Abstract
Huntington’s disease (HD) is a devastating, genetic neurodegenerative disease caused by a tri-nucleotide expansion in exon 1 of the huntingtin gene. HD is clinically characterized by chorea, emotional and psychiatric disturbances and cognitive deficits with later symptoms including rigidity and dementia. Pathologically, the cortico-striatal pathway is severely dysfunctional as reflected by striatal and cortical atrophy in late-stage disease. Brain-derived neurotrophic factor (BDNF) is a neuroprotective, secreted protein that binds with high affinity to the extracellular domain of the tropomyosin-receptor kinase B (TrkB) receptor promoting neuronal cell survival by activating the receptor and down-stream signaling proteins. Reduced cortical BDNF production and transport to the striatum have been implicated in HD pathogenesis; the ability to enhance TrkB signaling using a BDNF mimetic might be beneficial in disease progression, so we explored this as a therapeutic strategy for HD. Using recombinant and native assay formats, we report here the evaluation of TrkB antibodies and a panel of reported small molecule TrkB agonists, and identify the best candidate, from those tested, for in vivo proof of concept studies in transgenic HD models.
Introduction
Huntington’s disease (HD) is a devastating and fatal, autosomal dominant neurodegenerative disease whose etiology is simple but poorly understood. Early HD is characterized by chorea and psychiatric mood and cognitive disturbance deficits, followed by rigidity and dementia later in disease progression, with fatality occurring within 15–20 years of clinical diagnosis [1]–[6].
HD is caused by a tri-nucleotide expansion (cytosine, adenosine and guanosine, (CAG)) in exon 1 of the huntingtin gene [7]. The CAG codon encodes for the expression of the amino acid glutamine (Gln or Q); expansion of the polyglutamine (polyQ) chain on the N-terminus of the huntingtin (HTT) protein beyond 39 repeats affords a mutant form (mHTT) which leads to the onset of disease with complete penetrance. This expanded polyQ mutant form of HTT misfolds and aggregates, which occurs concomitantly with disease progression [8], [9]. However, although HD neuropathology reveals the presence of huntingtin protein inclusions in the nucleus and the cytosol of neurons as well as neuropil [10], it is unclear whether these aggregates confer a neuroprotective or neurotoxic effect [11], [12]. There is no current HD therapeutic that modifies the degenerative process. Current treatments are symptomatic and include neuroleptics, antipsychotics and antidepressants, with motor symptoms being treated with the only approved HD drug, tetrabenazine, a vesicular monoamine transporter (V-MAT) inhibitor.
Tropomyosin-receptor kinase (Trk) receptors (TrkA, TrkB and TrkC) are a family of kinase signaling receptors which regulate the peripheral and central nervous system through their interaction with the neurotrophins that include β-nerve growth factor (NGF), NT3, NT4 and brain-derived neurotrophic factor (BDNF). NGF is the preferred ligand for TrkA, BDNF and NT4 are preferred for TrkB, and NT3 for TrkC; NT3 can also bind TrkA and TrkB with reduced affinity [13]. All neurotrophins bind with lower affinity to the structurally distinct p75 receptor; p75 is reported to contribute to divergent cellular functions which include neuronal apoptosis [14], [15]. Binding of BDNF to TrkB induces receptor dimerization and leads to multiple tyrosine trans-phosphorylation events between the juxtaposed kinase domains that modulate catalytic activity (Tyr706/707) and form adapter protein docking sites (Tyr516, Tyr816) needed for pro-survival signal transduction pathways through the PI3K, PLCγ and MAPK pathways [16].
In HD, reduced levels of BDNF and TrkB mRNAs and proteins have been reported in human and mouse model brain cortices; a consequential reduction in neurotrophic support for the striatum has therefore been implicated in disease pathogenesis [17]–[19]. Forebrain knock-out of BDNF in mice results in a striatal expression profile that closely mirrors human HD striatal gene expression [20]. Indeed, over-expression of BDNF in the forebrain reduces the HD phenotype in YAC128 transgenic mice [21]. Poor bioavailability of intrathecally administered BDNF (BDNF precursor protein is 247 amino acids; mature BDNF is 119 amino acids) may underlie the lack of efficacy in clinical studies [22]. We therefore propose that enhancement of TrkB signaling with an exogenously administered TrkB agonist or positive allosteric modulator (PAM) may slow or reverse HD progression. In order to validate and characterize putative TrkB modulators, we applied a number of cell-based assays to measure proximal and distal cell signaling effects ( Figure 1A and 1B ). We also used a TrkB receptor-responsive, rodent primary neuronal HD model of neurodegeneration to confirm that active agents also work on native receptors. As part of our study, we evaluated a comprehensive panel of purported TrkB small molecule agonists that have been published in recent years [23] and compared their functionality with two TrkB monoclonal antibody (mAb) agonists [24], [25].
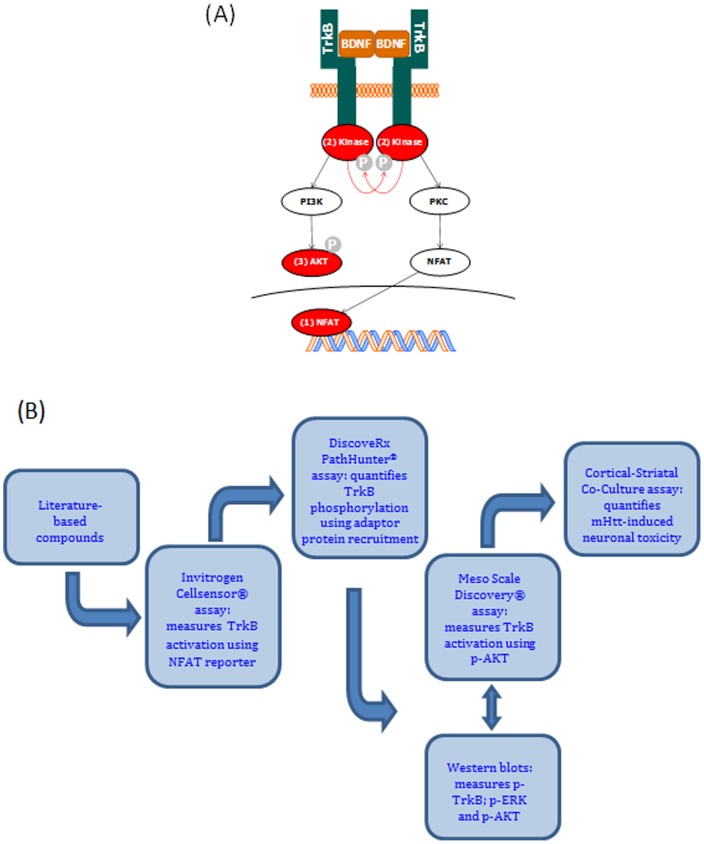
Figure 1. TrkB signaling and assay cascades.
(A) TrkB signalling cascade showing proximal and distal assay measurement points. Red arrows represent BDNF-induced trans-phosphorylation events within the intracellular tyrosine kinase domains. Changes in TrkB phosphorylation/activation detected by (1) Invitrogen CellSensor®; (2) DiscoveRx PathHunter®; and (3) MSD® pAKT assays. (B) Screening cascade used to characterize TrkB modulators.
Materials and Methods
Ethics Statement
Rat embryos were isolated at The Netherlands Organization for Applied Scientific Research (TNO; Leiden, The Netherlands) before isolation of neurons at Galapagos (Leiden, The Netherlands). Animal experiments were approved by the Institutional Animal Care and Use Committee of TNO (Dierexperimentencommissie TNO), and were in compliance with European Community specifications regarding the use of laboratory animals. All procedures involving animals were conducted humanely and were performed by or under the direction of trained and experienced personnel.
Culture Media
1x D-MEM with GlutaMax (31966-021), 1x MEM (31095-029), D-MEM/F12 (21331-020), Dialysed Foetal Bovine Serum (dFBS) (26400-044), Horse serum (16050-155), 1M HEPES solution (15630), NEAA (11140), L-glutamine (25030-024), Blasticidin (R210-01), Zeocin (R250-01), Hygromycin B (10687-010), Geneticin (10131-027), Pen/Strep (15140-122), Trypsin/EDTA (25300-054), PBS containing Ca2+ and Mg2+ (14040) and without Ca2+ and Mg2+ (14190-094) were obtained from Life Technologies. HyClone II fetal bovine serum (FBS) (SH30066-03) was obtained from PerBio.
Cell Culture
CellSensor® TrkA-NFAT-bla (K1516), TrkB-NFAT-bla (K1491), TrkC-NFAT-bla (K1515) and NFAT-bla (K1534) CHO-K1 cell lines were obtained from Life Technologies. CellSensor® cells were routinely cultured in growth medium (D-MEM with GlutaMax) supplemented with 10% dFBS, 1x MEM NEAA, 20 mM HEPES, 5 µg/mL Blasticidin and 200 µg/ml Zeocin (No antibiotics for NFAT-bla control line).
