Abstract
Centromeres are essential for chromosome inheritance and genome stability. Centromeric proteins, including the centromeric histone CENP-A, define the site of centromeric chromatin and kinetochore assembly. In many organisms, centromeres are located in or near regions of repetitive DNA. However, some atypical centromeres spontaneously form on unique sequences. These neocentromeres, or new centromeres, were first identified in humans, but have since been described in other organisms. Neocentromeres are functionally and structurally similar to endogenous centromeres, but lack the added complication of underlying repetitive sequences. Here, we discuss recent studies in chicken and fungal systems where genomic engineering can promote neocentromere formation. These studies reveal key genomic and epigenetic factors that support de novo centromere formation in eukaryotes.
Keywords: CENP-A, transcription, replication, heterochromatin, histone, gene conversion
Eukaryotes exhibit a range of centromeres
Preserving genome integrity is a major goal of cell division, as genetic information is passed from mother to daughter cells. The centromere is essential to faithful chromosome segregation and genome stability. It is generally recognized that both genomic and epigenetic pathways are critical for establishing and maintaining functional centromeres. Centromeres are often defined by repetitive DNA, but unique sequences are present at endogenous centromeres of Schizosacchromyces pombe, Candida albicans, and Gallus gallus. Centromeres can be small and similar in size and sequence, such as the 125bp “point” Sacchromyces cerevisiae centromere. Centromeres in larger eukaryotes are regional; the site of kinetochore assembly occurs at variably sized genomic regions, ranging from 40 kilobases to five megabases. In Caenorhabditis elegans, the chromosomes are holocentric, in that the centromere is formed along the length of each chromosome [1]. Sometimes, chromosomes contain two centromere regions. These dicentrics are usually products of chromosome fusion. Dicentrics are typically unstable during cell division; the activity of one centromere is suppressed so that dicentric segregation occurs in the manner of a monocentric chromosome [2]. Inactive centromeres represent a class of centromeres that remains to be fully characterized.
Neocentromeres are an intriguing type of centromere arising at atypical chromosomal sites, including chromosome arms or telomeres (reviewed by [3, 4]) (Box 1). They are unique models for studying de novo centromere formation because they usually form on non-repetitive DNA, yet recruit centromere proteins, and generally segregate faithfully during cell division. Neocentromeres were first described in humans in 1993, and since then, over 100 have been identified. They are usually ascertained due to their presence on chromosomes associated with abnormal phenotypes. These include marker chromosomes that have been deleted or duplicated from endogenous chromosomes [5–7] or native or marker chromosomes in which the normal centromere has been repressed [8, 9]. Although neocentromeres originating from nearly every human chromosome have been described, some appear to cluster in similar locations such as the long arms of chromosomes 3, 4, 8, 13, and 15 [4, 10]. These are not “hotspots” per se, because precise mapping of centromere protein binding regions showed that the different neocentromeres form on distinct DNA sequences, even within the same genomic interval [11, 12]. Furthermore, the sizes of the CENP-A domains on neocentromeres in the same genomic region can range four-fold (~100–400kb), emphasizing the plasticity of centromere assembly.
Box 1. Glossary of terms used.
- CENP-A
histone H3 variant that replaces canonical H3 at centromeres
- Centromere
chromosomal locus at which the kinetochore is assembled and spindle microtubules attach
- HJURP/Scm3
the chaperone protein that assembles CENP-A into chromatin
- Immature/Incomplete Centromere
a chromosomal locus that is contains CENP-A at low levels and/or fails to recruit a full complement of centromere/kinetochore proteins
- Kinetochore
the multi-protein structure that is assembled on centromeric DNA and facilitates chromosomal connection to spindle microtubules
- mardel(10)
one of the first human neocentromeres to be described and characterized; it is a marker chromosome derived from the long arm of chromosome 10 on which a neocentromere formed on non-centromeric DNA
- Neocentromere
a centromere that forms at a non-typical genomic region and usually at sequences that differ from endogenous centromeres
Understanding human neocentromere formation has been limited by the retrospective nature of many analyses. At the time of study, human neocentromeres are already stabilized in the karyotype. Mechanisms of their formation can only be insinuated by their structure and chromosomal origin, thus underscoring the need for strategies to induce neocentromere formation experimentally. In this review, we discuss exciting, recent studies of controlled neocentromere formation that have extended understanding of genomic and epigenetic factors that govern de novo centromere formation.
Centromere Specification through Unique Chromatin Assembly
The diversity of eukaryotic centromeric DNAs contrasts with the common chromatin organization that is largely independent of the underlying DNA sequence. Within centromeric chromatin the histone H3 variant Centromere Protein A (CENP-A) fully replaces canonical histone H3 in a subset of nucleosomes, so that centromeres contain a mixture of H3 nucleosomes and CENP-A nucleosomes [13, 14]. Replenishment of CENP-A during each cell cycle is critical to centromere stability. New CENP-A is loaded into chromatin by the CENP-A specific chaperone, HJURP (Holliday Junction Recognition Protein) (Scm3 in fungi, CAL1 in Drosophila). Tethering HJURP to non-centromeric sites can seed a de novo centromere [15] that persists following HJURP disassociation, emphasizing the important role for CENP-A in centromere specification.
