Abstract
Primary HIV-1 isolates are relatively resistant to neutralization by antibodies commonly induced after infection or vaccination. This is generally attributed to masking of sensitive epitopes by the V1/V2 domain and/or glycans situated at various positions in Env. Here we identified a novel masking effect mediated by subtype C-specific V3 sequences that contributes to the V1/V2-independent and glycan-independent neutralization resistance of chimeric and primary Envs to antibodies directed against multiple neutralization domains. Positions at several conserved charged and hydrophobic sites in the V3 crown and stem were also shown to affect neutralization phenotype. These results indicated that substitutions typically present in subtype C and related V3 sequences influence the overall conformation of native Env in a way that occludes multiple neutralization targets located both within and outside of the V3 domain, and may reflect an alternative mechanism for neutralization resistance that is particularly active in subtype C and related isolates.
Keywords: HIV-1, Subtype C, V3 domain, Conformational effect, Epitope masking
Introduction
The V3 region is a critical component of the co-receptor binding site and a major determinant of viral tropism (Hartley et al., 2005; Hwang et al., 1991; Shioda et al., 1992). Despite the diversity in the V3 domain, this region retains conserved structural features that presumably are required for these functions. Common features in the V3 domain include the presence of a disulfide link at the base of the loop, a relatively conserved length of approximately 35 amino acids, the presence of N-linked glycosylation sites external to one or both of the cysteines, an additional N-linked glycosylation site at the sixth residue of the loop, a number of conserved hydrophobic and basic residues, an overall positive charge, and the presence of an α or β turn at the tip of the loop most commonly mediated by a GPGR/Q motif. The recognition of such conserved features contributes to the broad cross-reactivity of a number of monoclonal V3-specific antibodies isolated from patients infected with HIV-1 (Jiang et al., 2010).
The crown of the V3 loop is an immunodominant region of HIV-1 Env, and high affinity antibodies specific for epitopes in this region are common in sera of almost all infected subjects (Baillou et al., 1993; Davis et al., 2009b; Krachmarov et al., 2005; LaRosa et al., 1990; Spear et al., 1994) and are readily induced by a variety of HIV-1 Env-based immunogens (Letvin et al., 2001; Li et al., 2006; Smith et al., 1998; Wu et al., 2006; Zolla-Pazner et al., 2008). Consistent with the strong immunogenicity of this region, anti-V3 antibodies tend to be closely related to germ-line Ig sequences (Andrabi et al., 2013; Gorny et al., 2009). This is in contrast to more recently isolated mAbs possessing broad and potently neutralizing activities (Moore et al., 2011; Walker et al., 2009, 2011; Wu et al., 2010), which are typically found only after several years of infection and require high levels of somatic mutations and unusually large CDR3 regions (Klein et al., 2013; Pancera et al., 2010; Pejchal et al., 2010; Walker et al., 2011; Zhou et al., 2010). Whereas the binding specificities of some anti-V3 mAbs are limited to certain strains and/or structural motifs, many V3-specific antibodies induced by infection in humans possess broad reactivities that extend across multiple isolates and subtypes (Gorny et al., 1997, 2002, 2004). Early studies showed that certain laboratory-cultured strains were highly susceptible to neutralization by anti-V3 antibodies (Carrow et al., 1991; Javaherian et al., 1990), and many V3-specific mAbs neutralized such isolates considerably more potently than mAbs against epitopes located in conserved domains of the gp120 molecule (Krachmarov et al., 2005). However, the majority of primary isolates are resistant to neutralization by anti-V3 mAbs (Bou-Habib et al., 1994; Spenlehauer et al., 1998; Vancott et al., 1995), and this is now understood to be largely due to masking of the V3 domain, primarily mediated by the V1/V2 domain (Krachmarov et al., 2006; Pinter et al., 2004) and by N-linked glycans located at other positions in Env (Wei et al., 2003).
HIV-1 can be divided into multiple subtypes and recombinant forms, which differ in their sequences and geographical distribution (Taylor et al., 2008). The initial HIV-1 isolates were from subtype B, which is dominant in the Americas and Europe, and much of the available information about HIV Env structure and immunology is based on this subtype. However, subtypes A, C and D account for 65% of worldwide HIV-1 infections, with subtype C alone being responsible for almost half of all global infections (Hemelaar et al., 2006), and it is therefore important to better understand the structural and immunological properties of these Envs. Analysis of available V3 sequences revealed that the subtype C V3 domain is more conserved than the subtype B V3 domain (Gaschen et al., 2002; Korber et al., 1994; Patel et al., 2008).
The subtype B and C consensus sequences differ at five positions within the V3 loop, and in the presence or absence of a potential N-linked glycosylation site flanking the N-terminal cysteine of the loop. A highly characteristic feature of subtype B is the presence of an Arg residue at position 18 at the tip of the V3 loop (GPGR) versus a Gln residue (GPGQ) for subtype C and other subtypes. The potent binding affinity of some subtype B-derived V3-specific mAbs (e.g. 447-52D) is dependent on the presence of Arg18 (Zolla-Pazner et al., 2004). These mAbs bind to many subtype B sequences with high affinities and neutralize a small percentage of such viruses, but possess little reactivity for most non-B-derived HIV isolates. This fact contributed to the perception that V3 mAbs were type-specific and possessed limited cross-reactivities. However, many V3-specific antibodies are less or not at all dependent on the residue at the tip of the loop, and consequently possess broader cross-reactivities (Gorny et al., 2006; Krachmarov et al., 2005). This is particularly the case for mAbs induced upon infection with non-B HIV-1 subtypes.
Although V1/V2 masking is a major determinant of resistance to neutralization by anti-V3 antibodies (Krachmarov et al., 2006; Pinter et al., 2004), this does not preclude the possibility that regions outside of V1/V2 can also induce neutralization resistance through alternate masking mechanisms. It has been reported that substituting the subtype B and C consensus V3 sequences into the SF162 Env backbone, which has relatively little V1/V2 masking activity, resulted in substantial differences in neutralization sensitivity to anti-V3 antibodies, with the consensus B V3 (V3ConB) chimera being much more sensitive to neutralization by polyclonal immune sera and a broad range of V3-specific mAbs than the consensus C V3 (V3ConC) chimera (Almond et al., 2012; Krachmarov et al., 2006). It has also been reported that some subtype B derived anti-V3 mAbs bound to both subtype B and C-derived peptides but only to subtype B gp120, suggesting a difference in conformation between the subtype C peptide and gp120 forms, and between subtype B and subtype C gp120s (Patel et al., 2008). To better understand the contribution of V3 sequence variations towards the immunoreactivities and neutralization sensitivities of HIV-1 Env, the native subtype B and C V3 domains were expressed as soluble fusion glycoproteins by joining the complete V3 coding domain to the N-terminus of a gene coding for a secreted form of the rabbit Fc sequence. This vector allows for correct folding and glycosylation of the V3 domain, consistent with previous studies showing that these recombinant V3 antigens more accurately display native V3 antigens than corresponding peptides (Davis et al., 2009a, 2009b). The two V3 sequences were also expressed in two common gp120 backbones, to allow equivalent binding assays in the context of the gp120 protein. Binding and neutralization studies were performed in parallel with a large panel of V3-specific mAbs derived from human subjects infected with HIV-1 isolates of various subtypes. These studies revealed a general discrepancy between the binding and neutralizing activities of these mAbs against the subtype C consensus V3 sequence, and the mechanistic basis of this discrepancy was explored by the studies described below.
Results
The subtype C V3 sequence is relatively resistant to neutralization by V3-specific antibodies
The effect of V3 sequence on neutralization sensitivity was initially examined in a series of neutralization assays using poly-clonal HIV-1 immune sera from chronically-infected subjects. The neutralization activities of these sera were compared for two viral pseudotypes containing SF162 Envs in which the V3 region was replaced with either the subtype B or subtype C consensus sequences. The V3ConC chimeric Env was substantially less sensitive to neutralization than the V3ConB Env, with a median ND50 for a panel of 25 immune sera of 1/781 compared to 1/38,000 for the V3ConB chimeric Env, corresponding to an almost 50-fold mean ratio of ND50 for the two pseudoviruses (Supplemental Fig. 1). A possible explanation for the preferential neutralizing activity of the sera for the V3ConB Env was that this was mediated by the presence of V3-specific antibodies that preferentially recognized the subtype B V3 sequence. To model this, the neutralization sensitivities of the two chimeric Envs were examined against a large panel of V3-specific mAbs derived from HIV-1-infected subjects from different geographical areas including US subjects infected with subtype B isolates, African subjects predominantly infected with CRF02_AG viruses and Indian subjects infected with subtype C and other non-B subtypes (Gorny et al., 2006, 2009).