PathHunter® U2OS TrkB cell line (93-0463C3) was obtained from DiscoveRx; cells were routinely cultured in growth medium (1x MEM) supplemented with heat inactivated FBS (Hyclone II), 100 µg/mL Hygromycin B and 200 µg/mL Geneticin.
SH-SY5Y human neuroblastoma cells were obtained from ATCC (CRL-2266); cells were routinely cultured in growth medium (D-MEM) supplemented with 10% heat inactivated FBS, 10 mM HEPES, 100 units Pen/strep.
Reagents
BDNF (PHC7074) and ToxBLAzer reagent (K1136, P36400) were obtained from Life Technologies. NGF-β (N1408), NT-3 (N1905), NT-4 (N1780), and Thapsigargin (T9033) were obtained from Sigma-Aldrich. Tool small molecules: LM22A-4, amitryptiline, 7,8-dihydroxyflavone and N-acetyl serotonin were supplied by CHDI and used after passing QC analysis; BAG peptide was synthesized by Peptide Synthetics; tool antibody panel was sourced commercially (BD mAb #610102; Millipore pAb #07-225) or from Pfizer Inc (38B8 and 29D7 mAbs); a pan-Trk kinase inhibitor was synthesized at BioFocus [26]. All-trans retinoic acid (R2625) and BSA (A0281) were obtained from Sigma-Aldrich. PathHunter® detection kit (93-0001) was obtained from DiscoveRx. DMSO (D/4121/PB17), Triton-X100 (10102913) and Halt Protease and Phosphatase Inhibitor Cocktail (100X) (78444) were obtained from ThermoFisher Scientific. Recombinant pAKT1 (31145) for Meso Scale Discovery studies was obtained from Active Motif; whole cell lysate kit – Phospho (Ser473)/ Total AKT assay (K11100D-2) was obtained from Meso Scale Discovery.
Western Blotting
Clarified cell lysates were resolved by SDS-PAGE, blotted onto nitrocellulose membranes and immunoprobed with the relevant primary and secondary antibody combinations. Anti-pTrkB Tyr516 (4619), anti-pTrkB Tyr706/707 (4621), anti-pTrkB, Tyr816 (4168), anti-pERK1/2 (4051), anti-pAKT (4051), anti-TrkB (4603), anti-AKT (4685) and anti-ERK1/2 (4695) were sourced from Cell Signalling Technologies. Anti-GAPDH (IMG-5143A) was obtained from Imgenex; Goat anti-rabbit HRP conjugate (PO448) and Goat anti-mouse HRP conjugate (PO447) were sourced from Dako. Goat anti-mouse 800CW conjugate and Goat anti-rabbit 680 conjugate were sourced from LI-COR. Immunoreactive signals were detected using enhanced chemiluminescence reagent (Amersham) or using the Odyssey infra-red reader.
Invitrogen CellSensor® Trk reporter gene assays
The assays were configured using Invitrogen CHO-K1 cell lines stably expressing a β-lactamase gene under the control of a NFAT promoter region and the relevant Trk (A, B or C) receptor (except for the Trk null cell line). The activity of the β-lactamase enzyme was quantified via FRET with a fluorescent substrate containing a β-lactam ring, according to manufacturer’s provided protocol. All CellSensor® NFAT-bla CHO-K1 cell lines were seeded into black 384w assay plates in CellSensor® plating media (growth media without antibiotics) at 5,000 cells in a volume of 25 µL per well; cells were incubated at 37°C, 5% CO2 overnight. Then, the assay plating media was removed and replaced with 32 µL of assay media (plating media without serum) and returned to the incubator for 1 hour. Test compounds were initially titrated in 100% DMSO, then diluted in DMSO/assay media so as to result in a final assay concentration of 0.5% (v/v) DMSO, previously demonstrated not to adversely affect the performance of the assay readouts (data not shown). Test compounds were added (4 µL) and cells were returned to the incubator for 1 hour; dependent on assay mode (agonist, antagonist or positive allosteric modulator) assay media alone (agonist mode), assay media containing EC80 neurotrophin (antagonist mode) or assay media containing EC20 neurotrophin (PAM mode) were then added to the cells (4 µL) for a further 4 hour incubation. ToxBLAzer detection reagent was reconstituted according to the manufacturer’s instructions and 8 µL/well added to the assay plates, these were then incubated at room temperature protected from light for 2 hours. Levels of β-lactamase activity were then quantified by measuring the fluorescent intensity ratio between EX400nm/EM535nm and EX400nm/EM460nm and cytotoxicity quantified by measuring the fluorescence intensity at EX590nm/EM665nm using a Perkin Elmer EnVision.
DiscoveRx PathHunter® TrkB Assay
The assay was configured using DiscoveRx PathHunter® U2OS cell line stably co-expressing two cDNAs: a ProLink™ (PK) tagged TrkB receptor and an Enzyme Acceptor (EA) tagged SH2 domain. Activation of the TrkB-PK receptor induces receptor dimerization and trans-phosphorylation, leading to SH2-EA recruitment, and thereby forcing complementation of the two β-galactosidase enzyme fragments (EA and PK). The resulting functional enzyme hydrolyzes substrate to generate a chemiluminescent signal. U2OS-TrkB PathHunter® cells were seeded into white 384w assay plates in plating media (1x MEM; 0.5% v/v horse serum) at 10,000 cells in a volume of 20 µL per well, cells were incubated at 37°C, 5% CO2 overnight to adhere to assay plates. Test compounds were added (5 µL) to the cells and the plates were returned to the incubator for 1 hour; dependent on assay mode (agonist, antagonist or positive allosteric modulator) assay media alone (agonist mode), assay media containing EC80 BDNF (antagonist mode) or assay media containing EC20 BDNF (PAM mode) were then added to the cells (5 µL) for a further 4 hour incubation at room temperature. PathHunter® detection reagent was then added (15 µL/well) and incubated at room temperature protected from light for 1 hour. Levels of β-galactosidase activity were then quantified by measuring the chemiluminescent signal obtained using a Perkin Elmer EnVision.
Meso Scale Discovery Phospho-AKT Assay
The assay was configured using SH-SY5Y human neuroblastoma cells differentiated with all-trans retinoic acid to induce TrkB expression [27] in combination with the Meso Scale Discovery (MSD) phospho/total AKT detection reagent (K11100D-2). SH-SY5Y cells were seeded at 20,000 cells into Greiner 96-well tissue culture plates in growth media and were incubated at 37°C, 5% CO2 for 24 hours. The growth medium was then removed and replaced with growth medium (200 µL) containing 5 µM all-trans retinoic acid and returned to the incubator for an additional 48 hours. The growth medium was then removed and replaced with growth media (200 µL) containing 5 µM all-trans retinoic acid and returned to the incubator for an additional 72 hours. On the day of assay, growth media was removed and replaced with serum-free growth media (90 µL) containing 5 µM all-trans retinoic acid; cells were incubated for 1 hour at 37°C, 5% CO2. Test compounds or BDNF contained in serum-free media were added (10 µL) to the cells and the plates were returned to the incubator for 20 minutes; the plate was then placed on ice and 25 µL 5x lysis buffer (100 mM Tris pH 7.4, 250 mM NaCl, 5% v/v Triton X100, 25 mM EDTA, 1x HALT protease/phosphatase inhibitor) was applied to each well. Following a 20 minute incubation on ice the resulting cell lysates (50 µL) were transferred to the MSD AKT assay plate and the manufacturer’s methodology was followed to allow for phospho/total AKT signal detection using the MSD Sector Imager 6000.
Primary Neuronal mHTT-Induced Cell Death Assay
Cortical and striatal neurons were isolated from E18 rat embryos and separately transfected with a HTT exon1 fragment containing 73 CAG repeats (HTTN90Q73), together with a plasmid encoding for either a green AcGFP or a red AsRed fluorescent reporter before being plated on glial cells and treated with candidate pharmacological agents [28]. After 5 days in culture, the number of fluorescent cells remaining was counted using an automated high content imaging and analysis platform (GE Healthcare InCell 2000) and any changes in neuronal survival quantified.
Solubility Assay
To align with the functional data, all compound samples and dilutions were prepared using the CellSensor® assay conditions that include: assay buffer, plates, sample volumes and the same timeframe. The assay incubation time was 7 hours. 0, 3 and 7 hour samples were analysed, in triplicate, in the final assay buffer (DMEM/Glutamax + NEAA + 20 mM Hepes pH7.4 + 0.01% BSA, final DMSO concentration 0.5%) at nominal compound concentrations of 5, 20 and 50 µM. 0 hours to assess the solubility at the start of the assay, 3 hours as this is the time-frame over which the compounds exert their activity and 7 hours for the final concentration in the assay. The 7 and 3 hour samples were prepared at times which allowed simultaneous filtering and sample analysis. Samples were analysed by HPLC-UV with confirmation of the peak of interest by LC-MS. All concentrations were determined using a 7 point calibration line prepared in DMSO. Hydrocortisone (good solubility) and reserpine (poor solubility) were included as kinetic solubility assay standards and treated in a similar manner to samples.