In addition to CENP-A containing chromatin, eukaryotic centromeres are also enriched for other types of chromatin. CENP-A chromatin forms the centromeric core and is surrounding by chromatin marked by H3K9 and H3K27 tri-methylation [16, 17]. CENP-A nucleosomes within the centromeric core of metazoans are interspersed with H3 nucleosomes methylated at K4 and K36 [18, 19]. Such distinct chromatin domains exist at centromeres ranging from fungi to plants to humans, suggesting that chromatin organization is fundamentally important for centromere specification and/or function.
Surprisingly, many neocentromeres lack common chromatin features. At the mardel(10) neocentromere, CENP-A-containing subdomains are interspersed with histone H3 subdomains, indicating shared chromatin organization with endogenous centromeres [20]. However, 13q neocentromeres lack interspersed H3 nucleosome and are defined by one major and one minor CENP-A domain [12]. Some neocentromeres contain varying amounts of heterochromatin while others lack heterochromatin altogether [11]. The absence of a consistent chromatin environment raises questions about genomic and epigenetic features that influence neocentromere formation.
Targeting CENP-A to certain non-centromeric sites can promote de novo centromere formation and recruitment of centromere proteins [21]. Yet despite the requirement for CENP-A at functional centromeres, the presence of CENP-A is not always sufficient for its continued maintenance. Studies in Drosophila and human cultured cells have shown that global, ectopically expressed CENP-A/CID incorporates at several different genomic sites [22, 23]. However, a complete protein repertoire of a fully functional centromere is not always recruited to every ectopic loci. Similar “immature” or incomplete centromeres have been observed at sites of where HJURP and CENP-A have been tethered [21]. The presence of the endogenous centromere might inhibit maturation of additional centromeres elsewhere on the same chromosome. But a more likely explanation is that certain chromatin environments favor CENP-A incorporation and new centromere formation/maturation [24] (see below).
Neocentromeres arise near sites of former centromere function
What makes certain genomic regions particularly amenable to centromere assembly is unclear. Inferences of mechanism are confounded by potential selection bias for retention of human neocentromeres that are associated with the most viable, least deleterious phenotypes. Two recent studies in chicken cells and C. albicans support the notion that experimentally-derived neocentromeres form at specific genomic locations [25, 26]. In order to induce neocentromere formation in these organisms, an endogenous centromere was physically removed and replaced with a selectable marker (bleomycin in DT40 chicken cells and URA3 in C. albicans). Cells lacking the endogenous centromere but that could still grow in media containing G418 (chicken) or media lacking uracil (C. albicans) were identified as those that had formed neocentromeres.
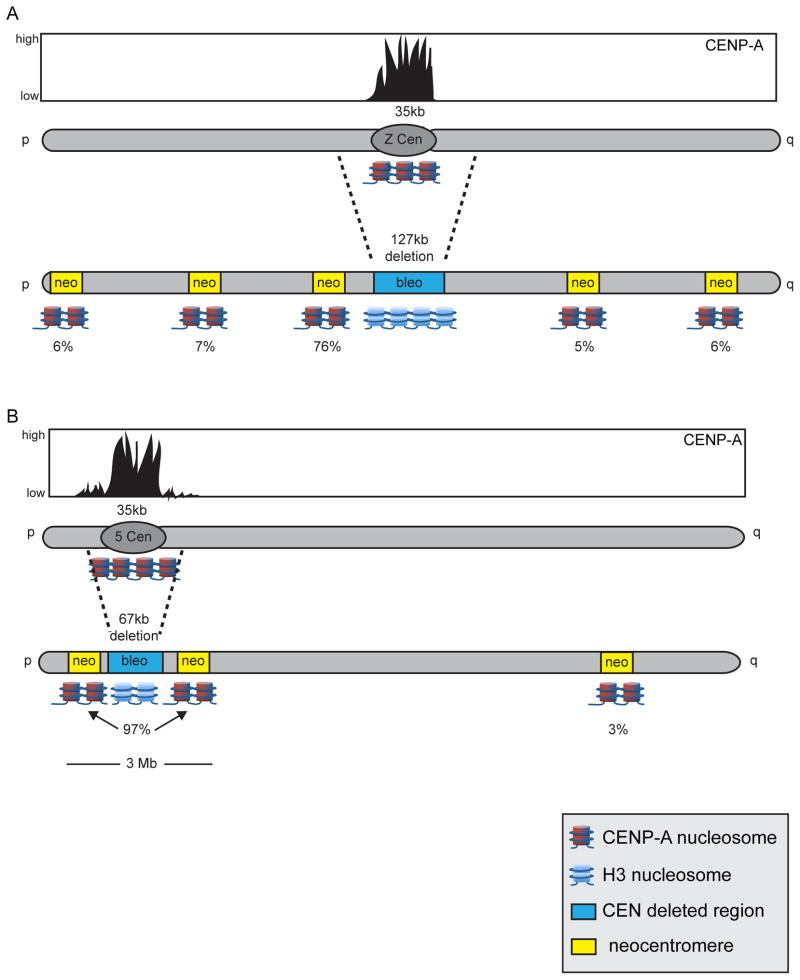
The centromeres of chicken chromosomes 5 and Z consist of non-repetitive DNA, and their CENP-A regions span 30–40kb, like other chicken centromeres. When a large (127kb) portion of the Z centromere was conditionally deleted, neocentromeres formed in several locations, ranging from near either chromosome end to the middle of the chromosome arm [25] (Figure 1A). Neocentromere formation occurred most frequently near the original Z centromere. Although endogenous chicken centromeres have ~35kb regions of concentrated CENP-A accumulation, a much larger region (~2Mb) surrounding the centromere region contains small amounts of non-kinetochore-associated CENP-A [25]. CENP-A enrichment in the flanking regions was low but still more enriched compared to the rest of genome. The preference for non-random neocentromere formation near the endogenous centromere was thought to be due to the presence of CENP-A in the flanking regions. Indeed, deletion of a smaller region (67kb) of centromere 5 resulted in 97% of neocentromeres forming within a 3Mb region near the original centromere (Figure 1B).
