Abstract
Acetylation of the RelA subunit of NF-κB at lysine-310 regulates the transcriptional activation of NF-κB target genes and contributes to maintaining constitutively active NF-κB in tumors. Bromodomain-containing factor Brd4 has been shown to bind to acetylated lysine-310 and to regulate the transcriptional activity of NF-κB, but the role of this binding in maintaining constitutively active NF-κB in tumors remains elusive. In this study, we demonstrate the structural basis for the binding of bromodomains of Brd4 to acetylated lysine-310 and identify bromodomain inhibitor JQ1 as an effective small molecule to block this interaction. JQ1 suppresses TNF-α-mediated NF-κB activation and NF-κB-dependent target gene expression. Additionally, JQ1 inhibits the proliferation and transformation potential of A549 lung cancer cells and suppresses the tumorigenicity of A549 cells in severe combined immunodeficiency (SCID) mice. Furthermore, we demonstrate that depletion of Brd4 or treatment of cells with JQ1 induces the ubiquitination and degradation of constitutively active nuclear form of RelA. Our results identify a novel function of Brd4 in maintaining the persistently active form of NF-κB found in tumors, and they suggest that interference with the interaction between acetylated RelA and Brd4 could be a potential therapeutic approach for the treatment of NF-κB-driven cancer.
Keywords: Brd4, cancer, degradation, JQ, NF-κB, ubiquitination
Introduction
Transcription factor NF-κB/Rel family proteins are master regulators of immune and inflammatory responses. The prototypical NF-κB, a heterodimer of p50 and RelA, is sequestered in the cytoplasm by its association with the inhibitor IκBα in unstimulated cells. A variety of stimuli, including proinflammatory cytokines, T or B cell signals, and bacterial or viral products, activate NF-κB by triggering the activation of IκB kinases, leading to the phosphorylation and degradation of IκBα and the nuclear translocation of NF-κB (1–3). Once in the nucleus, NF-κB binds to the cognate κB enhancers and stimulates the expression of genes involved in immune and inflammatory responses. In addition to its essential role in immunity, NF-κB is also a key player in the initiation and progression of human cancer (4–6). Many oncogenic proteins activate NF-κB, and constitutively active NF-κB is found in a variety of solid tumors and hematologic malignancies (6–9). Various animal models of cancer in which NF-κB signaling is disrupted by genetic approaches also demonstrate the promoting role of NF-κB in inflammation-linked cancer (10). In fact, many of the key hallmarks of cancer, including cell proliferation, apoptosis, inflammation, angiogenesis, and metastasis, are known to be regulated by target genes of NF-κB (8, 11).
Posttranslational modifications of histone and non-histone proteins, including phosphorylation, acetylation, methylation and ubiquitination, have been shown to be critically involved in transcription regulation and cancer development (12, 13). Emerging evidence suggests that reversible acetylation of the RelA subunit of NF-κB plays an important role in modulating the pathophysiologic functions of NF-κB (14, 15). Stimulus-coupled acetylation of RelA by p300/CBP at lysine-310 enhances the transcriptional activity of NF-κB (16, 17). Conversely, deacetylation of lysine-310 by histone deacetylases (HDACs) reduces transcriptional activity of NF-κB and sensitizes cells to TNF-α-induced apoptosis (16, 18). Hyperacetylated RelA has also been found in many cancer cells, and acetylation accounts for constitutively active NF-κB in cancer cells by prolonging NF-κB nuclear retention in tumors (19, 20). We have recently shown that acetylated lysine-310 of RelA is specifically recognized by bromodomain-containing protein 4 (Brd4) via its two bromodomains (21). Binding of Brd4 to acetylated lysine-310 recruits and activates CDK9 of the positive transcription elongation factor b (P-TEFb) complex to phosphorylate RNA polymerase II for the transcriptional activation of a subset of NF-κB target genes (21). However, it is not clear whether binding of Brd4 to acetylated lysine-310 also contributes to the persistence of constitutively active NF-κB in tumors.