Consistent with the effect seen for the polyclonal sera, mAbs derived from subjects in the US all possessed highly potent neutralizing activities for the V3ConB chimeric Env, with IC50s below 0.006 μg/ml (Fig. 1). These antibodies had much weaker activities for the V3ConC chimeric Env, with IC50s ranging from 0.4 μg/ml to >20 μg/ml, corresponding to >100-fold to >18,000-fold higher IC50 end-points for the V3ConC chimeras. However, most of the mAbs derived from West African subjects were also quite potent against the V3ConB Env pseudotype, with IC50s below 0.01 μg/ml for 9 of 11 of these mAbs. With one exception (2182), these mAbs neutralized the V3ConC Env considerably less strongly (although more effectively than the US-derived mAbs), with IC50s in the range of 0.1–1.8 μg/ml, corresponding to 36 to 350-fold higher concentrations than required for the V3ConB chimera. 2182 had no detectable activity against the V3ConC pseudotype. This mAb was derived from an unusual subtype A sequence that contained the GPGR sequence, and was previously shown to be extremely dependent on the presence of R18 at the tip of the V3 loop (Gorny et al., 2006; Krachmarov et al., 2006). A different pattern was seen for most of the mAbs derived from Indian patients. While one of these mAbs (3792) preferentially neutralized the V3ConB Env (with a 57-fold lower IC50), the other mAbs either possessed similarly weak activities for both chimeric Envs, or preferentially neutralized the V3ConC chimera by 4 to 14-fold.
Fig. 1.
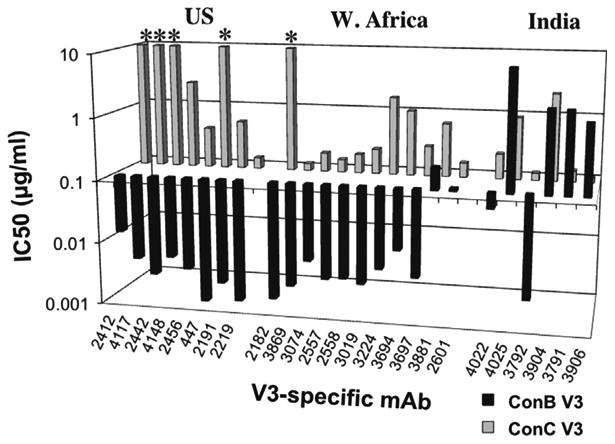
IC50s of V3-specific mAbs for SF162 Env with ConB and ConC V3s. IC50s of V3-specific mAbs for pseudovirions containing chimeric SF162 Envs with V3 sequences corresponding to various subtype consensus sequences. The antibodies are divided according to their origins. Values marked by * indicate that 50% neutralization was not reached at the highest concentration tested (10 μg/ml).
Lack of correlation between neutralizing activities of anti-V3 mAbs and binding to native V3 fusion glycoproteins
The fact that most of the West African-derived mAbs also possessed preferential neutralizing activity for the V3ConB chimeric Env showed that the greater sensitivity of this Env was not limited to subtype B-derived antibodies, suggesting that this effect could not simply be explained by the binding specificities of these antibodies. To explore this issue more extensively, the relative binding affinities of a number of V3-specific mAbs were compared for the V3ConB and V3ConC domains expressed as fusion glycoproteins, and compared to their neutralizing activities for viral pseudotypes containing the corresponding chimeric Envs. These V3 fusion glycoproteins contained the disulfide bond at the base of the V3 loop and the associated N-linked glycans located within the V3 loop and immediately flanking the two cysteines at the base, and were fused at their C-terminus to a dimeric rabbit Fc sequence. These fusion glycoproteins have previously been shown to absorb all of the V3-specific neutralizing activities of mAbs and polyclonal sera from infected and vaccinated subjects whereas synthetic V3 peptides with related sequences were not able to do so (Davis et al., 2009a, 2009b), and thus are considered to more accurately model the native V3 structure.
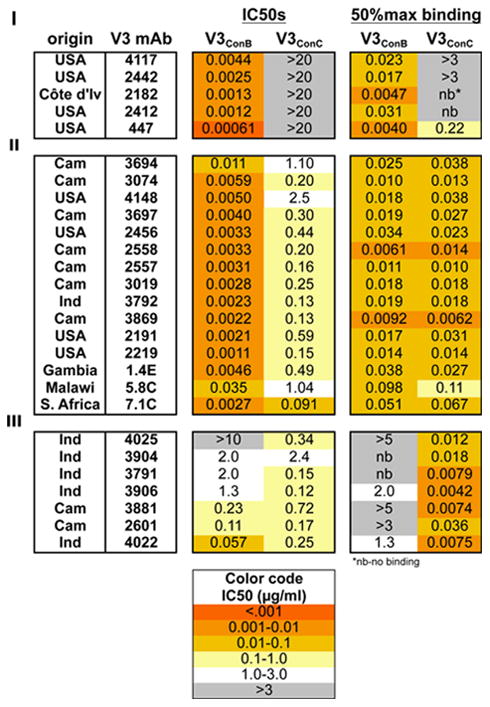
Binding and neutralization patterns for a total of 27 V3-specific mAbs of different origins are tabulated in Table 1, with representative patterns shown in Fig. 2. Three different binding/neutralization patterns were observed for these antibodies. The group I mAbs (represented by 2182 and 2412 in Fig. 2-I) selectively bound and neutralized the subtype B chimeric Env, but had neither activity against the subtype C chimera, even at the highest concentration tested. It was therefore concluded that for this group of mAbs resistance to neutralization was due to the absence of the epitope. The mAbs in group II (3074, 2456 and 2191 in Fig. 2-II), had identical binding affinities for the two fusion glycoproteins but nevertheless preferentially neutralized the V3ConB chimeric Env over the V3ConC chimeras (IC50 ratios ranging from 30 to 500-fold). This group represented the majority of antibodies tested, and included antibodies derived from infection with different HIV-1 subtypes. This showed that the greater resistance of the V3ConC chimera to neutralization by these mAbs was not due to a reduced binding affinity to the isolated epitope, but rather to some other effect. The group III mAbs (represented by 2601 and 3881 in Fig. 2-III) bound preferentially to the subtype C protein but weakly neutralized the two groups with relatively small differences in potency (1 to 13-fold differences in IC50, with occasional preferences for the V3ConC chimeric Env). Thus, for this group as well, there was discordance between binding affinity of the mAbs and their neutralizing potencies for the V3ConC chimera.
Table 1.
IC50s and 50% maximal binding concentrations for 27 V3-specific mAbs versus V3ConB and V3ConC. IC50s (μg/ml) of V3-specific mAbs are categorized according to the binding properties for the subtype consensus V3 fusion glycoproteins (some of the neutralization data is from (Krachmarov et al., 2006)), and 50% maximal binding concentrations were determined for the V3 fusion glycoproteins. For cases when binding did not reach a plateau level, 50% values were estimated based on the plateau levels obtained for the same antibody against the more sensitive fusion glycoprotein. For antibodies with no detectable binding, ‘nb’ (no binding) is indicated. MAbs in group I selectively bound the V3ConB fusion glycoproteins, those in group II bound the two subtypes equally well and those in group III preferentially bound the subtype C fusion protein. The mAbs were isolated from patients in the USA, Africa (Cameroon, Côte d'Ivoire, Gambia, Malawi or S. Africa) and India. For more accurate comparisons, the V3ConC chimera contained Asn295, introducing an N-linked glycan common at this position for subtype B isolates but typically absent for subtype C isolates.
|
nb-no binding
Fig. 2.
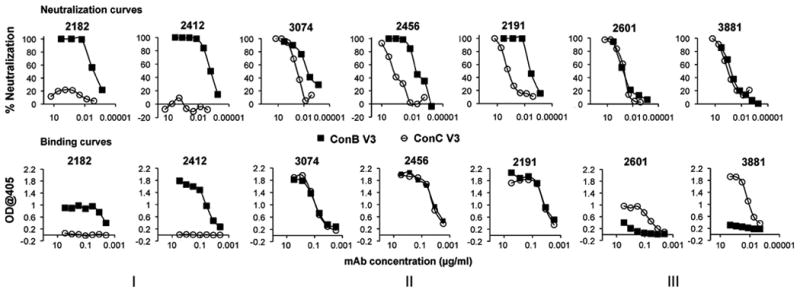
Comparison of neutralization sensitivity and binding affinity of V3 mAbs for V3B and V3C sequences. Neutralization assays (upper row) were performed for various V3 specific mAbs against HIV virions pseudotyped with SF162 Env containing either the V3ConB sequence (closed squares) or the V3ConC sequence (open circles). Corresponding binding assays were performed for the same mAbs against the full-length V3ConB (closed squares) and V3ConC (open circles) fusion glycoproteins.