Results
TrkB Recombinant and Native Cell-Based Screening Cascade
To test and characterize our panel of literature-based TrkB agonists, we established a panel of cell-based assays, shown in Figure 1B . We selected the Cellsensor® CHO-K1 TrkB NFAT (nuclear factor of activated T-cells) β-lactamase reporter gene assay [29] as our primary screening assay, which provided a distal transcriptional readout for receptor activation [30]. Trk-null NFAT, TrkA NFAT and TrkC NFAT CHO-K1 cell lines were also used to determine specificity and selectivity properties of the proposed pharmacological tools.
As an orthogonal assay format, we applied the TrkB enzyme fragment complementation (EFC) assay which provided a more proximal readout for receptor activation than the Cellsensor® assay (i.e receptor phosphorylation following activation results in enzyme complementation and activity readout; DiscoveRx PathHunter® assay). We also used additional assay formats to determine TrkB receptor activation status, which included: a) direct measurement of signal transduction phosphorylation events by western blot (phospho/total AKT, phospho/total ERK1/2 and phospho/total TrkB), b) Meso Scale Discovery assay for quantitative measurement of phospho/total AKT in the human neuroblastoma cell line, SH-SY5Y, which expresses endogenous TrkB upon retinoic acid differentiation, and c) an HD relevant primary neuronal mHTT-induced cell death assay, where BDNF is neuroprotective for striatal neurons. All of the assay formats were functionally validated with cognate neurotrophin(s) and a pan-Trk kinase domain inhibitor (data not shown) [26].
Literature-Based TrkB Functional Antibody Assessment
A panel of four reported TrkB modulating antibodies were tested and showed functional activity in the TrkB NFAT reporter gene assay, agreeing with previously published data ( Table 1 ) [24], [25], [31]. The TrkB antibody mediated modulation (agonism and antagonism) observed in this TrkB-dependent NFAT reporter gene assay system further validated the assay format as a viable screening platform for the characterization of putative small molecule TrkB modulators.
Table 1. TrkB antibody panel and associated activities.
| Antibody | Epitope | Description | Source | Reactivity# | Published hTrkB activity | TrkB NFAT reporter activity |
| 47/TrkB (610102) | Human extracellular subdomains d4 and d5 (TrkB amino acids 156–322) | Mouse monoclonal | Becton Dickinson | Human, Rodent | Partial antagonist; IC50 8.6 nM [31] | IC50 56 nM |
| pAb (07-225) | Entire rat extracellular domain | Rabbit polyclonal | Millipore | Human, Rodent | Partial agonist; EC50 33 nM [31] | EC50 159 nM |
| mAb (38B8) | Entire human extracellular domain | Mouse monoclonal | Pfizer Inc | Human, Rodent | Agonist; EC50 5 nM [24] | EC50 34 pM |
| mAb (29D7) | Entire human extracellular domain | Mouse monoclonal | Pfizer Inc | Human, Rodent | Agonist; EC50 200 pM [25] | EC50 64 pM |
Reactivity as described by suppliers.
Characterization of Two TrkB Monoclonal Antibody Agonists
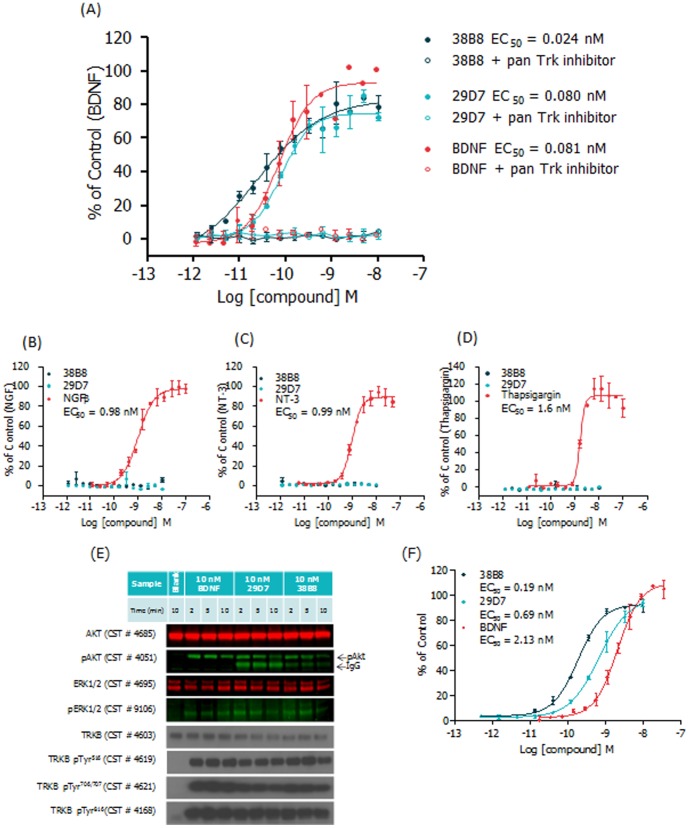
A concentration-dependent increase in NFAT reporter gene activity, equipotent to BDNF, was observed for the Pfizer mAbs ( Figure 2A ; representative data). Furthermore, induction of NFAT activity by TrkB mAbs was completely prevented by co-incubation with a pan-Trk tyrosine kinase inhibitor [26], confirming that the mAb-mediated increase in NFAT reporter gene activity requires TrkB receptor activation ( Figure 2A ).
Figure 2. Activation of TrkB signaling pathway by TrkB mAbs.
(A) CellSensor® TrkB-NFAT-bla CHO-K1 cells were stimulated with BDNF, mAb 38B8 and mAb 29D7 ± pan-Trk inhibitor (100 nM) over the indicated concentration range for 5 hours before β-lactamase assay was performed as described in Methods. % of control (maximal BDNF concentration = 9 nM) values were plotted for the indicated concentrations of each ligand (n = 2 ± SEM for each data point). The TrkB mAbs activated TrkB-dependent NFAT signaling that was inhibited by the pan-Trk kinase inhibitor; (B) CellSensor® TrkA-NFAT-bla, (C) CellSensor® TrkC-NFAT-bla, and (D) CellSensor® Trk null-NFAT-bla CHO-K1 cells were stimulated with mAb 38B8, mAb 29D7 and their respective cognate ligands (TrkA:NGFβ; TrkC:NT-3; NFAT:thapsigargin) over the indicated concentration range for 5 hours before β -lactamase assay was performed as described in Methods. % of control (maximal [NGF/NT-3/thapsigargin]) values were plotted for the indicated concentrations of each ligand (n = 2 ± SEM for each data point). Data suggest that the TrkB agonist mAbs are selective and specific; (E) CellSensor® TrkB-NFAT-bla CHO-K1 cells were stimulated with 10 nM BDNF, mAb 38B8 or mAb 29D7 over a short time course (2, 5 and 10 minutes). Cell lysates were resolved by SDS-PAGE, and phosphorylation levels of TrkB, AKT and ERK1/2 were detected using phospho-specific antibodies (pTrkB Tyr516, CST#4619; pTrkB Tyr706/707, CST#4621; pTrkB, Tyr816, CST#4168; pERK1/2, CST#4051; pAKT, CST#4051). Total levels of TrkB, AKT and ERK1/2 were also assessed (TrkB, CST#4603; AKT, CST#4685; ERK1/2, CST#4695). Relevant secondary antibodies were applied and signal was detected with ECL reagent (TrkB) or using the Odyssey infra-red reader (AKT, ERK). Data suggest activation of TrkB phosphorylation signal transduction cascade by TrkB mAbs; and (F) PathHunter® U2OS TrkB cells were stimulated with BDNF, mAb 38B8 and mAb 29D7 over the indicated concentration range for 5 hours before β-galactosidase assay was performed as described in Methods. % of control (maximal BDNF concentration = 8 nM) values were plotted for the indicated concentrations of each ligand (n = 2 ± SD for each data point). Data indicate activation of TrkB-dependent SH2 recruitment by TrkB mAbs using the TrkB EFC assay.
Using the TrkA, TrkC and Trk null NFAT reporter gene cell lines, we confirmed that both Pfizer mAbs were selective for TrkB receptors and that NFAT reporter gene activity was not modulated in the absence of the TrkB receptor, demonstrating specificity ( Figure 2B, C, D ). Validity of the TrkA, TrkC and Trk null NFAT reporter gene cell lines was confirmed using the cognate ligands, NGFβ, NT-3 and thapsigargin (an indirect NFAT activator, mediated by increase in cytosolic calcium), respectively.