The multi-functional Brd4 belongs to the BET family of proteins that contain two tandem bromodomains and an extra terminal (ET) domain (22–24). Brd4 binds to papillomavirus E2 proteins and tethers the viral DNA to host mitotic chromosomes for segregation of its genome into daughter cells (25). Brd4 also regulates gene transcription by recruiting different transcription components, such as Mediator and P-TEFb, to selective target genes by binding to acetylated histone H3 or H4 (26, 27). Recent studies have suggested that Brd4 might have additional function in tumorigenesis. Chromosome translocation involving Brd4 is found in NUT (nuclear protein in testis) midline carcinoma, and the resulting fusion Brd4-NUT oncoprotein is responsible for the pathogenesis of this rare human cancer (28). Brd4 has also been shown to be required for the maintenance of acute myeloid leukemia (29). The essential role of Brd4 in cancer development is further demonstrated by the recent findings that small molecules targeting bromodomains of Brd4 possess strong anti-tumor activities (29–31). These small molecules displace BET bromodomains from histones by competitively binding to the acetylated lysine recognition pocket (30, 32, 33). One of these BET inhibitors (BETi), JQ1, induces rapid differentiation and growth arrest of cells from NUT midline carcinoma and displays anti-tumor effects in several hematologic malignancies, including acute myeloid leukemia, multiple myeloma, and Burkitt’s lymphoma (29–31, 34). Interestingly, another BETi, I-BET, suppresses inflammatory gene expression and protects mice from lipopolysaccharide-induced endotoxic shock and bacteria-induced sepsis (32). Brd4 functions as a coactivator of NF-κB, which plays an essential role in regulating the inflammatory response and in the development of cancer, raising the intriguing question of whether the anti-tumor and anti-inflammatory effects of BETi might be partially derived from dislocation of Brd4 from the acetylated RelA subunit of NF-κB.
In an effort to understand the function of Brd4 binding to acetylated RelA in cancer cells, we found that Brd4 maintained the nuclear NF-κB levels by preventing its ubiquitination and degradation. Blockage of the interaction between Brd4 and RelA with JQ1 inhibits NF-κB activation and suppresses the proliferation and tumorigenicity of A549 lung cancer cells. Our results reveal a mechanism by which binding of Brd4 to acetylated RelA contributes to constitutively active NF-κB in cancer cells and suggest possible therapeutic approaches for the treatment of NF-κB-driven cancer by targeting the interaction between NF-κB and Brd4.
Results
Structural basis for the binding of Brd4 to acetylated lysine-310 of RelA
We have previously shown that Brd4 coactivates transactivation of NF-κB by binding, via its two bromodomains (BDs), to acetylated lysine-310 of the RelA subunit of NF-κB (21). In order to understand the molecular basis of this interaction, we sought to determine the three-dimensional structure of each of the two bromodomains in complex with a peptide encompassing the acetylated lysine-310 of RelA. The co-crystal structure of BD1 was solved to 1.5 Å resolution and that of BD2 to 2.0 Å resolution.
Although the identical peptide was used for the co-crystallization with each of the two bromodomain structures, the binding orientation of the peptide in each structure is distinct. Our structural data reveal that acetylated lysine-310 (AcLys310) directly interacts with the highly conserved asparagine (Asn) residues in both of the bromodomains (Asn140 in BD1 and Asn433 in BD2) (Figure 1A& 1B). Specifically, AcLys310 forms a hydrogen bond through the carbonyl oxygen of the acetylated side chain to the nitrogen atom of the respective asparagine residue in each structure (Figure 1A & 1B). Apart from the conservation of this notable interaction, each bromodomain engages the peptide in markedly different manners. In the BD1 co-crystal structure, the peptide adopts a helical conformation and AcLys310 is located at the base of the helix where it points into a binding cleft (Figure 1A). There are no additional hydrogen bond interactions between BD1 and the peptide, although there are a number of van der Waals contacts. In contrast, in the BD2 co-crystal structure, the peptide adopts a slightly more extended conformation. Most notably, histidine-437 (His437) is located immediately adjacent to the AcLys310 binding site, where it engages in hydrogen bonding interactions with both threonine-308 (Thr308) and serine-311 (Ser311) of RelA (Figure 1B). This residue is unique to BD2, and, in the BD1 co-crystal structure the corresponding residue (Asp144) is diverted away from the binding site and does not make any contacts with the RelA peptide (Figure 1A). The additional interactions between His437 of BD2 and Thr308 and Ser311 of RelA may account for ligand specificity.