V3 sequences also affect the sensitivity of sites in the CD4-binding domain to neutralization
A plausible explanation for the lack of correlation between the binding and neutralization activities described above is that replacement of the subtype B V3 domain with the subtype C sequence stabilized a conformation of the native Env complex which resulted in a reduced exposure of the V3 region on the surface of virions. If so, this might also be expected to affect other conformationally sensitive regions of Env, such as the CD4-binding domain (CD4-bd). To examine this possibility, the V3 domain of SF162 Env was substituted with the consensus V3 sequences of five subtypes, and the relative exposure of sites in the CD4-bd examined by neutralization assays with soluble CD4 (sCD4), and with 5145A and 1125H, two CD4-bd-specific mAbs that were sensitive to masking by the V1/V2 region (Pinter et al., 1993a, 2004; Tilley et al., 1991). Control antibodies tested included b12, a mAb directed against a CD4-bd epitope that was relatively resistant to V1/V2 masking (Pinter et al., 2004), and a pool of anti-V3 mAbs derived from subjects infected with subtype A viruses (αV3A pool).
Significant differences in neutralization sensitivities of the V3 chimeras were seen for all three of the CD4-bd ligands, with the V3ConB chimera being the most sensitive to neutralization, the V3ConC the most resistant, and the other V3 chimeras exhibiting intermediate neutralization sensitivities (Fig. 3A). These effects were greater for the two mAbs than for sCD4. The IC50 ratio for these V3ConB and V3ConC chimeras was 20-fold for sCD4, but exceeded 140- and 250-fold for the two mAbs, compared to 110-fold for the pooled V3 mAbs (Fig. 3B). The different V3 sequence exerted only a 3-fold effect on the neutralization activities of the control mAb, b12, and the rank of sensitivity towards this mAb did not correlate with those for the mAbs against V3 and the standard CD4-bd epitopes. These results indicated that ConC V3 sequence was the most effective at inducing the neutralization-resistant phenotype. The fact that this effect was limited to epitopes that were sensitive to conformational masking was consistent with an effect of V3 sequence on the overall conformation of Env, and not due to overall changes in neutralization sensitivity of Env due to more general factors such as changes in viral fitness.
Fig. 3.
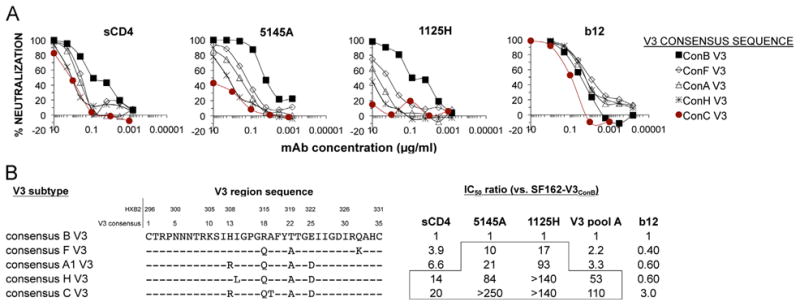
Neutralization efficacy of mAbs targeting the CD4-binding domain. (A) Neutralization curves of sCD4 and mAbs directed against various neutralization domains for HIV-1 pseudotypes with chimeric SF162 Envs containing V3 domains corresponding to consensus sequences of 5 viral subtypes. (B) Consensus V3 sequences for subtypes, and changes in IC50 ratios for sCD4 and mAbs against the chimeric SF162 Envs containing the various V3 domains. Higher ratios reflect reduced neutralization sensitivities.
Lack of effect of V3 sequence on binding of anti-V3 mAbs to monomeric gp120s
The results described above suggested that the increased neutralization resistance towards mAbs specific for the V3 and CD4-binding domains seen upon substitution of the V3ConC sequence may be due to a reduced exposure of these domains in the native trimeric Env complexes present on the surface of infectious viral particles. A previous study had reported a greater exposure of V3 epitopes in a soluble gp120 from a subtype B isolate than in a subtype C gp120 (Patel et al., 2008). That study used two unrelated gp120s and a limited set of subtype B-derived anti-V3 mAbs, and thus the mechanistic basis for this result was not clear. To determine whether the masking activity of the V3ConC sequence described earlier in this study was dependent on the trimeric structure of Env complexes or whether it was also manifested in monomeric gp120, binding studies were performed with matched pairs of monomeric gp120 proteins that differed only in their V3 sequences. Both SF162 and JRFL were used as backbones for the consensus V3 sequences in order to evaluate the effect of the differing levels of V1/V2 masking in these Envs on the exposure of V3 in the monomeric gp120s. These were generated by replacing the V3 region of SF162 with the ConB or ConC sequence and that of JR-FL gp120, which is identical to the V3ConB sequence, with the V3ConC sequence. The purified monomeric gp120 proteins were directly bound to an ELISA plate and analyzed against the set of mAbs shown to bind with equal affinities to the corresponding V3 fusion glycoproteins of which six are shown as representatives in Fig. 4. These antibodies all bound to the V3ConB and V3ConC chimeric gp120s with similar affinities. These results indicated that, contrary to the previous conclusion (Patel et al., 2008), the masking effect of the V3ConC sequence reflected in viral neutralization assays occurred only in the context of the native trimer, and was not seen for monomeric gp120s. Furthermore, this experiment shows that the masking activity of the JR-FL V1/V2 domain on neutralization sensitivity to V3-specific antibodies (Pinter et al., 2004) did not affect the exposure of the V3 domain in soluble, monomeric gp120.
Fig. 4.
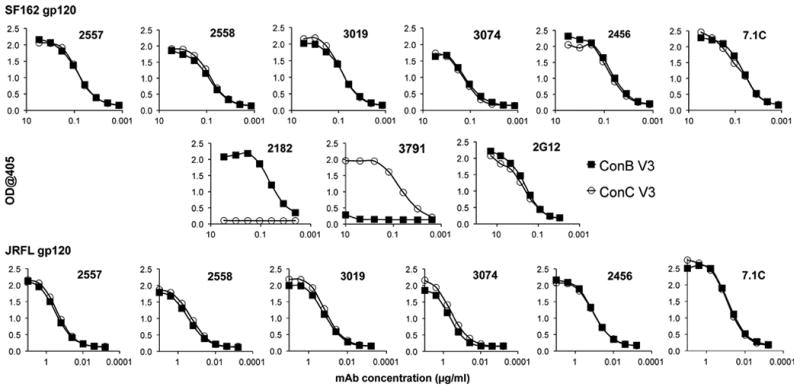
Binding of V3-specific mAbs to monomeric Env proteins containing V3ConB and V3ConC. The reactivity of six representative V3-specific mAbs which bound with equivalent affinities to both ConB and ConC V3-fusion glycoproteins to recombinant SF162 (top row) and JR-FL (bottom row) chimeric gp120 proteins containing either the V3ConB or V3Conc sequence, was measured by ELISA. V3ConB gp120 proteins are represented by closed squares, while V3ConC gp120 proteins are represented by open circles. The middle row shows curves obtained for the positive control, 2G12, and control MAbs specific for the V3ConB (2182) and V3Conc (3791) sequences.