As both Pfizer mAbs showed potential to become in vivo proof of concept tools and, ultimately, HD therapeutic agents, we used our established in vitro assay portfolio to further validate them. To provide direct evidence that the mAb-induced NFAT β-lactamase signal was mediated through TrkB, we evaluated proximal TrkB signalling using two methods: a) western blot detection of TrkB phosphorylation status using the TrkB-NFAT-bla CHO-K1 cell line (part of the Invitrogen CellSensor® system) and b) TrkB EFC detection of SH2 recruitment to activated TrkB receptor (part of the DiscoveRx PathHunter® system). TrkB-NFAT-bla CHO-K1 cells treated with Pfizer mAbs or BDNF over a short time course were evaluated by western blot; a panel of total and phospho-specific TrkB, AKT and ERK1/2 antibodies were applied ( Figure 2E ). The immunoblot data clearly show that, as with BDNF, the Pfizer mAbs induced robust phosphorylation of TrkB (Tyr516/Tyr706/707/Tyr816), and the downstream effector kinases AKT (Ser473) and ERK1/2 (Thr202/Tyr204). The ability of the Pfizer mAbs to induce concentration-dependent increases in the proximal TrkB EFC assay signal confirmed their agonistic mode of action ( Figure 2F ). Collectively, these activity profiles suggest that both mAbs are slightly more potent than BDNF, with mAb 38B8 displaying greater potency than mAb 29D7; however, their maximal responses are slightly below that achieved by BDNF (∼70–80% of BDNF response). A similar profile was observed when TrkB-mediated calcium mobilization in TrkB-NFAT-bla CHO-K1 cells (FLIPR-based assay; data not shown) and TrkB-mediated AKT phosphorylation in human neuroblastoma SH-SY5Y cells were measured (MSD-based assay; Figure S1). As seen in the TrkB NFAT reporter gene and EFC assay formats, mAb 38B8 was the most potent agonist; in the case of mAb 29D7, our AKT phosphorylation data substantiated functional data published by Qian et al. showing 29D7 mediated neurite outgrowth (EC50 470 pM) and survival in SH-SY5Y cells [25].
Both TrkB mAbs (38B8 and 29D7) Protect Primary Striatal Neurons from mHTT-Induced Cell Death
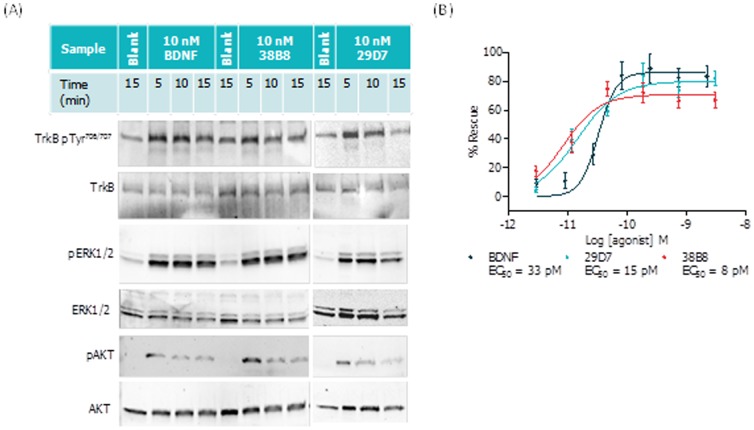
BDNF has been shown to protect neurons in a HD mouse model in vivo [21], in rat primary neurons expressing HTT in vitro [28] and in human HD iPSC-derived neurons in vitro [32]. In order to evaluate the TrkB agonist mAbs in an HD context, we assessed their pharmacology using primary neurons in a mutant HTT phenotypic assay. Initially, we confirmed the ability of the TrkB mAbs to activate TrkB and downstream pathways by Western blot in a neuronal system ( Figure 3A ), namely rat primary cortico-striatal co-cultures, as modified from Kaltenbach et al. ([28]; also see Materials and Methods).
Figure 3. Functional activity of TrkB mAbs in the primary cortico-striatal neuronal co-culture system.
Rat primary cortico-striatal co-cultures were stimulated with BDNF, mAb 38B8 and mAb 29D7 and profiled by (A) western blot; agonists tested at 10 nM over the indicated incubation times using non-transfected co-cultures; or (B) mHTT-induced co-culture protocol over the indicated concentration range, as described in the Materials and Methods. For western blot, lysates were prepared and run on SDS-PAGE gels and transferred to membranes that were then hybridized to the corresponding primary antibodies, as described in the Methods and Materials. For mHTT-induced co-culture assay, % Rescue (Normalized to in-plate controls; 0.22 nM BDNF (100% Rescue) and vehicle (0% Rescue)) values were plotted for striatal neurons over the indicated concentrations of each ligand (n = 6 ± SD for each data point). Data demonstrates activation of TrkB phosphorylation signal transduction cascade and rescue of mHTT-induced neuronal toxicity by TrkB mAbs.
Next, we used the same co-culture system, but expressing expanded repeat huntingtin (mHTT; see Methods) to assess the neuroprotective potential of the TrkB agonist mAbs ( Figure 3B ; representative data). In this co-culture system, mHTT expression, in either the striatal or cortical populations, results in neurotoxicity, as measured by the reduction of the number of surviving, viable cells. As observed with BDNF, both TrkB mAbs reproducibly demonstrated neuroprotection (low picomolar) in striatal neurons ( Figure 3B ), as measured by a significant reduction in cell loss. In contrast, cortical neuronal rescue was less clear for both BDNF and the TrkB mAbs, with protective effects being more variable and showing less potency than observed in striatal neurons (data not shown). We postulated that this may reflect differential TrkB expression between these two neuronal subpopulations. Antibody effects in striatal neurons were confirmed to be on target, as they were reversed in the presence of the pan-Trk kinase inhibitor (data not shown).
TrkB mAb Mechanism of Action Studies Reveal the Requirement for Antibody Bivalency
Due to the dimeric nature of BDNF and its reported role in mediating TrkB receptor dimerization, we sought to establish whether our functional mAbs required bivalency to exhibit agonism; such a requirement would be consistent with subsequent induction of receptor dimerization being involved in agonism or activation. In order to evaluate this hypothesis, we defragmented the complete IgG1 antibody (38B8) into Fab and F(ab’)2 fragments using a commercially available kit (Mouse IgG1 Fab and F(ab´)2 Preparation Kit, following manufacturer’s protocol; Thermo Scientific #44980). If our hypothesis for the mechanism of activation is correct, then the bivalent F(ab’)2 fragment derivative would behave as an agonist, whereas the monovalent Fab fragment would be inactive, with the potential to antagonize BDNF-mediated receptor activation should the binding sites overlap. Using the Pierce kit, we prepared the derivative Fab and F(ab’)2 fragments from the parent IgG1 antibody (38B8) (Figure S2) and assessed their activities in the TrkB NFAT reporter gene assay ( Figure 4 ). The bivalent F(ab’)2 derivative fragment retained a similar agonist activity ( Figure 4A ) to that of the parent complete IgG1, suggesting that the Fc domain was not required for receptor activation. While the monovalent Fab fragment did not display TrkB agonism, it did, however, show antagonism for BDNF-mediated TrkB activation at low micromolar concentrations ( Figure 4B ), suggesting that bivalency was required for agonism (as reflected by the F(ab’)2 fragment) and that the binding site for the antibody overlaped, at least in part, with that of BDNF (as reflected by the Fab fragment).
Figure 4. Functional activity of digested 38B8 mAb in the TrkB NFAT reporter gene assays.
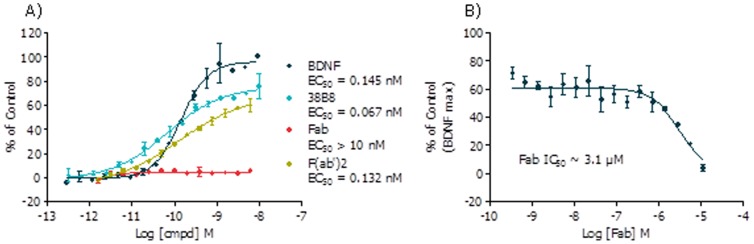
CellSensor® TrkB-NFAT-bla CHO-K1 cells were stimulated with (A) BDNF, mAb 38B8, 38B8 Fab or 38B8 F(ab’)2 over the indicated concentration range for 5 hours (agonist mode) or (B) 38B8 Fab for 1 hour prior to 0.3 nM BDNF stimulation for 4 hours (antagonist mode) before beta-lactamase assay was performed as described in Methods. % of control (maximal BDNF concentration = 9 nM) values were plotted for the indicated concentrations of each ligand (n = 2 ± SD for each data point).