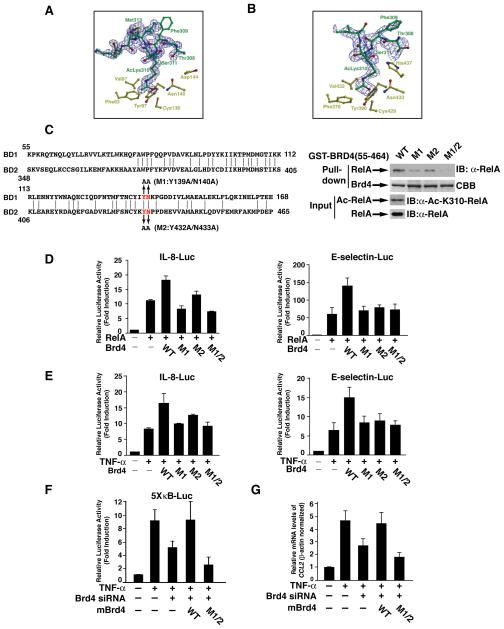
Figure 1. Structural basis for the binding of Brd4 to acetylated lysine-310 of RelA.
(A) Stereo view of BD1 ligand binding site showing the residues that are in the vicinity of the AcLys310 of RelA. Superimposed is a difference Fourier electron density map (contoured at 2.5σ over background in blue and 8σ over background in red) calculated with coefficients |Fobs| - |Fcalc| and phases from the final refined model with the coordinates of peptide deleted prior to one round of refinement. (B) Stereo view of BD2 ligand binding peptide showing equivalent residues. The superimposed difference Fourier electron density map was calculated as described in (A). (C) Sequence alignment of BD1 and BD2 of Brd4 and the description of various mutations within bromodomains (left panel). Recombinant proteins of GST-Brd4 bromodomains or its various mutants were used to pull down cell lysates containing acetylated RelA (right panel). (D) IL8-luciferase (IL-8-Luc) or E-selectin-luciferase (E-selectin-Luc) reporter plasmids were co-transfected with RelA expression vector, alone or in combination with WT Brd4 or its various mutants, into HEK293T cells. Luciferase activity was measured 30 h after transfection. Results represent the average of three independent experiments +/−SD. (E) HEK293T cells were co-transfected with IL-8-Luc or E-selectin-Luc reporter plasmid alone or in combination with expression vectors for WT Brd4 or its various mutants. Twenty-four hours after transfection, cells were treated with TNF-α (20 ng/ml) for 5 h and luciferase activity was measured as described in (D). (F) HEK293T cells transfected with Brd4 siRNA for 24 h were transfected with 5X-κB-Luc reporter plasmid alone or in combination with expression vectors for mouse WT or mutant Brd4. Twenty-four h after transfection, cells were treated with TNF-α and luciferase activity was measured as described in (D). (G) A549 cells transfected with Brd4 siRNA were reconstituted with mouse WT or mutant Brd4 followed by stimulation for 1 hrs with TNF-α and gene expression was analyzed by quantitative RT-PCR.
To further confirm the importance of the key amino acids (Asn140 in BD1 and Asn433 in BD2) in the interaction with acetylated lysine-310, we mutated these amino acids together with the adjacent highly conserved tyrosines to alanines (Figure 1C) and investigated the effect of these mutations on the interaction with the acetylated lysine-310 peptides. In the GST pull-down experiment, GST-Brd4 fusion proteins containing both bromodomains were used to pull down acetylated RelA at lysine-310 (Figure 1C, right panels). Bromodomains of Brd4 successfully pulled down acetylated RelA (Figure 1C). Mutation of the highly conserved asparagine and tyrosine within BD1 (designated as M1) or within BD2 (designated as M2) decreased the interaction with acetylated RelA (Figure 1C, right panels). Simultaneously mutation of these amino acids (designated as M1/2) abolished the interaction with acetylated RelA (Figure 1C, right panels). These mutagenesis studies further confirm that amino acids identified from our structural study are indeed involved in the interaction with acetylated RelA.
Since Brd4 coactivates transcriptional activity of NF-κB by binding to acetylated lysine-310 of RelA (21), we next assessed the abilities of these Brd4 mutants to coactivate NF-κB. Wild-type Brd4 enhanced transcriptional activity of RelA in the κB-luciferase reporter assay (Figure 1D). However, mutation of these key amino acids to alanines within BD1 or BD2 or both BD1 and BD2 impaired the coactivation function of Brd4 (Figure 1D). Mutation of these amino acids also compromised Brd4’s ability to coactivate TNF-α-induced endogenous NF-κB activation (Figure 1E). Furthermore, when siRNA resistant mouse Brd4 was re-introduced into Brd4 knockdown HEK293T cells, WT but not mutant Brd4 was able to recover reduced TNF-α-induced NF-κB activation in κB-luciferase reporter assay (Figure 1F). Similarly, in Brd4 knockdown A549 cells, WT but not mutant Brd4 rescued TNF-α-induced NF-κB target gene expression (Figure 1G). These results support the notion that binding of Brd4 to acetylated lysine-310 via its two bromodomains is critical for the coactivation function of Brd4 for NF-κB target gene expression.