Figure 1. Engineered neocentromeres in DT40 chicken cells arise non-randomly near the original centromere.
Endogenous chicken chromosomes Z and 5 contain CENP-A chromatin regions (black-filled curves; reddish-blue nucleosomes) that are ~35kb in size. (A) Removal of the 127kb of the centromere region of chromosome Z, including the 35kb CENP-A domain, and replacement with a bleomycin selectable marker cassette (blue box) using Cre-lox P genome engineereing led to neocentromere formation (yellow boxes) at various sites along chromosome Z. The location of neocentromere formation was preferentially skewed, with 76% of neocentromeres forming proximal to the original centromere. (B) Low levels of CENP-A were detected by ChIP-seq in a 2Mb region surrounding the endogenous centromere of chromosome 5. To test if these regions of more modest CENP-A incorporation are capable of nucleating a centromere in the absence of the adjacent, more enriched CENP-A domain, a smaller (67kb) region of centromere 5 was deleted. Nearly all neocentromeres (97%) formed adjacent to the original centromere, suggesting that in chicken cells, non-kinetochore CENP-A-enriched chromatin can seed neocentromere formation in the absence of the original centromere. Drawings are not drawn precisely to scale.
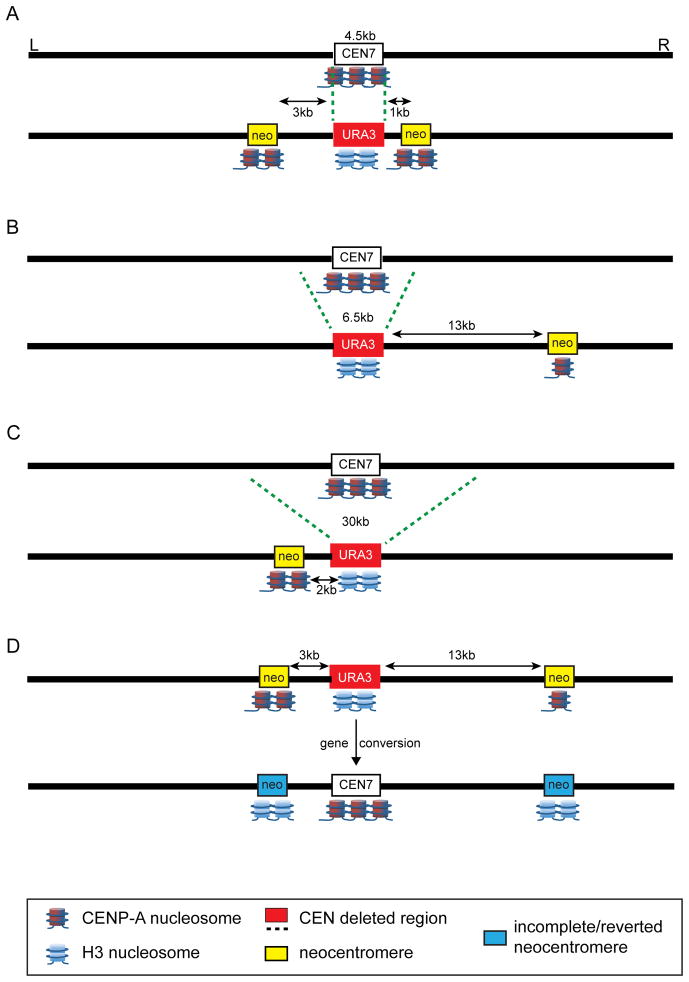
Similar experiments in C. albicans, a pathogenic yeast in which each centromere is ~4.5kb in size and defined by unique, non-repetitive sequences [27], support the notion that centromere-proximal sites are highly amenable to neocentromere formation. Varying amounts (4.5kb – 30kb) of endogenous centromere regions (CEN 1, CEN5, and CEN7) were deleted and replaced with a selectable reporter gene (Figure 2), and neocentromeres formed both proximal and distal to the centromere [26], agreeing with a previous study of neocentromere formation on chromosome 7 [28]. In the most recent study, the majority of neocentromeres preferentially formed between 1kb and 13kb from the location of the original centromere (Figure 2A–2C) [26,]. Interestingly, the neocentromeres that formed farther from the site of CEN7 contained 35% the amount of CENP-A compared to endogenous CEN7 levels, yet they were still viable. These findings agree with studies in humans indicating that centromeres with <20% the normal amount of CENP-A retain almost normal function [18, 29]. Although none of the yeast neocentromere strains exhibited significant chromosome loss, the neocentromeres located 13kb away from the deleted endogenous centromere were the only ones to show a low level of chromosome loss. These finding suggest that despite reduced CENP-A enrichment at these distal neocentromeres, a generally functional kinetochore was formed.