Literature-Based TrkB Functional Small Molecule Identification
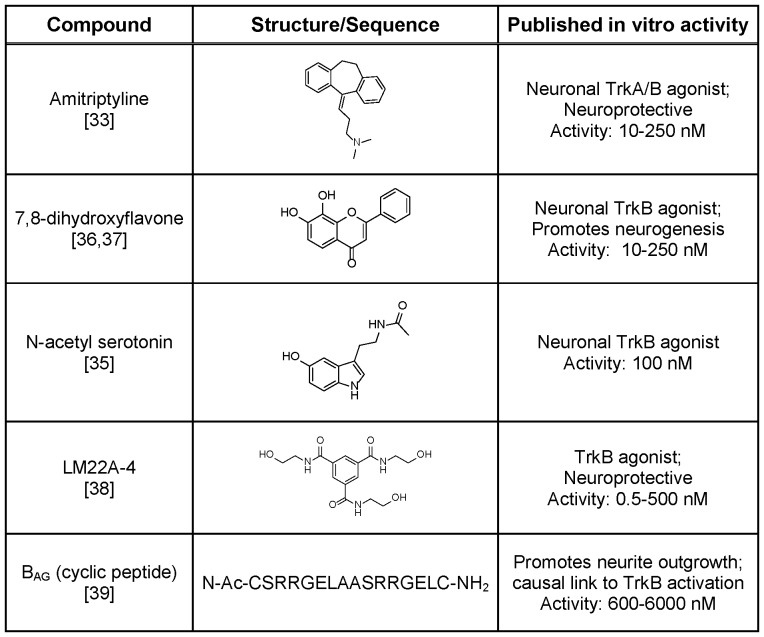
We identified literature reported TrkB small molecule agonists ( Figure 5 ) for validation and characterization. With the exception of the cyclic peptide (BAG; synthesized by Peptide Synthetics), the small molecules were either synthesized using literature routes or procured, where available. The purity of all compounds was assessed by HPLC and 1H NMR prior to testing and all showed >95% purity.
Figure 5. Literature-reported TrkB small molecule agonists.
Literature-reporteded TrkB small molecule agonists tested were amitriptyline [33], 7,8-dihydroxyflavone [36], [37], N-acetyl serotonin [35], LM22A-4 [38], and BAG (cyclic peptide [39]. Compound structure or sequence as well as corresponding reported properties and activities are indicated.
Amitriptyline is a tricyclic anti-depressant which has been reported as a potent TrkA and TrkB agonist that promotes receptor heterodimerization [33]. Jang et al reported robust TrkB phosphorylation in hippocampal neurons following treatment with 500 nM amitriptyline and state that its activity is ∼100-fold less potent than the cognate neurotrophin. Although known to inhibit the serotonin (SERT) and norepinephrine (NET) transporters, the pharmacology of amitriptyline also encompasses 5-HT1A, 5-HT2A, H1, α-1 and α-2 and muscarinic receptors [34] representing significant off-target activity. N-Acetyl serotonin is a natural metabolite accessed through the activity of N-acetyl transferase and is a precursor to melatonin, a regulator or circadian rhythms [35]. It is postulated to work in co-operation with BDNF to promote synaptic plasticity and neuroprotection and is reported to activate the TrkB receptor in primary cortical neurons in a dose dependent manner [35]. Jang et al report TrkB phosphorylation in hippocampal neurons following treatment with 100 nM N-acetyl serotonin. Similarly, 7,8-dihydroxyflavone, identified in a TrkB-dependent apoptosis assay, was reported as being a TrkB agonist activating AKT and ERK downstream kinases in hippocampal neurons; Liu et al report TrkB phosphorylation in primary rat cortical neurons following treatment with 500 nM 7,8-dihydroxyflavone [36], [37]. LM22A-4 was reported as a small molecule BDNF mimetic which prevented neuronal degeneration in rodents; this compound was identified following an in silico screen of small molecule libraries with a pharmacophore modelled on the loop 2 domain of BDNF [38]. LM22A-4 (500 nM) was described to selectively activate the TrkB receptor (native and recombinant), as well as the downstream kinases AKT and ERK, in mouse E16 hippocampal neurons and NIH3T3 TrkB-expressing cells. In addition, LM22A-4 was reported to match BDNF efficacy in prevention of neuronal death and improve motor learning after traumatic brain injury. BAG represents a designed 16-mer cyclic peptide based on the SRRGE motif at the amino terminus of NT-4. This neurotrophin mimetic was reported to increase cerebellar neurite outgrowth (maximal effect observed at ∼6 µM) and the activity was attributed to its ability to activate the TrkB receptor [39].
Putative Small Molecule TrkB Agonist Assessment
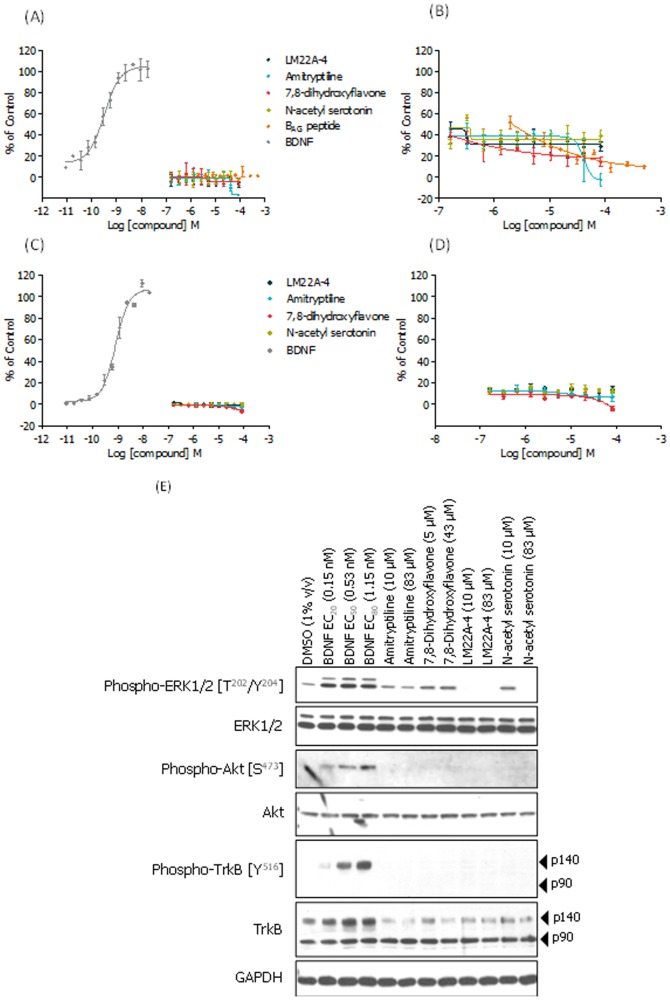
Initial activity assessment of the literature-based small molecule panel was carried out using the TrkB NFAT reporter gene assay and compounds were assessed in agonist and PAM modes ( Figure 6A and 6B , respectively). None of the compounds showed TrkB agonism or PAM activity across a concentration-range that spanned their reported activities. In fact, contrary to its reported activity, amitriptyline showed concentration-dependent TrkB inhibition ( Figure 6B ) indicating an ability to negatively modulate BDNF-induced NFAT reporter gene activation in this assay format.
Figure 6. TrkB activation assays: small molecule pharmacology.
Literature-claimed TrkB small molecule agonists tested in TrkB agonist (A, C) and PAM (B, D) modes in the CellSensor® NFAT reporter gene (A, B) and PathHunter® EFC (C, D) assays; for PAM mode the compounds are tested in the presence of ∼EC20 concentration of BDNF (40 pM, in (B) and 250 pM in (D). For agonist mode, BDNF potency (EC50) was 323 pM (A) and 823 pM (C), thereby validating the assay systems. No agonist or PAM activity was observed for the literature-claimed molecules. Curves n = 2 +/– SD, representative data where all experiments were performed in duplicate or triplicate (% of control based on maximal BDNF response). (E) Western blot assessment of ERK1/2, AKT and TrkB phosphorylation status. The CellSensor® TrkB NFAT-bla CHO-K1 cell line was treated for 5 hours using two concentrations of each test compound, as described above. As a positive control for TrkB, ERK1/2 and AKT phosphorylation BDNF was applied at three concentrations (0.15, 0.53 and 1.15 nM). Experiment also configured with a 1 hour treatment regimen; as with 5 hour data, no compound-mediated phosphorylation effects were observed at this earlier time point (data not shown).
In order to further investigate the activities (or lack thereof) of the putative literature-claimed TrkB agonist molecules, we also assessed them using the TrkB EFC assay (DiscoveRx PathHunter®), which represents a more proximal readout in the TrkB signalling pathway than the TrkB NFAT reporter gene assay, again employing both agonist and PAM modes (BAG was not characterized further due to having insufficient material). As was observed using the TrkB NFAT reporter gene assay, we did not observe any compound-mediated TrkB agonism or positive allosteric modulation ( Figure 6C, D ).
We were unable to reproduce the reported TrkB activity of any of these literature-based molecules in the TrkB EFC and NFAT reporter gene cell-based assays, reflecting proximal and distal TrkB signalling activities, respectively. In order to assess these molecules further, we used direct measurement of TrkB phosphorylation, and the down-stream kinases ERK1/2 and AKT as an approach to confirm the absence of activity of the TrkB signalling pathway by these literature-based molecules. The TrkB NFAT-bla CHO-K1 cell line was used as a suitable recombinant system. Once again, in agreement with the reporter gene activity, we did not observe molecule-mediated pathway activation ( Figure 6E ) while BDNF treatment induced phosphorylation of TrkB, ERK1/2 and AKT. We speculate that the absence of immunoreactive bands corresponding to basal phospho-ERK1/2 at the low (10 µM) and high (83 µM) concentrations of LM22A-4 and high (83 µM) concentration of N-acetyl serotonin ( Figure 6E ) represents compound-mediated and concentration-dependent protein modulation and/or cell toxicity.