Mapping the contribution of individual substitutions to the masking effect of V3ConC
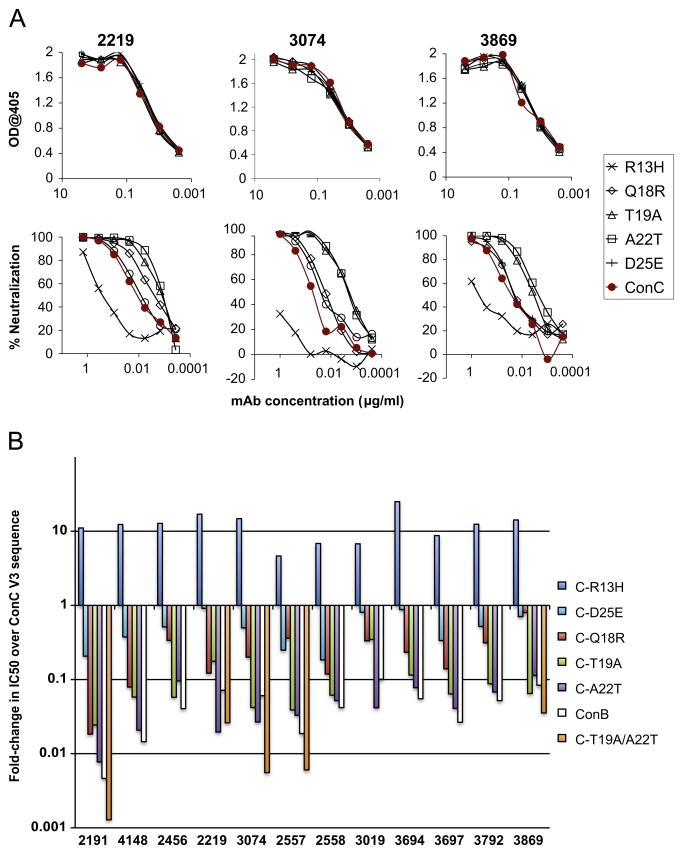
The ConC V3 sequence differs at five positions from the ConB domain (Fig. 3B). The contribution of each of these changes towards the neutralization resistance of the ConC chimera was determined by examining the effect of individual mutations at each of these positions on binding to V3 fusion glycoproteins and neutralization of pseudoviruses with the SF162-V3ConC Envs. Results are shown for a series of V3-specific mAbs that bound equally well to the ConB and ConC V3 fusion glycoproteins but preferentially neutralized the ConB chimera (Fig. 5). Consistent with their equivalent binding activity to the V3ConB and V3ConC fusion glycoproteins, these mAbs all bound with equal affinity to V3 fusion glycoproteins containing the single substitutions (Fig. 5A, top panel), but differed in their neutralization potencies for the mutant Envs (Fig. 5A, bottom panel). The T19A and A22T mutations resulted in significant increases in neutralization sensitivity for all of the mAbs, while the R13H mutation led to a reduction in sensitivity compared to the parental ConC sequence. The extent of the change in IC50 due to these mutations is quantitated in Fig. 5B for a larger set of the anti-V3 mAbs that bound equally well to the V3ConB and V3ConC fusion glycoproteins. The increase in IC50s over the parental V3ConC sequence due to the R13H mutation ranged from 5 to 25-fold, while the decrease in IC50s ranged from 3 to 42-fold for T19A and from 10 to 125-fold for A22T. The combination of the T19A and A22T mutations was usually additive, and the V3ConC double mutant was more sensitive than the V3ConB sequence to neutralization for the five mAbs tested. The D25E mutation had at most a small effect, ranging from no effect to a 5-fold increase in sensitivity, while the Q18R was more variable, ranging from no effect (mAb 3869) to as much as a 55-fold increased sensitivity (mAb 2191).
Fig. 5.
Effect of point mutations on binding and neutralizing activities of V3-specific mAbs. (A) The binding (top panel) and neutralizing (lower panel) activity of three V3-specific mAbs against the wild type V3ConC sequence and five point mutants in which individual ConC-specific residues were converted to those present in the V3ConB sequence. Binding was measured against the mutant V3-Fc fusion glycoproteins, and neutralization activity was determined for the corresponding SF162-V3ConC chimeras. The chimeric Envs used for these experiments did not have a glycosylation site at position 295, unlike in the above experiments, in order to more accurately represent native subtype C sequences. (B) Quantitation of effect of ConC V3 point mutations on neutralization sensitivity. Ratios of IC50s for the SF162-V3ConB chimeric Env or mutant SF162-V3ConC Envs containing one or two substitutions in V3 over the IC50s for the wild-type SF162-V3ConC Env are shown for 12 mAbs that bound with equal affinity to the corresponding V3 fusion glycoproteins. Values are also shown for five mAbs that were tested against the T19A/A22T double mutation.
Effect of specific ConC V3 sequences on exposure of epitopes in other domains of Env
The effect of the V3ConC to V3ConB point mutations on the exposure of epitopes located outside of the V3 region was also examined. Reagents tested included sCD4 and antibodies targeting the CD4-binding domain, CD4-induced (CD4i) epitopes, and two mAbs directed against epitopes located in the V1 domain of SF162 Env (Table 2; Supplemental Fig. 2). Similar to their effect on V3 antibodies, introducing the T19A and A22T mutations in the SF162-V3ConC chimeric Env strongly increased sensitivity to sCD4 and 5145A, and to reagents targeting the CD4-induced epitopes and epitopes in V1. Q18R had a modest unmasking effect on the CD4-bd-specific mAb, 5145A, and strongly unmasked the CD4i-specific ligands, while R13H led to modest decreases in neutralization sensitivity against all of these reagents. These mutations had minimal effects on the control mAb, 2F5. These results were consistent with a global conformational effect dominated by the subtype C-specific T19 and A22 residues that resulted in masking of neutralization targets in multiple domains, including V1, V3, the CD4-binding domain and CD4-induced epitopes.
Table 2.
IC50s for non-V3 mAbs against ConC and ConB chimeras compared to ConC point mutation chimeras. IC50 data (μg/ml) for sCD4, and mAbs targeting the CD4-bd (5145A), CD4i epitopes (X5, scFv M6) and V1 epitopes (58E1/B3 and 64B9/A6) against four of the V3 point mutations compared to V3ConC and V3ConB. MAb 2F5 was used as a control. All the subtype C Envs used for these experiments retained the Val at position 295.
| IC50s (μg/ml) | CD4-bd | CD4i | V1 linear | gp41 | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| V3 sequence/mAbs | sCD4 | 5145A | X5 | M6 | 58E1/B3 | 64B9/A6 | 2F5 |
| ConC | 0.19 | 2.9 | 0.29 | 0.091 | 3.8 | 1.5 | 0.52 |
|
| |||||||
| C-R13H | 2.8 | >20 | 0.62 | 0.22 | >10 | 4.5 | 0.95 |
| C-Q18R | 0.29 | 0.30 | 0.058 | 0.041 | 5.0 | 1.9 | 0.62 |
| C-T19A | 0.050 | 0.20 | 0.033 | 0.029 | 0.24 | 0.19 | 0.56 |
| C-A22T | 0.059 | 0.17 | 0.027 | 0.021 | 0.14 | 0.10 | 0.64 |
|
| |||||||
| ConB | 0.019 | 0.030 | 0.012 | 0.0087 | 0.24 | 0.10 | 0.85 |
The T19A/A22T unmasking mutations impact the neutralization sensitivity of primary isolates
The studies described above demonstrating the importance of subtype C consensus residues at positions 19 and 22 of the V3 loop were performed in the SF162 backbone. The wt SF162 Env is a highly sensitive tier 1 virus, and thus it can be argued that results obtained with these chimeric Envs may not be representative of primary Envs circulating in infected subjects. To explore this question, mutations at key V3 positions were introduced into the ConC Env and into two clonally related subtype C primary Envs, 55FPB4a and 55FPB28a, which were isolated early after infection from a Zambian subject and possessed more typical neutralization sensitivities (Derdeyn et al., 2004; Rong et al., 2007). Introducing the T19A/A22T mutations into both 55F clones led to significant increases in neutralization sensitivity against V3 mAbs and sCD4, with differences in IC50s ranging from 43 to greater than 1000-fold over that for the corresponding wild type Env (Fig. 6). Introducing the two subtype B-specific V3 mutations into the ConC sequence also increased the sensitivity of this Env to neutralization by anti-V3 mAbs, although by a much more modest extent (2 to > 5-fold). These results show that the V3-dependent masking mechanism mediated by residues at positions 19 and 22 contributes to the neutralization sensitivity of some primary subtype C viruses, suggesting that this mechanism may play a role in determining the neutralization phenotype of viruses circulating in infected people.
Fig. 6.
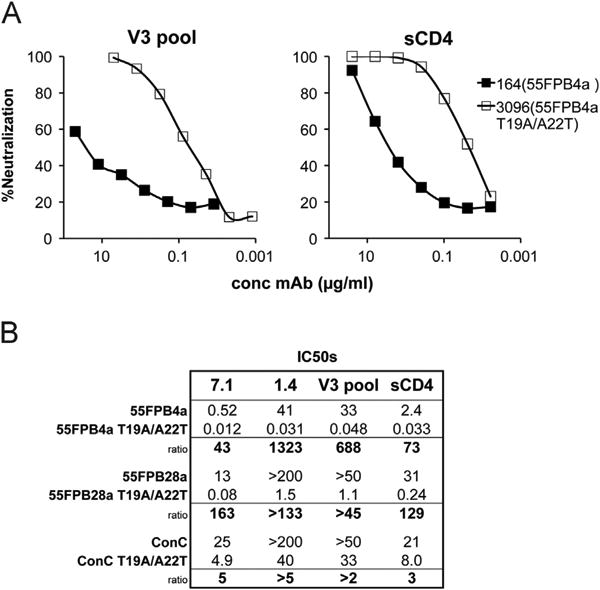
Effect of the two unmasking mutations, T19A and A22T, on neutralization sensitivity of primary isolates. (A) Representative curves of neutralizing activity of a pool of V3 mAbs, all of which bind to subtype B and C with equal affinity, and sCD4 against wild-type primary isolates and the T19A/A22T mutants in those Env backbones. (B) Average IC50s and ratios for two V3 mAbs, the V3 pool and sCD4 against 3 primary isolates and their corresponding mutants.