Figure 2. Induced neocentromeres in C. albicans form at high frequency near the original centromere.
(A) Replacement of the 4.5kb C. albicans CEN7 with a URA3 marker (red box) resulted in neocentromere formation (yellow boxes) within 1–3kb on either side of the original centromere. The amount of CENP-A (reddish-blue nucleosomes) at the neocentromeres relative to the amount at the original CEN7 is denoted by the number of cartoon CENP-A nucleosomes (1 = reduced to 3 = normal amount at the endogenous centromere). (B, C) Larger deletions (6.5kb or 30kb) of the CEN7 region produced neocentromeres that were located 2–13kb from the original centromere. Notably, neocentromeres that formed farther from the original centromere contained lower amounts of CENP-A. (D) In a subset of neocentromere-containing strains, the neocentromeres disappeared and the endogenous CEN7 was restored by gene conversion. In these strains, the neocentromeres contained lower amounts of CENP-A (denoted schematically by the blue boxes and few CENP-A nucleosomes), suggesting that the amount of CENP-A may mark the completeness of a centromere, or its probability of being reverted by gene conversion. Drawings are not drawn precisely to scale.
At least two interesting distinctions have emerged from the recent C. albicans and chicken neocentromere studies. First, C. albicans does not exhibit an obvious correlation between size of the deleted centromere region and the centromere-proximal location of neocentromeres. Second, all of the chicken neocentromeres were comparable in size to endogenous centromeres, whereas C. albicans neocentromere sizes varied among the different chromosomes. Neocentromeres formed from deletion of CEN1 or CEN5 were 2–4 times larger (6–12kb) than the endogenous centromeres (3–5kb). This variability in neocentromere size is more similar to that observed for human neocentromeres [12]. The observed plasticity in neocentromere size could simply reflect the absence of genomic features that repress de novo centromere formation, given that Candida neocentromeres frequently form in large, intergenic regions [28]. Targeting CENP-A to intergenic regions that are variable in size and chromatin enrichment, while simultaneously deleting the endogenous centromere, could address this question.
Centromere assembly and replication timing: cause or effect?
An intriguing property of centromeres is that they replicate at a different time than bulk DNA. In the yeasts, S. cerevisiae, S. pombe, and C. albicans, centromeres replicate early in S. phase. Such early replication of centromeres appears to be crucial for proper kinetochore assembly in S. cerevisiae [30] and in S. pombe where it is regulated by the centromere protein Swi6 [31]. In fungi at least, early replicating domains may be preferred sites of neocentromere formation over late replicating domains. CENP-A loading in early S might drive neocentromere formation at early replicating sites. However, neocentromere formation at a late replicating domain in C. albicans created a replication shift to early S phase [32], suggesting that replication timing alone is not a primary determinant of de novo centromere assembly. More recent studies corroborate this finding in S. cerevisiae. The repositioning of the chromosome XIV centromere from its endogenous locus to a late replicating domain not only results in a functional centromere, but also shifts timing of replication to early S phase [33]. Thus, it appears that in these organisms, replication timing is an inherent property of endogenous centromeres that can be transferred to neocentromeres.
In contrast to fungal centromeres, replication of centromeres in vertebrates and other multicellular organisms occurs in mid-to late S phase [34–36]. Perhaps CENP-A loading at neocentromeres in yeast is linked to replication timing. This might explain why early replicating regions are preferred sites of neocentromere formation, especially in C. albicans [32]. In the DT40 neocentromere studies, one neocentromere formed at an already late replicating domain, and did not alter replication timing [25]. However, two other neocentromeres formed in early replicating domains that shifted to late upon neocentromere formation. Similarly, human neocentromere formation on chromosome 10 shifts replication timing of the region to a later time [37]. The mechanism by which centromere assembly alters replication timing - either late to early in fungi or early to late in metazoans – remains unclear. Engineered neocentromeres in fungi and chicken provide controllable experimental systems to now explore the effects of replication timing on centromere assembly and vice versa.
De Novo Centromeres and Transcription
Because they are typically embedded in pericentric heterochromatin, sites of kinetochore assembly were historically presumed to lack transcriptional activity. However, on the heels of discoveries that pericentric heterochromatin domains can be transcriptionally active in fission yeast [38, 39], landmark studies in maize demonstrated that DNA interwoven with CENP-A-containing nucleosomes is permissive to RNA Polymerase II (RNAPII) mediated transcription [40]. Over the past decade, transcripts homologous to the primary sequence underlying native kinetochore assembly sites have been identified in the yeasts S. cerevisiae [41] and S. pombe [42], rice [43, 44], mouse [45, 46], tammar wallaby [47], and humans [48]. Furthermore, centromeres present on human artificial chromosomes (HACs) are likewise transcriptionally active [18, 49, 50]. Defining the types and properties of centromere-derived transcripts, including both endogenous genes and non-coding RNAs (ncRNAs), is the next challenge in understanding centromeric transcription [40, 43–45, 47].