JQ1 inhibits the binding of Brd4 to acetylated lysine-310 and suppresses NF-κB activation
Bromodomain inhibitor JQ1 specifically binds to the asparagines of the bromodomains of Brd4 and prevents the binding of acetylated histone peptides to the binding pocket (30). Our structural and mutagenesis analysis of the bromodomains of Brd4 also reveals that these highly conserved asparagines are critically involved in the direct interaction with acetylated lysine-310 of RelA (Figure 1), raising the possibility that JQ1 might interfere with the binding of acetylated RelA to the bromodomains of Brd4. To test this possibility, we first employed an in vitro peptide-binding assay to examine the effect of JQ1 on the binding of Brd4 bromodomains to acetylated lysine-310 RelA peptides. In this binding assay, biotin-labeled RelA peptides containing acetylated lysine-310 were used to pull-down recombinant GST-fusion proteins of the bromodomains of Brd4. Acetylated lysine-310 peptides were found to associate with a substantial amount of bromodomains of Brd4 (Figure 2A). Notably, the interaction was inhibited by the addition of JQ1 (Figure 2A), suggesting that JQ1 inhibits the binding of acetylated lysine-310 RelA peptides to the bromodomains of Brd4. To further confirm this, we examined the effect of JQ1 on the binding of acetylated RelA to full-length Brd4 in an in vivo co-immunoprecipitation assay. Consistent with our previous findings (21), the interaction between RelA and Brd4 was enhanced by the co-expression of p300, which acetylates RelA at lysine-310 (Figure 2B). However, the interaction was gradually reduced with the addition of increasing amount of JQ1 (Figure 2B). Furthermore, when the effect of JQ1 on the interaction between endogenous nuclear RelA and Brd4 was examined in A549 cells, we found that JQ1 inhibited the interaction of RelA and Brd4 (Figure 2C). All together these results indicate that JQ1 indeed attenuates the binding of Brd4 to RelA.
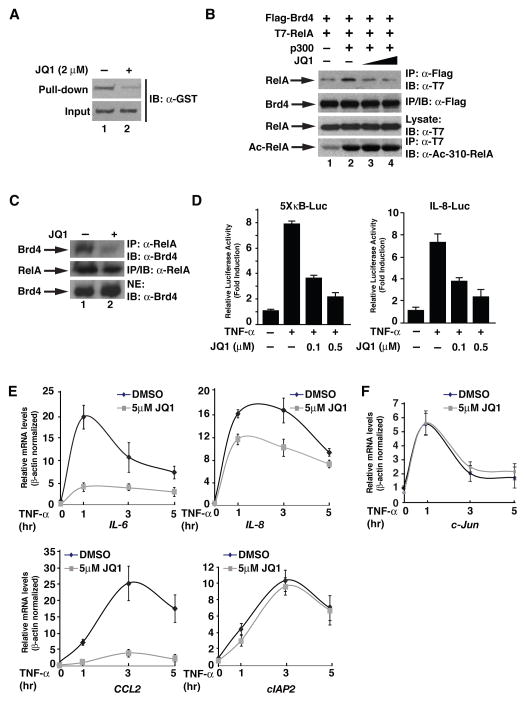
Figure 2. JQ1 inhibits the binding of Brd4 to RelA and suppresses NF-κB activation.