Conserved residues in V3 also contribute to V3 masking
A common feature in the V3 domain of HIV-1 is a cluster of hydrophobic residues that flank the turn at the tip of the loop (Ile12, Ile14 and Phe20) and have been implicated as binding sites for some V3-specific mAbs (Jiang et al., 2010; Stanfield et al., 2006). It was observed that a subtype C Env mutant with atypical residues at positions 12 (Met in place of Ile) and 14 (Leu in place of Ile) was more sensitive to neutralization by sCD4 than a related isolate with the consensus residues at these positions (Lynch et al., 2010). This suggested that mutations at these positions may also affect the conformation of Env complexes containing the V3ConC sequence. This was examined by introducing mutations at these positions in the context of SF162-V3ConC Env. The double mutation at positions 12 and 14 (I12M/I14L) was also introduced into the V3ConC Fc fusion protein, to control for the effect of these substitutions on binding of V3-specific mAbs.
Consistent with the important role of these residues on immunoreactivity, binding studies with the mutant V3ConC-Fc fusion glycoprotein showed that the double I12M/I14L mutation inhibited recognition of 10 of 14 V3-specific mAbs tested, including six mAbs derived from subtype B infections and four mAbs derived from non-subtype B infections, which either did not recognize the mutant sequence at all, or bound to it considerably less strongly than to the wild type sequence (Supplemental Fig. 3). This was consistent with crystallographic evidence that these residues formed critical van der Waals and hydrogen bond contacts with a number of V3-specific Fabs (Jiang et al., 2010; Stanfield et al., 2006; Stanfield and Wilson, 2005). However, four of the non-subtype B-derived mAbs (2601, 3697, 3074 and 3791) bound equally well to the wild type and mutant sequences, and these were used to examine the effect of modifications at positions 12 and 14 in the V3 loop on neutralization sensitivity of the SF162-V3ConC chimeric Env.
Both the I12M and the I12M/I14L double mutants were considerably more sensitive to neutralization by these V3-reactive mAbs than the wild-type Env, with increases in IC50 ranging from ∼50 to 350-fold for the I12M mutant, and ∼88 to over 1000-fold for the double mutation (Fig. 7; Supplemental Fig. 4).
Fig. 7.
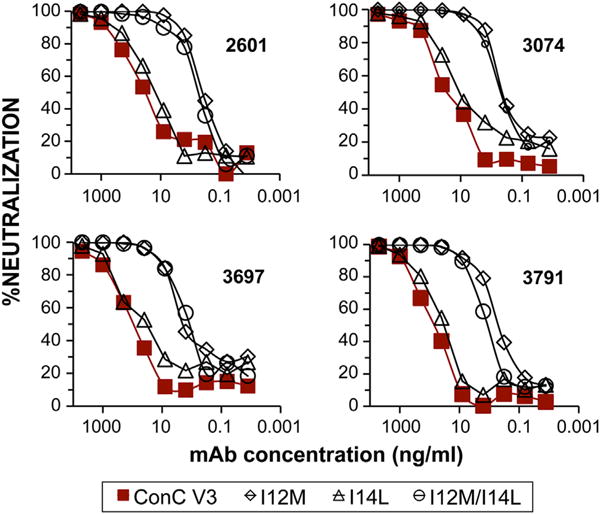
Effects of point mutations at conserved hydrophobic positions on neutralization. Differences in neutralization sensitivities of four V3-specific mAbs which bound equally well to the wild type V3ConC fusion protein and to the single and double mutants at positions 12 and 14. The N-terminal glycan was not present in these constructs.
The single I14L mutant was slightly more sensitive to neutralization than the wild type sequence. I12M and the I12M/I14L double mutant also significantly increased the sensitivity for several standard CD4-bd-specific mAbs (> 100 fold), and significant increases in sensitivity (13–50 fold) were also observed for several Fabs and an scFv directed against CD4-induced epitopes (Supplemental Fig. 4). To more accurately resemble the subtype C sequence, the Asn-linked glycan N-terminal to the V3 loop was removed from these constructs. Since this glycan slightly masks sites in V3, this resulted in a small decrease in IC50s for the wt V3ConC chimeric Env than reported previously in Table 1.
The importance of two conserved positively charged residues at positions 9 and 10 in the V3 loop towards the V3ConC masking effect was also examined. Arg9 and Lys10 of SF162-V3ConC were individually mutated to Ala, both as fusion glycoproteins to study binding, and as chimeric Envs to study effects on neutralization sensitivity. Both mutations reduced the binding affinity of several subtype B-derived V3-specific mAbs, also consistent with previously reported evidence that the basic side chains at these positions form critical interactions with a number of V3-specific Fabs (Jiang et al., 2010; Stanfield et al., 2006), but these substitutions did not affect the binding activity of two non-subtype B-derived mAbs, 3074 and 3791 (Fig. 8A). Despite the equivalent binding of the latter two mAbs, both mutations dramatically increased sensitivity to neutralization (Fig. 8B). The K10A mutation had the greater effect, with a 235-fold reduction in IC50 for 3074 and a 116-fold reduction for 3791. The R9A mutation also had a significant effect on the neutralizing activity of 3074 (68-fold), but only a modest 4-fold effect on 3791. These results illustrate that both the conserved hydrophobic patch and charged residues at positions 9 and 10 made important contributions towards maintaining the structure mediating the V3ConC-specific conformational masking of epitopes in the V3, CD4-bd and CD4i domains.
Fig. 8.
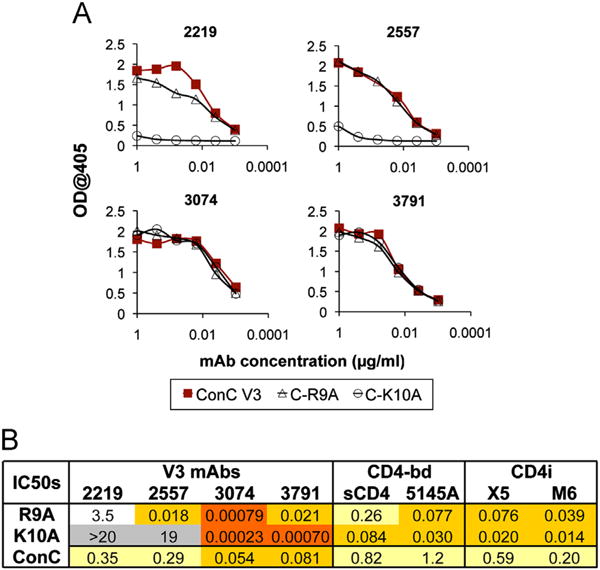
Effects of point mutations at conserved charged positions on binding and neutralization. Binding activity of four V3-specific mAbs against the V3ConC fusion glycoproteins with either the R9A or K10A mutations are shown as ELISA curves (A), while the IC50s for these mAbs against the corresponding pseudoviruses are tabulated in (B). The table also includes neutralization results for sCD4 and antibodies targeting the CD4-bd and CD4i epitopes. The R9A and K10A constructs contained the Asn at position 295, resulting in the presence of an N-linked glycosylation site N-terminal to the V3 loop typical of subtype B sequences. They also contained an additional D25E mutation, which as shown above does not influence neutralization sensitivity to anti-V3 mAbs.
Discussion
This study analyzed the contribution of specific V3 sequences towards the resistance of HIV-1 Envs to neutralization by poly-clonal immune sera and monoclonal antibodies directed against sites both in the V3 region and elsewhere in gp120. The results demonstrated that the V3ConC sequence induced a significant V1/V2-independent masking effect compared to the V3ConB sequence on epitopes located within the V3 region. This V3 “self-masking” was observed for mAbs derived from multiple subjects infected with different HIV-1 subtypes, including mAbs that bound with equal affinities to wild-type V3ConB and V3ConC fusion glycoproteins and recombinant gp120s. This showed that this effect was not related to changes in the inherent binding affinity of V3-specific antibodies to the isolated epitopes, and suggested instead that this reflected a different conformation of the trimeric Env structure induced by the V3ConC sequence.