There are important links between the level of RNAPII transcriptional activity at CENP-A-containing chromatin domains and centromere identity and function. An emerging “Goldilocks” model of centromeric transcription in both unicellular and multi-cellular eukaryotes posits that transcription that is too high or too low negatively affects centromere function. Instead, a “just right” amount is important for proper centromere assembly and chromosome segregation [51]. In humans, studies have taken advantage of easily manipulated HACs to demonstrate that targeting of transcriptional activators to a HAC core domain not only alters gene expression, but also modifies chromatin structure and HAC stability [49, 50]. When HAC transcriptional activity was reduced, CENP-A incorporation and mitotic stability were significantly compromised [18]. Where it has been studied, low transcriptional activity is also a feature of endogenous centromeres. For example, experimental manipulation of core domain transcription results in chromosome missegregation and lagging chromosomes in both S. cerevisiae and tammar wallaby [41, 47]. Likewise, treatment of mammalian cells with inhibitors of RNAPII compromises centromere function [52]. Studies of endogenous centromeres in S. pombe support and extend the conclusion that a low level of transcription is a normal feature of eukaryotic centromeres [42].
In light of these findings, it is not surprising that neocentromeres in both C. albicans and G. gallus frequently form adjacent to genes or predicted genes [25, 26, 28]. Furthermore, recent studies predict that ORFs associated with neocentromere formation are transcriptionally active. The steady state transcript level of neocentromere adjacent genes is strongly reduced upon neocentromere formation in yeast [26, 28]. Similarly, in S. pombe, neocentromere-adjacent genes that are typically induced by nitrogen starvation remain repressed upon nitrogen depletion [53]. In chicken cells, changes to gene transcription after neocentromere formation are less obvious, because neocentromeres form over both transcriptionally active and inactive genomic regions [25]. Unfortunately, at most loci in the chicken neocentromere study, the transcriptional activity of genes could not be ascertained due to technical limitations, although at one testable locus, transcription was down regulated.
Whether transcriptional effects are causes or consequences of neocentromere assembly remains an unanswered question. Intriguingly, C albicans neocentromeres assembled at or near the URA3 reporter gene can move locally in response to experimental manipulation of growth conditions that change the amount of URA3 transcription. Increased transcriptional activity prohibits CENP-A incorporation, whereas transcriptional repression results in CENP-A association at gene promoters [26, 28]. Of the few human neocentromeres that have been studied, the mardel(10) neocentromere showed a distinct correlation between centromere function and LINE-1 transcription [54]. Although it remains to be formally tested, the transcriptional activity associated with heterochromatin formation in S. pombe [55] may also contribute to the site of neocentromere formation.
Chromatin environments that favor new centromere formation
In fission yeast, neocentromeres rarely form adjacent to the excised endogenous centromere [53]. This is likely due to the nature of the specific engineered deletions that removed both CENP-A and pericentric heterochromatin domains in the neocentromere studies. Low levels of CENP-A and/or heterochromatin would not be expected outside of the excised regions, and as a result, neocentromeres might preferentially assemble at subtelomeric regions that do contain heterochromatin [53, 56]. These findings imply that a distinct chromatin environment promotes neocentromere assembly. Indeed, de novo centromere assembly on circular artificial chromosomes in S. pombe requires the presence of pericentric heterochromatin [57]. Similarly in Drosophila, genomic regions near or within heterochromatin are preferred sites of neocentromere formation [22, 24, 58, 59]. Even some human neocentromeres are located in or near heterochromatic regions, such as the acrocentric short arms [60]. Nevertheless, other human neocentromeres are formed in non-heterochromatic regions, and de novo centromeres in C. elegans are assembled in the absence of heterochromatin [61]. While heterochromatin may strongly promote or support neocentromere formation, it is not the only type of chromatin environment in which neocentromere assembly can occur. Thus, questions regarding the perfect environment for neocentromere formation remain to be experimentally addressed.
A recent study in S. pombe suggests that regions depleted for nucleosomes that contain H2A.Z are particularly suited for neocentromere formation [56]. Indeed, increased neocentromere formation in fission yeast was observed at regions lacking H2A.Z, suggesting that CENP-A and H2A.Z are typically not present in the same nucleosomes. These studies suggested that Scm3/HJURP has decreased affinity for nucleosomes containing H2A.Z, which consequently inhibits new CENP-A incorporation. Since heterochromatin contains little H2A.Z, a feasible model is that centromeric and telomeric heterochromatin promotes maturation of new centromere formation once CENP-A incorporation has occurred. If CENP-A is aberrantly loaded into sites that contain little or no H2A.Z or in regions that experience high histone turnover, neocentromere formation may be more easily seeded and reinforced by continued, efficient recruitment of Scm3/HJURP.
