cell cycle stabilize a slow-dividing macrophage state with high PU.1 levels? To address this issue, we constructed a stochastic single-cell dynamical model, where PU.1 inhibits the G1 to S cell cycle transition above a threshold concentration (Fig. 4A, top). This model exhibits bi-stability, supporting both a fast-dividing low-PU.1 state and a slow-dividing high-PU.1 state (Fig. 4A; Figs. S15-S16). In our simple model, G1 checkpoint release depends solely on PU.1 levels; during macrophage development, other regulatory factors also promote checkpoint release and thus regulate feedback engagement. Once the high PU.1 state is established, it is relatively stable compared to the corresponding state of a hypothetical pure transcriptional feedback system with similar parameters, which exhibits more frequent spontaneous switches between states due to larger, more rapid protein fluctuations [Fig. S17, see (22)]. Taken together, these results show how cell-cycle coupled feedback provides a simple mechanism to support multiple stable states that exhibit different rates of cell division.
Fig. 4. A cell-cycle coupled feedback loop stably maintains a slow-dividing differentiated state.
A) Schematic of a cell-cycle coupled positive feedback loop (top), and time traces from stochastic simulations of this circuit architecture (bottom), showing four cells with different initial PU.1 levels but identical rate constants. B) Schematic of a hybrid cell-cycle coupled/transcriptional positive feedback circuit (top), and stochastic simulations of this architecture (bottom), showing maintenance of three stable steady-states. C) Phase diagrams for the two circuit architectures, showing PU.1 synthesis rates (black – hybrid; gray – cell-cycle coupled), as well as dilution rate and cell division rate (same for both models) against PU.1 levels. Red, gray and blue circles denote B, progenitor and macrophage steady-states respectively, and arrows indicate flow of the system. A thorough analysis and discussion of all models is given in the mathematical appendix (22).
Besides cell-cycle coupled feedback, cells also contain PU.1 transcriptional feedback, which appears to take effect at lower PU.1 levels (Fig. S12). When both feedbacks are incorporated simultaneously, the model can generate three stable steady states with low, medium and high levels of PU.1 corresponding to the pro-B, progenitor and macrophage populations respectively (Fig. 4B–C). Because of its lower PU.1 threshold, transcriptional feedback allows developing B-cells to down-regulate PU.1 while maintaining similar rates of division, consistent with observations. We propose this dual feedback system as a working model for further study of PU.1 regulation during lymphoid and myeloid development.
Could cell-cycle coupled feedback operate in other systems? Interestingly, a similar type of bi-stability was recently observed in a bacterial synthetic cell-cycle coupled feedback circuit (29). In the context of cell differentiation, other fate regulators have also been shown to promote cell-cycle arrest (30). Although some transcription factors are notoriously short-lived (e.g. Fos), recent studies have found that many mammalian proteins are stable over multiple cell cycles (23, 24), and other regulatory proteins may resemble PU.1 in this respect. Moreover, induced cell-cycle lengthening is known to promote differentiation in other systems (31, 32), suggesting that mechanisms based on accumulation of stable fate regulators during cell-cycle arrest may be more prevalent. Where cell fate decisions depend on the balance between two factors with different stabilities – such as PU.1 and the unstable C/EBPα, for instance (33, 34) – cell cycle speed may act as a tiebreaker, with slowing favoring the more stable factor and acceleration favoring the less stable one. In general, our results imply a mutual regulatory relationship between the cell cycle and transcription factor activities in cell differentiation, and similar relationships may impact other processes that involve cell cycle length changes, such as cancer.
Supplementary Material
Acknowledgments
We thank R. Butler and S. Washburn for mouse care, and J. Verceles, J. Grimm and D. Perez of the Caltech flow cytometry facility for cell sorting. We also thank members of the Rothenberg and Elowitz labs, and L. Goentoro for insightful discussions. The data presented in this manuscript are tabulated in the main paper and the supplementary materials. This work was supported by a CRI/Irvington Postdoctoral Fellowship to H.Y.K.; an Australian Research Council Future Fellowship and the Victorian State Government Operational Infrastructure Support, National Health and Medical Research Council of Australia IRIIS to S.L.N, and NIH grants to E.V.R (RC2 CA148278, R33 HL089123, and R01 CA90233), the Albert Billings Ruddock Professorship, the Al Sherman Foundation, and the Louis A. Garfinkle Memorial Laboratory Fund.