Consistent with this interpretation, substitution of V3ConC into SF162 Env also masked targets elsewhere in gp120, including standard CD4-bd epitopes, CD4-induced epitopes, and epitopes located in the C-terminus of V1. Not affected by this mechanism were broadly neutralizing antibodies to CD4bs epitopes such as VRC01, which are not sensitive to masking (data not shown) and MPER targets. Since the antibodies sensitive to this effect are commonly induced by infection, this suggested that V3-dependent masking might be a contributing factor towards the resistance of subtype C viruses towards circulating antibodies in infected subjects.
Three of the five positions that distinguish the V3ConC from theV3ConB sequence strongly affected the V3ConC masking activity. Replacement of the polar (Thr) residue common at position 19 in subtype C by the hydrophobic (Ala) residue found at this position in subtype B sequences (T19A), and to an even greater extent, the reverse substitution at position 22 (A22T), strongly increased neutralization sensitivity, indicating that the ConC-specific residues at these positions were important for masking. For some mAbs, the Q18R substitution at the tip of the V3 loop also reduced masking, showing that the Q18 residue characteristic of subtype C further contributed to masking in these sequences, although to a smaller extent than the residues at positions 19 and 22. In contrast to these effects, replacement of the characteristic V3ConC Arg13 residue with the His residue frequently found in V3ConB further increased the masking activity of the ConC sequence, indicating that Arg13 modulated the extent of masking by the V3ConC sequence. Arg13 is highly dominant in subtype C (94%), whereas in subtype B His is the most common residue (64%) and Arg is rare (< 3%). The neutralization-resistant phenotype of the V3ConC chimeric Env indicated that the three masking residues at positions 18, 19 and 22 overrode the unmasking effect of His13, and accounted for the strong masking activity of the ConC sequence.
The combination of the three masking residues, Gln18, Thr19 and Ala22 is common in subtype C sequences (60%), but occurs less frequently in other subtypes (19% for subtype A) and was not found in any of the subtype B sequences analyzed (http://www.hiv.lanl.gov, 2011 alignment). Although masking may be modulated by substitutions at other V3 positions, the relative frequency of the Q18/T19/A22 signature in subtype C sequences suggests that the masking effect mediated by these residues is particularly important for this group of viruses.
Insertion of two of the V3 masking mutations, T19A and A22T, was shown to strongly influence the neutralization phenotypes of two subtype C primary isolates, showing that the effects seen with SF162 Env chimeras were relevant for natural Envs as well. 55FPB4a and 55FPB28a are two HIV-1 clones obtained shortly after transmission from a patient infected by male to female transfer (Derdeyn et al., 2004). These clones differed from each other at two positions in V2 and one position in the cytoplasmic domain of gp41, and differed somewhat in their neutralization sensitivity to autologous plasma, reportedly a result of the two V2 amino acid changes (Rong et al., 2007). The wild-type 55FPB28a clone was highly resistant to several anti-V3 antibodies and to sCD4, while the wild-type 55FPB4a clone was more sensitive, presumably due to reduced masking by the V1/V2 domain. The double V3 substitution led to significant increases in neutralization sensitivity in both clones, with increases in sensitivity for the 55FPB28a mutant of 1300-fold for V3-specific mAb 1.4 and ∼700-fold for the mAb pool. Increases in sensitivity were also observed when the double mutation was inserted into ConC Env, although the effect in this virus was much smaller. The wild type ConC Env was less sensitive than the 55F clones to neutralization by all of the reagents tested, and unpublished experiments from our lab showed that the ConC V1/V2 domain possessed stronger masking activity than the 55F V1/V2 domains. This suggests that the V3-specific masking effect described in this study may be particularly important for viruses that possess less effective V1/V2 masking.
In addition to the subtype-specific residues, mutations at several conserved positions in V3 were also found to affect the neutralization phenotype of Env. Substitutions at hydrophobic residues, Ile12 and Ile14, and at charged residues Arg9 and Lys10 increased the exposure of V3 and other neutralization domains, indicating that these positions were also involved in maintaining the overall Env conformation. This suggests that combinations of hydrophobic, charged and polar residues in V3 interact with additional regions within the native Env complex to regulate Env conformation.
A previous study utilizing ab initio folding calculations concluded that there was a greater structural rigidity and non-β-strand secondary protein structure in the V3 segment at positions 12–14 in the crown of the subtype C V3 loop than in the subtype B V3 loop, and it was postulated that this correlated with a greater resistance in neutralization (Almond et al., 2012). Those findings did not address the mechanism for neutralization resistance, and the effect of substitutions at positions 19 and 22 on β-strand structure was not examined in that study, so the relevance of those calculations on the neutralization effects described in the current study are unclear.
The observation that V3ConC-specific masking effects were not detected in monomeric gp120 proteins was also consistent with recent data (Andrabi et al., 2013) showing equivalent binding affinities for many anti-V3 mAbs for various subtype B and C-derived gp120s. However, these results contrasted with an earlier study that reported more effective binding of some V3-specific mAbs to a subtype B-derived gp120 than to a subtype C-derived gp120, despite equivalent binding to related V3 peptides (Patel et al., 2008). That study used a limited set of subtype B-derived mAbs and a mismatched pair of Envs (JR-FL and BR025), and while the gp120s were glycosylated, the V3 fusion proteins used to control for binding affinities were not. In addition, the subtype C Env used in that study, BR025, did not contain Thr19, which is dominant in subtype C sequences and which our studies have shown is an important determinant of conformation.
In conclusion, this paper describes a V3 structural motif common in subtype C viruses that results in greater occlusion of epitopes in V3 and more effective masking of other potential neutralization sites, including the CD4-binding and CD4-induced domains. The fact that the optimal V3 masking motif did not affect the accessibility of V3 epitopes in monomeric gp120 suggested that this involved the stabilization of an interaction of the V3 region with another site in a neighboring protomer of the native Env trimer. This is consistent with recent evidence from cryoelectron tomography studies suggesting that the V1, V2 and V3 domains are located at the apex of the unliganded Env trimer, and form a trimer-association domain (TAD) that shields critical sites in the CD4 and CCR5 receptor-binding domains (Mao et al., 2012, 2013). An intimate association between the V2 and V3 domains is also suggested by the contribution of sequences in these regions to the formation of quaternary epitopes, such as the type-specific 2909 and related macaques mAbs (Gorny et al., 2005; Krachmarov et al., 2011), and the broadly-reactive PG9, PG16 and related antibodies (Bonsignori et al., 2011; Moore et al., 2011; Walker et al., 2009, 2011). These results suggest that the V3-interaction site involved in masking may be located somewhere in the V1/V2 domain or in components of the adjacent bridging sheet known to affect global conformation.
A further implication of these results is that viruses or trimeric Env immunogens containing the V3ConC sequence would differ in structure and antigenicity from corresponding immunogens containing the V3ConB sequence, and thus may induce a different set of antibody specificities. Immunogens with an unmasked V3 domain would be likely to possess a more open structure in which epitopes in V3 and the CD4-bd are exposed and immunogenic. On the other hand, immunogens with a masked V3 domain would exist in a closed conformation in which V3, CD4-bd and CD4-induced epitopes are less accessible. This may reduce the immunogenicity of those targets, and instead, allow the immune response to focus on targets preferentially exposed on the native trimer, such as those dependent on quaternary epitopes and glycan-dependent epitopes, and perhaps enhance the immunogenicity of those targets. It would be useful to test this hypothesis in a vaccine trial in a relevant animal model.
Materials and methods
Generation of chimeric and mutant Envs
The V3 chimeric and mutant Envs described in this study were all generated either by Quickchange™ site-directed mutagenesis (Stratagene, Inc.), or by oligonucleotides substitutions at unique restriction sites inserted into various positions of the V3 region by silent mutations. To facilitate the oligonucleotide exchanges, Env chimeras were constructed that contained unique restriction sites inserted into the V3 region of either the subtype B or subtype C consensus sequence. The parental Envs were digested at restriction sites flanking the desired mutation site, and two complementary oligonucleotide pairs containing the desired mutations in the excised V3 sequence were inserted at these sites. To facilitate screening, the oligonucleotides contained additional silent mutations that either introduced or removed internal restriction sites. The Env vectors were dephosphorylated by treatment with Calf Intestinal Phosphatase (NEB) and the oligonucleotides inserts were phosphorylated by treatment with T4 polynucleotide kinase (NEB). Oligonucleotides and vector were mixed at a molar ratio of ∼5:1 and ligated with T4 DNA ligase (Roche). After transformation, colonies were screened by PCR with primers flanking the insertion site and the amplified product digested with the appropriate restriction enzyme and analyzed on gels. All oligonucleotides were obtained from Integrated DNA Technologies, Inc., and enzymes were obtained from New England Biolabs, Inc. We thank Dr. Cynthia A. Derdeyn (Emory Vaccine Center) for HIV-1 Env clones 55FPB4a and 55FPB28a.