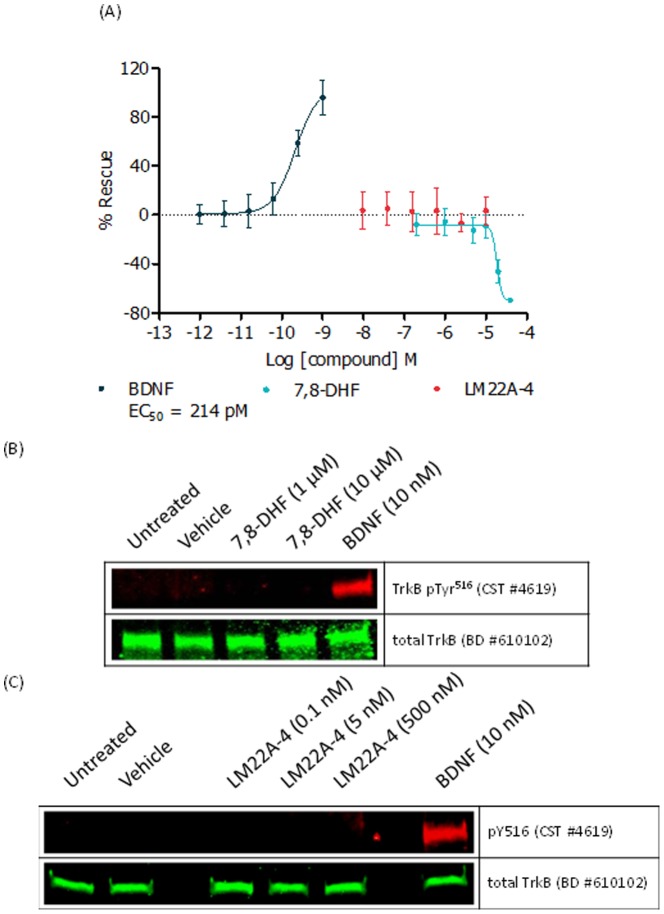
Given that the majority of the data previously described for these small molecules was generated in neuronal systems, we performed additional characterization studies of 7,8-dihydroxyflavone and LM22A-4 using our rodent primary neuron based assay system [28], [37], [38].
We assessed the ability of 7,8-dihydroxyflavone and LM22A-4 to protect rat neurons from mHTT-induced cell death in our cortico-striatal co-culture system and also assessed their ability to modulate TrkB phosphorylation. Both 7,8-dihydroxyflavone and LM22A-4 failed to mimic the ability of BDNF (and the Pfizer mAbs 38B8 and 29D7) to protect striatal neurons from mHTT-induced cell death ( Figure 7A ) or induce TrkB phosphorylation ( Figure 7B, C ); indeed, 7,8-dihydroxyflavone toxicity was evident at concentrations ≥20 µM. These neuronal data in combination with the recombinant TrkB cell line results suggested that these compounds fail to activate the TrkB receptor and don’t display agonist properties in these systems.
Figure 7. mHTT-induced neuronal toxicity: 7,8-dihydroxyflavone and LM22A-4 pharmacology.
Using our primary rat cortico-striatal co-culture system we stimulated with 7,8-dihydroxyflavone, LM22A-4 or BDNF over the indicated concentration range according to the mHTT-induced co-culture protocol (A), as described in Methods. % Rescue (Normalized to in-plate controls; 1 nM BDNF (100% Rescue) and vehicle (0% Rescue)) values were plotted for striatal neurons over the indicated concentrations of each ligand (n = 6 ± SD for each data point). Rat primary cortico-striatal cells (non-transfected) were stimulated with 7,8-dihydroxyflavone (B) or LM22A-4 (C) using the indicated concentrations for 15 minutes and profiled by western blot. BDNF- (10 nM) mediated TrkB phosphorylation validated the experimental system.
Discussion
A significant body of work indicates that the BDNF-TrkB axis is an attractive target to develop novel therapeutics for HD. One major drawback of BDNF as a therapeutic agent is its poor bioavailability and lipophilic nature [22]. We surmised that intrathecal delivery of BDNF using pumps may not result in broad distribution throughout the striatal target area [40] and elected instead to pursue alternative modalities that would activate TrkB receptors in vivo, which included purported small molecule and functional antibody activators of the TrkB receptor.
To enable our characterization of potential TrkB activators, we configured two reporter assay formats which allowed for independent orthogonal readouts of receptor activation. Our primary TrkB assay used a distal readout, an NFAT β-lactamase reporter gene assay, which was validated using commercially available BDNF and functional antibodies ( Table 1 ), all of which were reported to bind to the TrkB extracellular domain. Our secondary assay format used a more proximal readout, a TrkB functional cell line engineered to co-express a ProLink™ (PK) tagged TrkB receptor and an Enzyme Acceptor (EA) tagged SH2 domain (PathHunter®). Activation of the TrkB-PK tagged receptor induces trans-phosphorylation, leading to SH2-EA recruitment, and forcing complementation of the two β-galactosidase enzyme fragments (EA and PK). The resulting functional enzyme hydrolyzes substrate to generate a chemi-luminescent signal. Additional assay formats were also implemented to enable thorough characterization of TrkB modulation, including a primary neuronal system to allow for assessment of native receptors.
We further characterized the Pfizer mAbs using our assay portfolio, and confirmed they are selective and specific for TrkB. The selectivity of these antibodies may have crucial therapeutic advantages over the use of neurotrophins, given that the latter show an ability to also activate the ‘low affinity’ p75NTR receptor that is involved in neuronal cell death, representing an adverse on-target effect [41]. Indeed, we show here that downstream signal transduction for 29D7 and 38B8 mAbs was similar to BDNF, in that they induced TrkB phosphorylation (Y516, Y706,707, Y816), intracellular Ca2+ release (data not shown), ERK1/2 and AKT phosphorylation in native and/or recombinant receptor cell-based systems. Furthermore, using these TrkB-specific antibody agonists, we also showed that 29D7 and 38B8 rescued mHTT-induced cell death in a primary neuronal co-culture assay, supporting evaluation of these potentially therapeutic mAbs in a HD proof of concept study. As with BDNF, 29D7 and 38B8 showed preferential rescue of striatal neurons, adding further rationale for their application to HD where striatal cell loss is a major hallmark of the disease.
To characterize its activation mechanism, we proteolytically cleaved 38B8 to generate Fab (monovalent) and F(ab’)2 (bivalent) derivative fragments and assessed their functional properties using our TrkB NFAT β-lactamase reporter gene assay. The F(ab’)2 derivative fragment displayed agonism with similar potency to that of the parent complete IgG1 (38B8) whereas the monovalent Fab derivative fragment was unable to activate the receptor, highlighting the importance of antibody bivalency for activity and suggesting that the mAb mediates receptor dimerization through its ability to bridge two adjacent receptor molecules. Furthermore, we observed Fab inhibition of BDNF-mediated TrkB activation with potency in the low micromolar range, perhaps indicating that the Fab fragment has much lower binding affinity compared to its bivalent IgG1 and F(ab’)2 counterparts. These data also suggest that Fab (and 38B8) bind in a region which overlaps or partially overlaps with the BDNF binding site and such “partial” overlap may also contribute to successful albeit low potency antagonism that was observed.
As with BDNF, the proposed bridging mechanism for 38B8 thought to mediate receptor dimerization and activation is an extremely challenging function for monovalent small molecules. Nevertheless, alternative models of small molecule-mediated dimeric receptor activation have been put forward, including allosteric mechanisms reported for other receptors and support the possibility of similar mechanisms for TrkB [23]. Given the clear therapeutic delivery advantages of optimized small molecules over macromolecular neurotrophins and functional mAbs, we sought to assess a panel of literature-reported TrkB small molecules. We therefore assessed amitriptyline, 7,8-dihydroxyflavone, N-acetyl serotonin, LM22A-4 and a cyclic peptide (BAG) that had reported ability to agonise the TrkB receptor in recombinant and/or native cellular systems. Using our recombinant reporter assays, we were unable to detect TrkB agonist or positive allosteric modulatory activities in this panel of small molecules. In contrast to the TrkB agonist activity reported for amitriptyline, we observed inhibitory NFAT reporter gene activity in the micromolar range. To ensure compound availability in the assay, we characterized the solubility profiles in the exact solutions used in our NFAT reporter gene assay (Table S1). With the exception of 7,8-dihydroxyflavone, all compounds were soluble at the upper concentration ranges used in our study (5–50 µM). In the case of 7,8-dihydroxyflavone, the solubility profile indicated that only ∼20% of the compound remained in solution beyond the tested lower concentration of 5 µM; regardless, our tests allowed for predicted in-solution concentrations equivalent to and greater than those reported (10–250 nM), thereby suggesting that this compound was genuinely inactive in this TrkB reporter gene system.