Mechanisms that Counter Spontaneous Neocentromere Formation
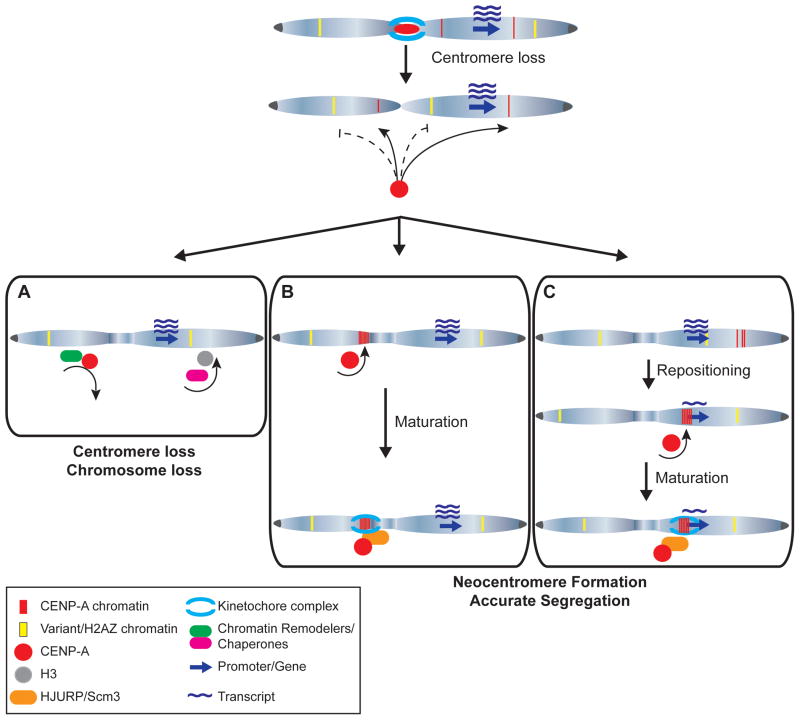
Low levels of CENP-A are found at non-centromeric sites in multiple organisms, including promoters, yet these regions do not mature to fully functional centromeres. And in instances in which CENP-A is over-expressed or tethered at specific genomic regions, only partial centromere assembly occurs [21–23, 59]. In fungi, transient neocentromeres can form that contain lower (<15%) amounts of CENP-A [28, 56]. However, these immature or incomplete centromeres disappear and relocate, either naturally or under stress conditions to more favorable genomic regions where they become more enriched for CENP-A [28, 56]. An open question, then, is why new centromeres do not arise regularly throughout the genome. Several lines of evidence indicate that multiple mechanisms protect the genome against de novo centromere formation (Figure 3).
Figure 3. The formation and fate of de novo centromeres arising at atypical genomic locations.
Non-centromeric chromosomal loci contain low levels of CENP-A (red) and histone variants, including H2A.Z (yellow). Upon centromere loss, CENP-A is preferentially incorporated at existing CENP-A loci, whereas H2A.Z may guard against CENP-A incorporation. (A) Chromatin remodeling complexes and histone H3 chaperones monitor local chromatin structure and evict misincorporated CENP-A, resulting in centromere loss. (B) Alternatively, following centromere loss, CENP-A is incorporated at loci already containing a low level of CENP-A or other chromatin structures permissive to neocentromere formation, such as heterochromatin. HJURP association enables maturation of incomplete centromeres, followed by recruitment of centromere and kinetochore proteins necessary for neocentromere function. (C) Failure to recruit a sufficient amount of CENP-A in diploid organisms can result in incomplete neocentromere formation, which may be corrected by repositioning that results in CENP-A incorporation at a more favorable site or by homologous recombination (not shown). Neocentromeres can form, perhaps preferentially, within or adjacent to genes, resulting in reduced transcriptional activity adjacent to the mature neocentromere.
CENP-A deposition at centromeres of eukaryotic centromeres corresponds with several events, including its own transcription, the availability of chaperones that load it into chromatin, and regulation of the CENP-A assembly machinery by cyclin-CDK complexes [62–64]. For instance, the Drosophila centromere protein CAL1 that shares homology with HJURP and Scm3 is present in limiting amounts during the cell cycle to ensure that CENP-A/CID assembly occurs appropriately [65]. In addition, chromatin remodelers participate in both the incorporation of CENP-A at centromeres [66] and in the preservation of H3 chromatin, thereby ensuring that CENP-A is not incorporated at non-centromeric sites [42]. At all times, the cell is surveying H3 chromatin and misincorporated CENP-A. Since promoter regions often contain higher than average amounts of H2A.Z, this variant histone may also help to prevent inappropriate CENP-A deposition [56]. Neocentromere formation may represent instances in which even slight perturbations in chromatin regulation or genome surveillance allow CENP-A to encroach into unauthorized genomic regions.
Although excess or inappropriately incorporated CENP-A can lead to partial or complete centromere formation, mechanisms exist to evict mislocalized CENP-A. Ubiquitin-mediation proteolysis has been demonstrated to prevent CENP-A misincorporation and effectively control normal CENP-A levels in several organisms [67–72]. If chromatin remodelers or E3 ubiquitin ligases are mutated or ineffective, a critical mass of misincorporated CENP-A may remain in certain genomic regions. Indeed when CENP-A/Cse4p is over-expressed in S. cerevisiae strains mutated for Psh1, a E3 ubiquitin ligase, misincorporated CENP-A/Cse4p is not removed from non-centromeric loci [73]. CENP-A misincorporation and/or failure in eviction may represent an early step in new centromere formation. As CENP-A persists in a new location, levels of H2A.Z or other restrictive chromatin marks may decrease, allowing the neocentromere to mature, perhaps in concert with enrichment for permissive chromatin, such as heterochromatin or H3K4me/H3K36me. The minimal level of CENP-A that can bypass or escape eviction and proteolysis remains to be tested, although some studies suggest that only a few molecules of CENP-A can maintain centromere function [18, 28, 29].