Monoclonal antibodies
Monoclonal anti-V3 mAbs were derived from human subjects infected with HIV-1 of multiple subtypes derived from human subjects in the US, West Africa (Cameroon, Côte d'Ivoire) and India, as previously described (Gorny et al., 2009; Pinter et al., 1993b). Three new V3 mAbs isolated in the Robinson laboratory, 5.8C, 1.4E and 7.1C, were included in this study. 5.8C was derived from an HIV-1 infected patient from Malawi, where the dominant serotype is subtype C. 1.4E was derived from Gambia patient N036703, where several CRF strains along with subtype C and A are common (de Silva et al., 2010). 7.1C was obtained from a subject in South Africa, where subtype C is dominant. Screening was done by ELISA using recombinant Env proteins as previously described (Robinson et al., 2005). Antibodies 5.8C and 1.4E were initially derived from EBV-transformed B cell lines that secreted the mAbs, while 7.1C was obtained from B cells stimulated with cytokines in the absence of EBV The heavy and light chain genes were subsequently cloned into expression vectors which were used to produce the recombinant antibodies.
Control mAbs were obtained from the NIH AIDS Research and Reference Reagent Program: IgG1b12 (Burton et al., 1994), directed against an epitope that overlaps the CD4-binding site, was contributed by Drs. Dennis Burton and Paul Parren; 2G12 (Trkola et al., 1996), directed against a conformational epitope involving high mannose glycans, and 2F5 (Muster et al., 1993), directed against an epitope in the ectodomain of gp41, were contributed by Dr. Hermann Katinger. The X5 Fab, M6 scFv and M9 scFv targeting CD4-induced epitopes (Moulard et al., 2002; Zhang et al., 2004) were provided by Dr. Dimiter S. Dimitrov at the NCI in Frederick, MD. MAbs 58E1/B3 and 64B9/A6, which target the V1/V2 domain, were isolated in our lab (He et al., 2002). Polyclonal HIV-1 immune sera were obtained from chronically infected patients followed at the Montefiore AIDS Clinic in the Bronx, N.Y.
Neutralization assays
Neutralization activity was determined as previously described (Krachmarov et al., 2001) with a single cycle infectivity assay using virions generated from the Env-defective luciferase-expressing pNL4-3.Luc.R-E- genome (Connor et al., 1995) pseudotyped with a molecularly cloned HIV Env of interest. Briefly, pseudotyped virions in culture supernatants from transfected 293T cells were incubated with serial dilutions of mAbs for 1 h at 37 °C, and then added to U87-T4-CCR5 target cells plated out in 96-well plates in the presence of polybrene (10 μg/ml). After 24 h, cells were fed with RPMI medium containing 10% calf serum and polybrene, and luciferase activity in lysed cells was measured 72–96 h post-infection with a microtiter plate luminometer (HARTA, Inc.), using assay reagents from Promega. IC50 values reported were determined by interpolation from neutralization curves or through the Excel Growth function which calculates predicted exponential growth, and are averages of at least three independent assays.
Recombinant antigens and binding assays
V3-Fc fusion glycoproteins were prepared by inserting the desired V3 sequence into the pFUSE-rFc2 (IL2ss) vector (Invivogen, Inc.). This plasmid contains the rabbit IgG Fc sequence comprising the CH2 and CH3 domains and the hinge region of the IgG heavy chain, and includes the sh ble gene which confers Zeocin resistance. The complete 35 residue disulfide-bonded V3 loop, together with four flanking N-terminal residues and three flanking C-terminal residues, including the N-linked glycosylation sites, was inserted in frame between the IL2 signal sequence and the Fc-coding sequence, using EcoRI and BglII sites present in the multiple cloning site located upstream of the rabbit IgG. The V3 insert sequences were constructed by PCR using oligonucleotides flanking the region of interest with the upstream primers containing the EcoRI restriction site and downstream primers containing the BglII restriction site. The ligated plasmid was transformed into DH5α cells (Invitrogen), in the presence of Zeocin and plasmid sequences were confirmed by PCR and sequencing. The V3-Fc glycoproteins were expressed by transfection of 293T cells, and supernatants containing the secreted proteins were harvested 3–5 days post-transfection.
Monoclonal antibody concentrations giving 50% maximal binding to individual V3 fusion glycoproteins were determined by ELISA, and used as a measure of relative affinity (Kayman et al., 1994). The V3-Fc antigens were captured for 1 h at 37 °C on an ELISA plate coated overnight with goat anti-rabbit IgG and blocked with 2% powdered milk/PBS for 30 min at 37 °C. The concentration of the antigen was normalized by titration with anti-rabbit IgG. Binding of anti-V3 mAbs was determined by adding serially diluted mAbs to the plates for 1 h at 37 °C, followed by washing and then incubation with alkaline phosphatase-conjugated goat anti-human IgG F(ab)2 (Jackson IR Laboratories; SouthernBiotech) for 45 min at 37 °C. The assay was developed using phosphatase substrate (Sigma) and the optical density was read at 405 nm at multiple time points.
Soluble SF162-gp120 was expressed by inserting stop codons into SF162-V3ConB and SF162-V3ConC Env plasmids after the gp120 coding sequence using Phusion™ site-directed mutagenesis (Finn-zymes-NEB). Full-length recombinant JR-FL gp120 was expressed from the syngp120-JRFL plasmid, which contained the codon-optimized gene (Andre et al., 1998). For JR-FL gp120-V3ConC, the V3 region (which corresponded to the ConB sequence) was replaced by the consensus subtype C sequence by inserting synthetic oligonucleotides expressing the corresponding sequence between two unique silent restriction sites, MluI and PshAI, which were engineered into the V3-coding region of syngp120-JRFL vector using Quickchange™ site-directed mutagenesis (Stratagene, Inc.). The four gp120s were expressed by transfection of Expi293™ cells (Gibco) according to the manufacturers protocol, and purified with agarose beads conjugated to Galanthus Nivalis lectin (Vector Labs). The relative affinity of various V3-specific mAbs antibodies was measured by coating purified gp120 (5-10 μg/ml) on wells of 96-well ELISA plates and incubating at 4 °C overnight. The wells were then blocked with 2% powdered milk/PBS for 30 min at 37 °C, washed, and incubated with serially diluted anti-V3 mAbs for 1 h at 37 °C, followed by washing and incubation with alkaline phosphatase-conjugated goat anti-human IgG F(ab)2 for 45 min at 37 °C. The assay was developed using phosphatase substrate (Sigma) and the optical density was read at 405 nm at multiple time points.
Supplementary Material
Acknowledgments
This study was supported in part by NIH grants U01 AI078410-01 and P01 AI088610-01 to AP, and a Collaboration for AIDS Vaccine Development (CAVD) grant from the Bill and Melinda Gates Foundation.
Footnotes
Appendix A. Supplementary material: Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2013.10.007.