To substantiate these initial findings, we assessed the phosphorylation status of the TrkB receptor and its downstream effector kinases following compound treatment in the CHO-K1 TrkB NFAT cell line. In agreement with our reporter assays we failed to observe TrkB, ERK or AKT phosphorylation, in contrast to that observed using BDNF and the 29D7 and 38B8 mAbs.
Given that the majority of reports using these putative TrkB agonists were undertaken in primary neurons, we assessed the activity of two of the most characterized compounds, 7,8-dihydroxyflavone and LM22A-4, in our primary neuronal systems. However, unlike BDNF and the Pfizer mAbs, we did not observe BDNF-TrkB pathway modulation using these small molecules; furthermore, we observed neuronal toxicity for 7,8-dihydroxyflavone at a concentration above 20 µM. Previously, in a TrkB-null cell line (Hippocampal HT-22), 7,8-dihydroxyflavone was reported to protect against glutamate-, hydrogen peroxide- and menadione-induced toxicity while increasing glutathione levels and reducing levels of reactive oxygen species, a profile consistent with an antioxidant effect [42], suggesting its action in any given cellular context may be multi-faceted and likely to involve pathways independent of TrkB. Furthermore, the HT-22 cell viability data presented by Chen et al. and our co-culture survival results show similar toxicity effects at concentrations in excess of ∼20 µM. It may have been through alternative pathways that positive effects with 7,8-dihydroxyflavone, were seen in N171-82Z HD mice, as described by Jiang et al. [43]. Although the authors demonstrated in vivo TrkB phosphorylation at a very modest level, our inability to observe TrkB phosphorylation with this compound is not completely understood but may reflect differences that exist between the complex in vivo setting and those described in both engineered tissue culture cells and primary neuronal cultures.
Previous reports indicate that LM22A-4 induced a TrkB signal transduction cascade distinct from that of BDNF [38], in that a modest induction of TrkB-Tyr516 phosphorylation (at ∼30% of the efficacy of BDNF) in hippocampal neurons stimulated a robust up-regulation of ERK and AKT phosphorylation when compared to levels induced by native neurotrophins. This phosphorylation profile was not apparent in our studies using native and recombinant systems. In contrast, we observed no induction of phospho-TrkB (pTyr516) in our rat cortico-striatal co-culture system and decreases in basal pERK1/2 in our recombinant NFAT reporter cell line. These latter data suggest that any compound-mediated modulation of pERK levels occur through a mechanism distinct from the BDNF-TrkB pathway and its action is dependent on cellular context and experimental conditions.
Our findings contradict previous reports that purport monovalent small molecules can act as TrkB neurotrophin mimetics or partial agonists and suggest that further research is required to design and identify molecules that will robustly achieve this therapeutic goal. Other possible mechanisms (and screening approaches) for small molecule TrkB receptor modulation could be considered; for example, molecules that increase neurotrophin concentration or receptor expression/trafficking.
Our characterization of reported TrkB small molecule and functional mAb agonists suggests that the 29D7 and 38B8 mAbs from Pfizer are the only candidates tested that, in our hands, induced receptor activation in a manner consistent with the activation profile of the cognate neurotrophin, BDNF. Furthermore, these mAbs showed efficacy in both recombinant and native cellular systems, with the latter system being configured in a HD-relevant context showing mAb-mediated protection of striatal neurons from mHTT-induced cell death.
In the absence of a robust brain penetrant, small molecule agonist, a proof-of-concept evaluation of the 29D7 and 38B8 mAbs in an HD relevant rodent model will be challenging due to the difficulty in delivering the antibody to the striatum. Peripheral mAb injection may also be assessed given a recent publication describing the beneficial effects of systemically administered BDNF in the R6/2 mouse model of HD [44]; however, the anorexigenic side-effect previously reported for peripherally-administered TrkB ligands will need to be carefully considered. Additionally, the mechanism by which peripheral application of BDNF induced striatal and cortex BDNF mRNA levels described was not defined [44] and requires investigation. We are currently considering bilateral microinjection of these mAbs into the striatum and other brain regions in an HD model. The proposed enhancement of TrkB signalling by these mAb TrkB agonists may be beneficial and will provide insight into the tractability of this therapeutic strategy for HD.
Supporting Information
Induction of AKT phosphorylation by TrkB mAbs using SH-SY5Y cells expressing endogenous TrkB. SH-SY5Y cells expressing endogenous TrkB. Retinoic acid-differentiated SH-SY5Y cells were stimulated with BDNF, mAb 38B8 and mAb 29D7 over the indicated concentration range for 20 minutes before measuring % phospho-AKT levels by MSD, as described in Methods. % phosphoprotein = ((2* Phospho signal)/(Phospho signal + Total signal)) *100 (n = 2 ± SEM for each data point).
(TIF)
IgG1 (38B8) digestion profile. Lane 1 & 8: MW Standards; lanes 2 & 5: IgG1; lanes 3 & 6: F(ab’)2; lanes 4 & 7: Fab. 38B8 IgG1 and IgG1 fragments (Fab and F(ab’)2) were analyzed by non-reducing (lanes 2–4) and reducing (lanes 5–7) SDS PAGE (4–12% Bis-Tris). Each well was loaded with 1.00–1.25 µg of protein. Coomassie InstantBLUE gel stain was used for detection. Expected bands under non-reduced conditions: Fab (45–50 kDa); IgG1 (150 kDa); F(ab’)2 (110 kDa). Expected bands under reduced conditions: Fab, F(ab’)2, IgG1 light chain (25 kDa); IgG1 heavy chain (50 kDa). Due to incomplete reduction (lane 5) we also observed a band at ∼100 kDa (most likely representing IgG1 heavy chain dimer).
(TIF)
Solubility assessment. Solubility analysis of the literature-based small molecules; solubility of the cyclic peptide (BAG) was not determined. Reserpine (poor solubility profile) and hydrocortisone (good solubility profile) were applied as calibration standards.
(DOCX)
Acknowledgments
The authors thank Ruth Gimeno, Ruslan Grishanin and John Lin from Pfizer for sharing their monoclonal antibody data, helpful discussions and providing the monoclonal antibodies for our studies. We also appreciate the work of Catherine O’Connell at BioFocus for collecting the solubility data, Jennifer Turney at BioFocus for confirming the digestion patterns of the 38B8 antibody fragments, Nick van den Berg for generating the primary neuronal western blot data, Alex Kiselyov for initial contributions, discussions and insight, Ramee Lee and Deanna Marchionini for helpful discussions and the assistance of Kelvin Chan, Maria Beconi, Evelyn Galstian, Mithra Mahmoudi and Simon Noble at CHDI.
Funding Statement
CHDI Foundation is a not-for-profit biomedical research organization exclusively dedicated to discovering and developing therapeutics that slow the progression of Huntington's disease. The research described was conducted by BioFocus (UK and Netherlands) under a fee-for-service agreement from CHDI Foundation. The funder, through CHDI Management, fully participated in study design, data collection and analysis, the decision to publish, and preparation of the manuscript.