Understanding the molecular switch between new centromere formation and centromere suppression is relevant beyond neocentromere biology. Similar mechanisms might underlie centromere inactivation in de novo dicentric chromosomes and, when ineffective or mutated, might explain why some dicentrics fail to inactivate the second centromere [2, 74–76]. CENP-A is also over-expressed in many cancers [77, 78]. It is tempting to speculate that surveillance/eviction machinery might be compromised in these cells, and neocentromeres may arise more often and contribute to genome instability that is a hallmark of cancers.
Finally, a new view of centromere maintenance has emerged from C. albicans in which genomic mechanisms related to centromere or chromosome pairing protect against new centromere formation [26]. Deletion of endogenous CEN7 led to neocentromere formation, but in a fraction of strains, the centromere was restored at the endogenous locus by gene conversion through recombination with CEN7 on the unaltered homolog. Notably, the C. albicans neocentromeres that “disappeared” contained lower amounts of CENP-A compared to the original centromere (Figure 2). These findings suggest that neocentromere formation in diploid organisms probably happens more often than appreciated. These events may go unobserved because incomplete/immature centromeres are reverted or removed, and endogenous centromere function is restored by recombination (Figure 3). Gene conversion has been reported at both budding yeast and maize centromeres [79, 80], and is thought to occur at human centromeres, although the latter has been more difficult to study. In light of the new findings in C. albicans, models of centromere stability now include recombination-based mechanisms that maintain centromere location, diversify centromeric DNAs, and suppress propagation of unfavorable or disadvantageous new centromeric locations, perhaps based on the amount or extent of CENP-A incorporation.
Final remarks
The ability to efficiently engineer and recover neocentromeres in both fungal and vertebrate wild-type cells represents a powerful strategy to study the establishment and maintenance of de novo centromeres. Recent studies have provided insight into some genomic and epigenetic factors that promote de novo centromere formation, but many intriguing questions still remain (see Box 2). It will be important to define roles for transcription, replication, and chromatin environment in neocentromere formation. Such studies have implications not only for basic centromere and chromosome biology, but also for developing strategies to create controllable centromeres or repress centromere function for therapeutic applications and disease treatments.
Box 2. Questions Outstanding in Neocentromere Research.
Does replication timing direct centromere specification or does centromere assembly trigger a change in replication dynamics?
Do neocentromeres preferentially assemble near origins of replication, at non-coding RNAs, and/or within domains enriched for cohesins?
Do neocentromeres non-randomly arise next to centromeres defined by repetitive DNA?
Can neocentromeres arise in organisms with genetically determined centromeres?
How do diseased cell states influence neocentromere formation?
Does primary incorporation of ectopic CENP-A occur at sites of DNA damage?
Do neocentromeres preferentially assemble within specific nuclear locations/territories?
What are the molecular mechanisms that control the maturation of centromeres from incomplete sites of CENP-A incorporation to fully functional centromeres?
Is there a molecular difference between incomplete and repressed (neo)centromeres?
Highlights.
Deletion of native centromeres induces neocentromeres in fungi and chicken cells
Neocentromeres are preferentially formed near original centromeres.
Low levels of CENP-A at non-kinetochore sites can seed neocentromere formation.
Some neocentromeres never mature to become fully functional centromeres.
Gene conversion in C. albicans can reverse neocentromere formation.
Acknowledgments
We apologize to our colleagues whose work on centromeres and neocentromeres was not acknowledged due to space constraints. We thank Megan Aldrup-MacDonald for critical reading of the manuscript. The Scott lab is supported by Duke Institute for Genome Sciences & Policy. Sullivan lab research is funded by grants R01 GM098500 (NIH) and #1-FY13-434 from the March of Dimes Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maddox PS, et al. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- 2.Stimpson KM, et al. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res. 2012;20:595–605. doi: 10.1007/s10577-012-9302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrack LS, Berman J. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res. 2012;20:607–619. doi: 10.1007/s10577-012-9296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 5.Depinet TW, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- 6.Voullaire L, et al. Trisomy 20p resulting from inverted duplication and neocentromere formation. Amer J Med Genet. 1999;85:403–408. [PubMed] [Google Scholar]
- 7.Voullaire LE, et al. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 8.Amor DJ, et al. Human centromere repositioning “in progress”. Proc Natl Acad Sci USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasson D, et al. Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres. Chromosoma. 2011;120:621–632. doi: 10.1007/s00412-011-0337-6. [DOI] [PubMed] [Google Scholar]
- 10.Marshall OJ, et al. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso A, et al. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alonso A, et al. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum Mol Genet. 2003;12:2711–2721. doi: 10.1093/hmg/ddg282. [DOI] [PubMed] [Google Scholar]
- 13.Kim SM, et al. Early-replicating heterochromatin. Genes Dev. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earnshaw WC, et al. Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant. Chromosome Res. 2013;21:101–106. doi: 10.1007/s10577-013-9347-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choo KH. Domain organization at the centromere and neocentromere. Dev Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 17.Partridge JF, et al. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 18.Bergmann JH, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struc Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chueh AC, et al. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum Mol Genet. 2005;14:85–93. doi: 10.1093/hmg/ddi008. [DOI] [PubMed] [Google Scholar]
- 21.Barnhart MC, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heun P, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Hooser AA, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 24.Olszak AM, et al. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- 25.Shang WH, et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell. 2013;24:635–648. doi: 10.1016/j.devcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur J, Sanyal K. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 2013;23:638–652. doi: 10.1101/gr.141614.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanyal K, et al. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci USA. 2004;101:11374–11379. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ketel C, et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLOS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fachinetti D, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamura E, et al. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi MT, et al. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol. 2009;11:357–362. doi: 10.1038/ncb1845. [DOI] [PubMed] [Google Scholar]