(A) Purified GST-bromodomains (1 μg) of Brd4 were incubated with a biotin-labeled RelA acetylated K310 peptide (21) bound to Streptavidin agarose beads. Binding of the bromodomains to the peptide in the presence or absence of JQ1 (2 μM) was detected by immunoblotting with anti-GST antibody (upper panel). (B) JQ1 inhibits the interaction between Brd4 and RelA in vivo. HEK293T cells were transfected with the indicated combination of expression vectors for Flag-Brd4, T7-RelA, and HA-p300. Brd4 immunoprecipitation was performed in the presence or absence of JQ1 (0, 10, 20 μM) and immunoprecipitates were immunoblotted with anti-T7 antibodies. (C) A549 cells were treated with 5 μM JQ1 or vehicle for 4 hr. Nuclear extracts were prepared and subject to immunoprecipitation with anti-Brd4 antibodies. Brd4 associated RelA was detected by immunoblotting the Brd4 immunoprecipitates with anti-RelA antibodies. Levels of Brd4 and RelA were shown in the lower panels. (D) JQ1 inhibits TNF-α-mediated activation of NF-κB. HEK293T cells were co-transfected with 5XκB-Luc or IL-8-Luc reporter plasmid. Twenty-four hours after transfection, cells were pretreated with different doses of JQ1 for one hour before treatment with TNF-α for 5 hr, and luciferase activity was measured as in Figure 1D. (E),(F) A549 cells were pretreated with JQ1 (5 μM) for one hr and stimulated with TNF-α for indicated time points. Quantitative RT-PCR was performed to analyze the TNF-α-induced expression of NF-κB (E) or AP1 (F) target genes. Results represent the average of three independent experiments +/−SD.
Since binding of Brd4 to acetylated RelA is essential for the transcriptional activation of NF-κB (21), we hypothesized that JQ1, by blocking the interaction between RelA and Brd4, might suppress the transcriptional activation of NF-κB. In the luciferase assay using 5X-κB-Luc or IL-8-Luc luciferase reporter, we observed that JQ1 inhibited TNF-α-stimulated NF-κB activation in a dose-dependent manner (Figure 2D). Furthermore, when we examined the effect of JQ1 on TNF-α-induced NF-κB target gene expression, we found that JQ1 compromised TNF-α-induced expression of IL-6, IL-8, and CCL2 mRNA (Figure 2E). However, TNF-α-induced expression of c-IAP2 was barely affected by JQ1, indicating that JQ1 selectively inhibits TNF-α-induced NF-κB target gene expression (Figure 2E). Interestingly, TNF-α-induced expression of c-Jun, which is activated by transcription factor AP1 but not by NF-κB (35), was not affected by JQ1 (Figure 2F). Collectively, these data demonstrate that JQ1 specifically inhibits the transcriptional activation of a subset of NF-κB target genes.
JQ1 suppresses the tumorigenicity of lung cancer cells
Since constitutively active NF-κB contributes to the proliferation of cancer cells and JQ1 suppresses the activity of NF-κB (Figure 2), we next examined the effect of JQ1 on the proliferation of A549 lung cancer cells. A549 cells treated with control vehicles alone exhibited a steady growth over the tested six days (Figure 3A). In contrast, the growth of A549 cells was reduced in the presence of JQ1 in a dose-dependent manner (Figure 3A), suggesting that JQ1 inhibits the proliferation of A549 cells.
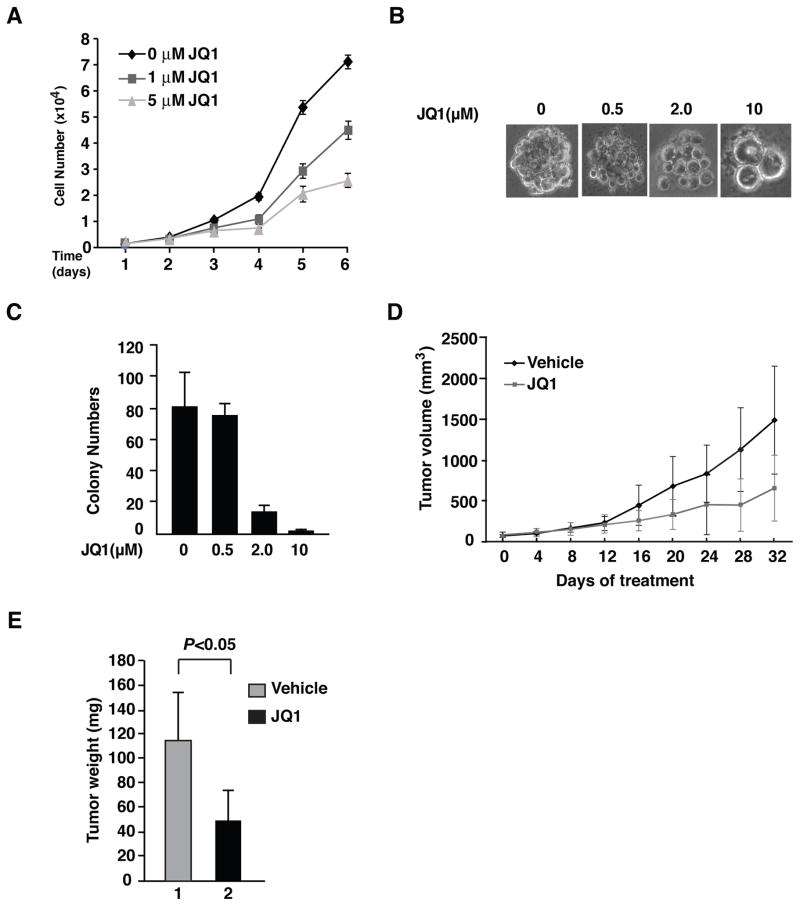
Figure 3. JQ1 suppresses the proliferation and tumorigenesis of A549 cells.