References
- Almond D, Krachmarov C, Swetnam J, Zolla-Pazner S, Cardozo T. Resistance of subtype C HIV-1 strains to anti-V3 loop antibodies. Adv Virol. 2012;2012:803535. doi: 10.1155/2012/803535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi R, Williams C, Wang XH, Li L, Choudhary AK, Wig N, Biswas A, Luthra K, Nadas A, Seaman MS, Nyambi P, Zolla-Pazner S, Gorny MK. Cross-neutralizing activity of human anti-V3 monoclonal antibodies derived from non-B clade HIV-1 infected individuals. Virology. 2013;439:81–88. doi: 10.1016/j.virol.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre S, Seed B, Eberle J, Schraut W, Bultmann A, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillou A, Brand D, Denis F, M'Boup S, Chout R, Goudeau A, Barin F. High antigenic cross-reactivity of the V3 consensus sequences of HIV-1 gp120. AIDS Res Hum Retroviruses. 1993;9:1209–1215. doi: 10.1089/aid.1993.9.1209. [DOI] [PubMed] [Google Scholar]
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O'Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao HX, Haynes BF. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Habib DC, Roderiquez G, Oravecz T, Berman PW, Lusso P, Norcross MA. Cryptic nature of envelope V3 region epitopes protects primary mono-cytotropic human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Stiehm ER, Bryson YJ, Cao Y, Moore JP, Ho DD, Barbas CF., 3rd Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Carrow EW, Vujcic LK, Glass WL, Seamon KB, Rastogi SC, Hendry RM, Boulos R, Nzila N, Quinnan GV., Jr High prevalence of antibodies to the gp120 V3 region principal neutralizing determinant of HIV-1MN in sera from Africa and the Americas. AIDS Res Hum Retroviruses. 1991;7:831–838. doi: 10.1089/aid.1991.7.831. [DOI] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Davis KL, Bibollet-Ruche F, Li H, Decker JM, Kutsch O, Morris L, Salomon A, Pinter A, Hoxie JA, Hahn BH, Kwong PD, Shaw GM. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J Virol. 2009a;83:1240–1259. doi: 10.1128/JVI.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, Montefiori DC, Graham BS, Keefer MC, Pinter A, Morris L, Hahn BH, Shaw GM. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology. 2009b;387:414–426. doi: 10.1016/j.virol.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva TI, Turner R, Hue S, Trikha R, van Tienen C, Onyango C, Jaye A, Foley B, Whittle H, Rowland-Jones SL, Cotten M. HIV-1 subtype distribution in the Gambia and the significant presence of CRF49_cpx, a novel circulating recombinant form. Retrovirology. 2010;7:82. doi: 10.1186/1742-4690-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. The v3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J Virol. 2004;78:2394–2404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Hioe C, Israel ZR, Michael NL, Conley AJ, Williams C, Kessler JA, 2nd, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J Immunol. 1997;159:5114–5122. [PubMed] [Google Scholar]
- Gorny MK, Wang XH, Williams C, Volsky B, Revesz K, Witover B, Burda S, Urbanski M, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S, Nadas A. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol Immunol. 2009;46:917–926. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76:9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Konings FA, Nadas A, Anyangwe CA, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol. 2006;80:6865–6872. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res Hum Retroviruses. 2005;21:171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- He Y, Honnen WJ, Krachmarov CP, Burkhart M, Kayman SC, Corvalan J, Pinter A. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human Ig loci. J Immunol. 2002;169:595–605. doi: 10.4049/jimmunol.169.1.595. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- Javaherian K, Langlois AJ, LaRosa GJ, Profy AT, Bolognesi DP, Herlihy WC, Putney SD, Matthews TJ. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990;250:1590–1593. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong XP. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat Struct Mol Biol. 2010;17:955–961. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- Kayman SC, Wu Z, Revesz K, Chen H, Kopelman R, Pinter A. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J Virol. 1994;68:400–410. doi: 10.1128/jvi.68.1.400-410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, Gnanapragasam PN, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, Nussenzweig MC. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber BT, MacInnes K, Smith RF, Myers G. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J Virol. 1994;68:6730–6744. doi: 10.1128/jvi.68.10.6730-6744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov C, Lai Z, Honnen WJ, Salomon A, Gorny MK, Zolla-Pazner S, Robinson J, Pinter A. Characterization of structural features and diversity of variable-region determinants of related quaternary epitopes recognized by human and rhesus macaque monoclonal antibodies possessing unusually potent neutralizing activities. J Virol. 2011;85:10730–10740. doi: 10.1128/JVI.00365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005;79:780–790. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80:7127–7135. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Kayman SC, Honnen WJ, Trochev O, Pinter A. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1737–1748. doi: 10.1089/08892220152741432. [DOI] [PubMed] [Google Scholar]
- LaRosa GJ, Davide JP, Weinhold K, Waterbury JA, Profy AT, Lewis JA, Langlois AJ, Dreesman GR, Boswell RN, Shadduck P, Holley LH, Karplus M, Bolognesi DP, Matthews TJ, Emini EA, Putney SD. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science. 1990;249:932–935. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- Letvin NL, Robinson S, Rohne D, Axthelm MK, Fanton JW, Bilska M, Palker TJ, Liao HX, Haynes BF, Montefiori DC. Vaccine-elicited V3 loop-specific antibodies in rhesus monkeys and control of a simian-human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate envelope. J Virol. 2001;75:4165–4175. doi: 10.1128/JVI.75.9.4165-4175.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J Virol. 2006;80:1414–1426. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch RM, Rong R, Li B, Shen T, Honnen W, Mulenga J, Allen S, Pinter A, Gnanakaran S, Derdeyn CA. Subtype-specific conservation of isoleucine 309 in the envelope V3 domain is linked to immune evasion in subtype C HIV-1 infection. Virology. 2010;404:59–70. doi: 10.1016/j.virol.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Wang L, Gu C, Herschhorn A, Desormeaux A, Finzi A, Xiang SH, Sodroski JG. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci USA. 2013;110:12438–12443. doi: 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat Struct Mol Biol. 2012;19:893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Sheward D, Madiga M, Ranchobe N, Lai Z, Honnen WJ, Nonyane M, Tumba N, Hermanus T, Sibeko S, Mlisana K, Abdool Karim SS, Williamson C, Pinter A, Morris L, Study C. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J Virol. 2011;85:3128–3141. doi: 10.1128/JVI.02658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, Zhang MY, Sidorov IA, Broder CC, Robinson J, Parren PW, Burton DR, Dimitrov DS. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci USA. 2002;99:6913–6918. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, Kwong PD. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure–function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MB, Hoffman NG, Swanstrom R. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J Virol. 2008;82:903–916. doi: 10.1128/JVI.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci USA. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, Racho ME, Tilley SA. A potent, neutralizing human monoclonal antibody against a unique epitope overlapping the CD4-binding site of HIV-1 gp120 that is broadly conserved across North American and African virus isolates. AIDS Res Hum Retroviruses. 1993a;9:985–996. doi: 10.1089/aid.1993.9.985. [DOI] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, Tilley SA. Conformational changes affecting the V3 and CD4-binding domains of human immunodeficiency virus type 1 gp120 associated with env processing and with binding of ligands to these sites. J Virol. 1993b;67:5692–5697. doi: 10.1128/jvi.67.9.5692-5697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JE, Elliott DH, Martin EA, Micken K, Rosenberg ES. High frequencies of antibody responses to CD4 induced epitopes in HIV infected patients started on HAART during acute infection. Hum Antibodies. 2005;14:115–121. [PubMed] [Google Scholar]
- Rong R, Bibollet-Ruche F, Mulenga J, Allen S, Blackwell JL, Derdeyn CA. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J Virol. 2007;81:1350–1359. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T, Levy JA, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Geisler SC, Chen AA, Resnick DA, Roy BM, Lewi PJ, Arnold E, Arnold GF. Human rhinovirus type 14:human immunodeficiency virus type 1 (HIV-1) V3 loop chimeras from a combinatorial library induce potent neutralizing antibody responses against HIV-1. J Virol. 1998;72:651–659. doi: 10.1128/jvi.72.1.651-659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear GT, Takefman DM, Sharpe S, Ghassemi M, Zolla-Pazner S. Antibodies to the HIV-1 V3 loop in serum from infected persons contribute a major proportion of immune effector functions including complement activation, antibody binding, and neutralization. Virology. 1994;204:609–615. doi: 10.1006/viro.1994.1575. [DOI] [PubMed] [Google Scholar]
- Spenlehauer C, Saragosti S, Fleury HJ, Kirn A, Aubertin AM, Moog C. Study of the V3 loop as a target epitope for antibodies involved in the neutralization of primary isolates versus T-cell-line-adapted strains of human immunodeficiency virus type 1. J Virol. 1998;72:9855–9864. doi: 10.1128/jvi.72.12.9855-9864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80:6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Wilson IA. Structural studies of human HIV-1 V3 antibodies. Hum Antibodies. 2005;14:73–80. [PubMed] [Google Scholar]
- Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley SA, Honnen WJ, Racho ME, Hilgartner M, Pinter A. A human monoclonal antibody against the CD4-binding site of HIV1 gp120 exhibits potent, broadly neutralizing activity. Res Virol. 1991;142:247–259. doi: 10.1016/0923-2516(91)90010-z. [DOI] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancott TC, Polonis VR, Loomis LD, Michael NL, Nara PL, Birx DL. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res Hum Retro-viruses. 1995;11:1379–1391. doi: 10.1089/aid.1995.11.1379. [DOI] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff P, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wu L, Yang ZY, Xu L, Welcher B, Winfrey S, Shao Y, Mascola JR, Nabel GJ. Cross-clade recognition and neutralization by the V3 region from clade C human immunodeficiency virus-1 envelope. Vaccine. 2006;24:4995–5002. doi: 10.1016/j.vaccine.2006.03.083. [DOI] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Shu Y, Rudolph D, Prabakaran P, Labrijn AF, Zwick MB, Lal RB, Dimitrov DS. Improved breadth and potency of an HIV-1-neutralizing human single-chain antibody by random mutagenesis and sequential antigen panning. J Mol Biol. 2004;335:209–219. doi: 10.1016/j.jmb.2003.09.055. [DOI] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cohen SS, Krachmarov C, Wang S, Pinter A, Lu S. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology. 2008;372:233–246. doi: 10.1016/j.virol.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, Gorny MK. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20:1254–1258. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.