Footnotes
“This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
References
- 1.Singh H, DeKoter RP, Walsh JC. PU.1, a shared transcriptional regulator of lymphoid and myeloid cell fates. Cold Spring Harb Symp Quant Biol. 1999;64:13–20. doi: 10.1101/sqb.1999.64.13. [DOI] [PubMed] [Google Scholar]
- 2.Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–31. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Back J, Allman D, Chan S, Kastner P. Visualizing PU.1 activity during hematopoiesis. Exp Hematol. 2005;33:395–402. doi: 10.1016/j.exphem.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Arinobu Y, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–27. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 5.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–41. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 6.Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–96. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 7.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–86. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carotta S, et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–41. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–17. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 10.Okuno Y, et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol. 2005;25:2832. doi: 10.1128/MCB.25.7.2832-2845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarnegar MA, Chen J, Rothenberg EV. Cell-type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol Cell Biol. 2010;30:4922–4939. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leddin M, et al. Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood. 2011;117:2827–38. doi: 10.1182/blood-2010-08-302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staber PB, et al. Sustained PU.1 Levels Balance Cell-Cycle Regulators to Prevent Exhaustion of Adult Hematopoietic Stem Cells. Mol Cell. 2013:1–13. doi: 10.1016/j.molcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 15.Choe KS, Ujhelly O, Wontakal SN, Skoultchi AI. PU.1 directly regulates cdk6 gene expression, linking the cell proliferation and differentiation programs in erythroid cells. J Biol Chem. 2010;285:3044–52. doi: 10.1074/jbc.M109.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook WD, et al. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood. 2004;104:3437–44. doi: 10.1182/blood-2004-06-2234. [DOI] [PubMed] [Google Scholar]
- 17.Rosenbauer F, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–30. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 18.Walter MJ, et al. Reduced PU.1 expression causes myeloid progenitor expansion and increased leukemia penetrance in mice expressing PML-RARa. Proc Natl Acad Sci U S A. 2005;102 doi: 10.1073/pnas.0504247102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metcalf D, et al. Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:1486–91. doi: 10.1073/pnas.0510616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgado MD, et al. Spi-1/PU.1 proto-oncogene induces opposite effects on monocytic and erythroid differentiation of K562 cells. Biochem Biophys Res Commun. 1998;252:383–91. doi: 10.1006/bbrc.1998.9587. [DOI] [PubMed] [Google Scholar]
- 21.DeKoter RP, Walsh JC, Singh H. PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J. 1998;17:4456–68. doi: 10.1093/emboj/17.15.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Materials and methods available in supplementary material.
- 23.Eden E, et al. Proteome half-life dynamics in living human cells. Science. 2011;331:764–8. doi: 10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- 24.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 25.Toogood PL, et al. Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J Med Chem. 2005;48:2388–406. doi: 10.1021/jm049354h. [DOI] [PubMed] [Google Scholar]
- 26.Kihara-Negishi F, et al. Down-regulation of c-myc and bcl-2 gene expression in PU.1-induced apoptosis in murine erythroleukemia cells. Int J Cancer. 1998;76:523–30. doi: 10.1002/(sici)1097-0215(19980518)76:4<523::aid-ijc14>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Bellon T, Perrotti D, Calabretta B. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor PU.1 correlates with negative regulation of the c-myb promoter. Blood. 1997;90:1828–39. [PubMed] [Google Scholar]
- 28.Franco CB, et al. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–8. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan C, Marguet P, You L. Emergent bistability by a growth-modulating positive feedback circuit. Nat Chem Biol. 2009;5:842–8. doi: 10.1038/nchembio.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buttitta LA, Edgar BA. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr Opin Cell Biol. 2007;19:697–704. doi: 10.1016/j.ceb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinman RA. Cell cycle regulators and hematopoiesis. Oncogene. 2002;21:3403–13. doi: 10.1038/sj.onc.1205325. [DOI] [PubMed] [Google Scholar]
- 32.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20:233–43. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Dahl R, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–36. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 34.Trivedi AK, et al. Proteomic identification of C/EBP-DBD multiprotein complex: JNK1 activates stem cell regulator C/EBPalpha by inhibiting its ubiquitination. Oncogene. 2007;26:1789–801. doi: 10.1038/sj.onc.1209964. [DOI] [PubMed] [Google Scholar]
- 35.Taghon TN, David ES, Zuniga-Pflucker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez RC, Woods RE. Digital Image Processing, 3rd Edition. Prentice Hall; New Jersey: 2007. [Google Scholar]
- 38.Veenman CJ, Reinders MJT, Backer E. Resolving motion correspondence for densely moving points. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2001;23:54–72. [Google Scholar]
- 39.Gubelmann C, et al. GETPrime: a gene- or transcript-specific primer database for quantitative real-time PCR. Database. 2011;2011:bar040. doi: 10.1093/database/bar040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–19. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamath MB, et al. Dose-dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B-cell identity. Leukemia. 2008;22:1214–25. doi: 10.1038/leu.2008.67. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Mortazavi A, Williams B, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–82. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
- 45.Savageau MA. A theory of alternative designs for biochemical control systems. Biomed Biochim Acta. 1985;44:875–80. [PubMed] [Google Scholar]
- 46.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–8. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.