References
- 1. Martin JB, Gusella JF (1986) Huntington's disease. Pathogenesis and management. N Engl J Med 315: 1267–1276. [DOI] [PubMed] [Google Scholar]
- 2. Vonsattel JP, Keller C, Cortes Ramirez EP (2011) Huntington's disease - neuropathology. Handb Clin Neurol 100: 83–100. [DOI] [PubMed] [Google Scholar]
- 3. McFarland KN, Cha JH (2011) Molecular biology of Huntington's disease. Handb Clin Neurol 100: 25–81. [DOI] [PubMed] [Google Scholar]
- 4. Shannon KM (2011) Huntington's disease - clinical signs, symptoms, presymptomatic diagnosis, and diagnosis. Handb Clin Neurol 100: 3–13. [DOI] [PubMed] [Google Scholar]
- 5. Vonsattel JP, Keller C, Del Pilar Amaya M (2008) Neuropathology of Huntington's disease. Handb Clin Neurol 89: 599–618. [DOI] [PubMed] [Google Scholar]
- 6. Harper PS (1999) Huntington's disease: a clinical, genetic and molecular model for polyglutamine repeat disorders. Philos Trans R Soc Lond B Biol Sci 354: 957–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. The Huntington's Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72: 971–983. [DOI] [PubMed] [Google Scholar]
- 8. Perutz MF, Johnson T, Suzuki M, Finch JT (1994) Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA 91: 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, et al. (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90: 549–558. [DOI] [PubMed] [Google Scholar]
- 10. DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, et al. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277: 1990–1993. [DOI] [PubMed] [Google Scholar]
- 11. Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431: 805–810. [DOI] [PubMed] [Google Scholar]
- 12. Sánchez I, Mahlke C, Yuan J (2003) Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature 421: 373–379. [DOI] [PubMed] [Google Scholar]
- 13. Barbacid M (1994) The Trk family of neurotrophin receptors. J Neurobiol 25: 1386–1403. [DOI] [PubMed] [Google Scholar]
- 14. Gentry JJ, Barker PA, Carter BD (2004) The p75 neurotrophin receptor: multiple interactors and numerous functions. Prog Brain Res 146: 25–39. [DOI] [PubMed] [Google Scholar]
- 15. Ibanez CF, Simi A (2012) p75 neurotrophin receptor signaling in nervous system injury and degeneration: paradox and opportunity. Trends Neurosci 35: 431–440. [DOI] [PubMed] [Google Scholar]
- 16. Pollack SJ, Harper SJ (2002) Small molecule Trk receptor agonists and other neurotrophic factor mimetics. Curr Drug Targets CNS Neurol Disord 1: 59–80. [DOI] [PubMed] [Google Scholar]
- 17. Gines S, Bosch M, Marco S, Gavalda N, Diaz-Hernandez M, et al. (2006) Reduced expression of the TrkB receptor in Huntington's disease mouse models and in human brain. Eur J Neurosci 23: 649–658. [DOI] [PubMed] [Google Scholar]
- 18. Zuccato C, Marullo M, Conforti P, MacDonald ME, Tartari M, et al. (2008) Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington's disease. Brain Pathol 18: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, et al. (2001) Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science 293: 493–498. [DOI] [PubMed] [Google Scholar]
- 20. Strand AD, Baquet ZC, Aragaki AK, Holmans P, Yang L, et al. (2007) Expression profiling of Huntington's disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci 27: 11758–11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie Y, Hayden MR, Xu B (2010) BDNF overexpression in the forebrain rescues Huntington's disease phenotypes in YAC128 mice. J Neurosci 30: 14708–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. BDNF Study Group T (1999) A controlled trial of recombinant methionyl human BDNF in ALS. Neurology 52: 1427. [DOI] [PubMed] [Google Scholar]
- 23. Longo FM, Massa SM (2013) Small-molecule modulation of neurotrophin receptors: a strategy for the treatment of neurological disease. Nat Rev Drug Discov 12: 507–525. [DOI] [PubMed] [Google Scholar]
- 24. Lin JC, Tsao D, Barras P, Bastarrachea RA, Boyd B, et al. (2008) Appetite enhancement and weight gain by peripheral administration of TrkB agonists in non-human primates. PLoS ONE 3: e1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qian MD, Zhang J, Tan X-Y, Wood A, Gill D, et al. (2006) Novel Agonist Monoclonal Antibodies Activate TrkB Receptors and Demonstrate Potent Neurotrophic Activities. The Journal of Neuroscience 26: 9394–9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyden TL, Guo F, Lagreca SD, Lorenz DA, Shanker RM, et al. (2007) Salts, prodrugs and formulations of 1-(5-(4-amino-7-isopropyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonyl)-2-methoxyphenyl)-3-(2,4-dichlorophenyl)urea. In: Organization WIP, editor.
- 27. Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, et al. (2000) Sequential Treatment of SH-SY5Y Cells with Retinoic Acid and Brain-Derived Neurotrophic Factor Gives Rise to Fully Differentiated, Neurotrophic Factor-Dependent, Human Neuron-Like Cells. J Neurochem 75: 991–1003. [DOI] [PubMed] [Google Scholar]
- 28. Kaltenbach LS, Bolton MM, Shah B, Kanju PM, Lewis GM, et al. (2010) Composite primary neuronal high-content screening assay for Huntington's disease incorporating non-cell-autonomous interactions. J Biomol Screen 15: 806–819. [DOI] [PubMed] [Google Scholar]
- 29. Wang J, Hancock MK, Dudek JM, Bi K (2008) Cellular assays for high-throughput screening for modulators of Trk receptor tyrosine kinases. Curr Chem Genomics 1: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Groth RD, Mermelstein PG (2003) Brain-derived neurotrophic factor activation of NFAT (nuclear factor of activated T-cells)-dependent transcription: a role for the transcription factor NFATc4 in neurotrophin-mediated gene expression. J Neurosci 23: 8125–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cazorla M, Arrang JM, Premont J (2011) Pharmacological characterization of six trkB antibodies reveals a novel class of functional agents for the study of the BDNF receptor. Br J Pharmacol 162: 947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. HD iPSC Consortium T (2012) Induced Pluripotent Stem Cells from Patients with Huntington's Disease Show CAG-Repeat-Expansion-Associated Phenotypes. Cell Stem Cell 11: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jang SW, Liu X, Chan CB, Weinshenker D, Hall RA, et al. (2009) The antidepressant amitriptyline is a TrkA and TrkB receptor agonist that promotes TrkA/TrkB heterodimerization and has potent neurotrophic activity. Chem Biol 16: 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Owens MJ, Morgan WN, Plott SJ, Nemeroff CB (1997) Neurotransmitter receptor and transporter binding profile of antidepressants and their metabolites. J Pharmacol Exp Ther 283: 1305–1322. [PubMed] [Google Scholar]
- 35. Jang SW, Liu X, Pradoldej S, Tosini G, Chang Q, et al. (2010) N-acetylserotonin activates TrkB receptor in a circadian rhythm. Proc Natl Acad Sci U S A 107: 3876–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jang SW, Liu X, Yepes M, Shepherd KR, Miller GW, et al. (2010) A selective TrkB agonist with potent neurotrophic activities by 7,8-dihydroxyflavone. Proc Natl Acad Sci U S A 107: 2687–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X, Chan CB, Qi Q, Xiao G, Luo HR, et al. (2012) Optimization of a small tropomyosin-related kinase B (TrkB) agonist 7,8-dihydroxyflavone active in mouse models of depression. J Med Chem 55: 8524–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Massa SM, Yang T, Xie Y, Shi J, Bilgen M, et al. (2010) Small molecule BDNF mimetics activate TrkB signaling and prevent neuronal degeneration in rodents. J Clin Invest 120: 1774–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams G, Williams EJ, Maison P, Pangalos MN, Walsh FS, et al. (2005) Overcoming the inhibitors of myelin with a novel neurotrophin strategy. J Biol Chem 280: 5862–5869. [DOI] [PubMed] [Google Scholar]
- 40. Ochs G, Penn RD, York M, Giess R, Beck M, et al. (2000) A phase I/II trial of recombinant methionyl human brain derived neurotrophic factor administered by intrathecal infusion to patients with amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis 1: 201–206. [DOI] [PubMed] [Google Scholar]
- 41. Huang EJ, Reichardt LF (2003) Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem 72: 609–642. [DOI] [PubMed] [Google Scholar]
- 42. Chen J, Chua KW, Chua CC, Yu H, Pei A, et al. (2011) Antioxidant activity of 7,8-dihydroxyflavone provides neuroprotection against glutamate-induced toxicity. Neurosci Lett 499: 181–185. [DOI] [PubMed] [Google Scholar]
- 43. Jiang M, Peng Q, Liu X, Jin J, Hou Z, et al. (2013) Small-molecule TrkB receptor agonists improve motor function and extend survival in a mouse model of Huntington's disease. Hum Mol Genet 22: 2462–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giampa C, Montagna E, Dato C, Melone MA, Bernardi G, et al. (2013) Systemic delivery of recombinant brain derived neurotrophic factor (BDNF) in the R6/2 mouse model of Huntington's disease. PLoS One 8: e64037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Induction of AKT phosphorylation by TrkB mAbs using SH-SY5Y cells expressing endogenous TrkB. SH-SY5Y cells expressing endogenous TrkB. Retinoic acid-differentiated SH-SY5Y cells were stimulated with BDNF, mAb 38B8 and mAb 29D7 over the indicated concentration range for 20 minutes before measuring % phospho-AKT levels by MSD, as described in Methods. % phosphoprotein = ((2* Phospho signal)/(Phospho signal + Total signal)) *100 (n = 2 ± SEM for each data point).
(TIF)
IgG1 (38B8) digestion profile. Lane 1 & 8: MW Standards; lanes 2 & 5: IgG1; lanes 3 & 6: F(ab’)2; lanes 4 & 7: Fab. 38B8 IgG1 and IgG1 fragments (Fab and F(ab’)2) were analyzed by non-reducing (lanes 2–4) and reducing (lanes 5–7) SDS PAGE (4–12% Bis-Tris). Each well was loaded with 1.00–1.25 µg of protein. Coomassie InstantBLUE gel stain was used for detection. Expected bands under non-reduced conditions: Fab (45–50 kDa); IgG1 (150 kDa); F(ab’)2 (110 kDa). Expected bands under reduced conditions: Fab, F(ab’)2, IgG1 light chain (25 kDa); IgG1 heavy chain (50 kDa). Due to incomplete reduction (lane 5) we also observed a band at ∼100 kDa (most likely representing IgG1 heavy chain dimer).
(TIF)
Solubility assessment. Solubility analysis of the literature-based small molecules; solubility of the cyclic peptide (BAG) was not determined. Reserpine (poor solubility profile) and hydrocortisone (good solubility profile) were applied as calibration standards.
(DOCX)