- 32.Koren A, et al. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLOS Genet. 2010;6:e1001068. doi: 10.1371/journal.pgen.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohl TJ, et al. Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae. PLOS Genet. 2012;8:e1002677. doi: 10.1371/journal.pgen.1002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hultdin M, et al. Replication timing of human telomeric DNA and other repetitive sequences analyzed by fluorescence in situ hybridization and flow cytometry. Exp Cell Res. 2001;271:223–229. doi: 10.1006/excr.2001.5391. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan B, Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J Cell Biol. 2001;154:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ten Hagen KG, et al. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol Cell BIol. 1990;10:6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo AW, et al. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 2001;20:2087–2096. doi: 10.1093/emboj/20.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volpe T, et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 39.Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Letters. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 40.Topp CN, et al. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohkuni K, Kitagawa K. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr Biol. 2011;21:1695–1703. doi: 10.1016/j.cub.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi ES, et al. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLOS Genet. 2012;8:e1002985. doi: 10.1371/journal.pgen.1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan H, et al. Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell. 2006;18:2123–2133. doi: 10.1105/tpc.106.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan H, et al. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell. 2005;17:3227–3238. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouzinba-Segard H, et al. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferri F, et al. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucl Acids Res. 2009;37:5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carone DM, et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118:113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 48.Wong LH, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardinale S, et al. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20:4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano M, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohkuni K, Kitagawa K. Role of transcription at centromeres in budding yeast. Transcription. 2012;3:193–197. doi: 10.4161/trns.20884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan FL, et al. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci USA. 2012;109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishii K, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 54.Chueh AC, et al. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLOS Genet. 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 56.Ogiyama Y, et al. Epigenetically-induced paucity of histone H2A.Z stabilizes fission yeast ectopic centromeres. Nat Struc Mol Biol. 2013 doi: 10.1038/nsmb.2697. [DOI] [PubMed] [Google Scholar]
- 57.Kagansky A, et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maggert KA, Karpen GH. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mendiburo MJ, et al. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 60.Klein E, et al. Five novel locations of Neocentromeres in human: 18q22.1, Xq27.1 approximately 27.2, Acro p13, Acro p12, and heterochromatin of unknown origin. Cytogenet Genome Res. 2012;136:163–166. doi: 10.1159/000336648. [DOI] [PubMed] [Google Scholar]
- 61.Yuen KW, et al. Rapid de novo centromere formation occurs independently of heterochromatin protein 1 in C. elegans embryos. Curr Biol. 2011;21:1800–1807. doi: 10.1016/j.cub.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jansen LE, et al. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shelby RD, et al. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silva MC, et al. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 65.Mellone BG, et al. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLOS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warburton PE, et al. Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol. 1993;13:6520–6529. doi: 10.1128/mcb.13.10.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collins KA, et al. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 68.Gross S, et al. Centromere architecture breakdown induced by the viral E3 ubiquitin ligase ICP0 protein of herpes simplex virus type 1. PLOS ONE. 2012;7:e44227. doi: 10.1371/journal.pone.0044227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lermontova I, et al. Arabidopsis KINETOCHORE NULL2 Is an Upstream Component for Centromeric Histone H3 Variant cenH3 Deposition at Centromeres. Plant Cell. 2013;25:3389–3404. doi: 10.1105/tpc.113.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lomonte P, et al. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J Biol Chem. 2001;276:5829–5835. doi: 10.1074/jbc.M008547200. [DOI] [PubMed] [Google Scholar]
- 71.Moreno-Moreno O, et al. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucl Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thakur J, Sanyal K. A coordinated interdependent protein circuitry stabilizes the kinetochore ensemble to protect CENP-A in the human pathogenic yeast Candida albicans. PLOS Genet. 2012;8:e1002661. doi: 10.1371/journal.pgen.1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ranjitkar P, et al. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Earnshaw WC, et al. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 75.Stimpson KM, et al. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLOS Genet. 2010;6 doi: 10.1371/journal.pgen.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sullivan BA, Willard HF. Stable dicentric X chromosomes with two functional centromeres. Nat Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- 77.Tomonaga T, et al. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 78.Wu Q, et al. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012;77:407–414. doi: 10.1016/j.lungcan.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 79.Shi J, et al. Widespread gene conversion in centromere cores. PLOS Biol. 2010;8:e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Symington LS, Petes TD. Meiotic recombination within the centromere of a yeast chromosome. Cell. 1988;52:237–240. doi: 10.1016/0092-8674(88)90512-0. [DOI] [PubMed] [Google Scholar]