(A) A549 cells were plated in 96-well plates in DMEM and cultured for three days. Cells were then treated with or without JQ1 as indicated for up to 6 days. Cell proliferation was measured by using the MTS assay. Data represent the average of three independent experiments +/−SD. (B) A total of 5000 A549 cells were suspended in DMEM containing 0.35% SELECT Agar® (Invitrogen) and then plated in 6-well plates coated with an initial underlay of 0.5% SELECT Agar® (Invitrogen) in culture medium. Colony growth was scored after 14 days of cell incubation with or without JQ1 treatment as indicated. Representative photographs were taken at day 14 to show colonies. All the colony formation assays presented in this study were repeated in at least 3 independent experiments. (C) Nude mice bearing A549 xenografts were treated with vehicle or JQ1 daily for the indicated times with the dosage of 50 mg/kg (mpk). Mice (n=3 in each group) were killed when tumor volume reached 2,000 mm3. (D) Summary of the average weight of tumors from (C). Statistical analysis (P value) was performed using a student’s t-test. Data represent mean ± SD (n=3). All experiments involving mice were approved by the Institutional Animal Care and Use Committee (IACUC).
Next, we investigated the effect of JQ1 on the transformation potential of A549 cells by anchorage-independent growth assay. As expected, A549 cells formed colonies in soft agar when treated with control vehicles (Figure 3B). Treatment with different concentrations of JQ1 revealed a dose-dependent inhibitory effect on the size of the colonies formed from A549 cells, from a moderate effect with 0.5 μM of JQ1 to a remarkable effect with 10 μM of JQ1 treatment (Figure 3B). Additionally, we observed a similar inhibitory effect of JQ1 on the number of the colonies formed from A549 cells (Figure 3C). These data indicate that JQ1 impairs the transformation potential of A549 cells.
We then evaluated the effect of JQ1 on tumor formation and growth in a nude mouse xenograft model. Measurable tumors were grown in immune-deficient nude mice after subcutaneous injection with A549 cells for 2 weeks. A cohort of six mice bearing A549 xenograft were randomized into two groups with approximately equal tumor burden. One group was treated with daily subcutaneous injection of JQ1 for 32 consecutive days, whereas the control group was treated with control vehicles only. The A549 xenografts in mice treated with JQ1 grew significantly more slowly than those in mice treated with control vehicles after 12 days (Figure 3D). The average weight of the tumors formed from JQ1 treated mice was >60% lower than those treated with control vehicles (Figure 3E). All together, these data suggest that JQ1 is effective in halting tumor growth, likely by inhibiting the proliferation of the tumor cells.
Brd4 stabilizes nuclear NF-κB by preventing the ubiquitination of RelA
In cancer cells, the sustained nuclear expression of NF-κB accounts for constitutively active NF-κB, which is closely associated with the tumorigenicity of many cancer cells (6–9). Since Brd4 inhibitor JQ1 inhibits NF-κB activity and suppresses the tumorigenicity of A549 lung cancer cells (Figures 2&3), we hypothesized that Brd4 might play a role in regulating constitutively active NF-κB. To test this hypothesis, we first examined the effect of down-regulation of Brd4 using RNA interference on the levels of nuclear and cytoplasmic NF-κB in A549 cells. Notably, the decreased expression of Brd4 by Brd4 siRNA in A549 cells was associated with reduced nuclear but not cytoplasmic levels of RelA (Figure 4A). This down-regulation seems to be specific for RelA since knockdown of Brd4 had no effect on the nuclear and cytoplasmic levels of p50 (Figure 4A).
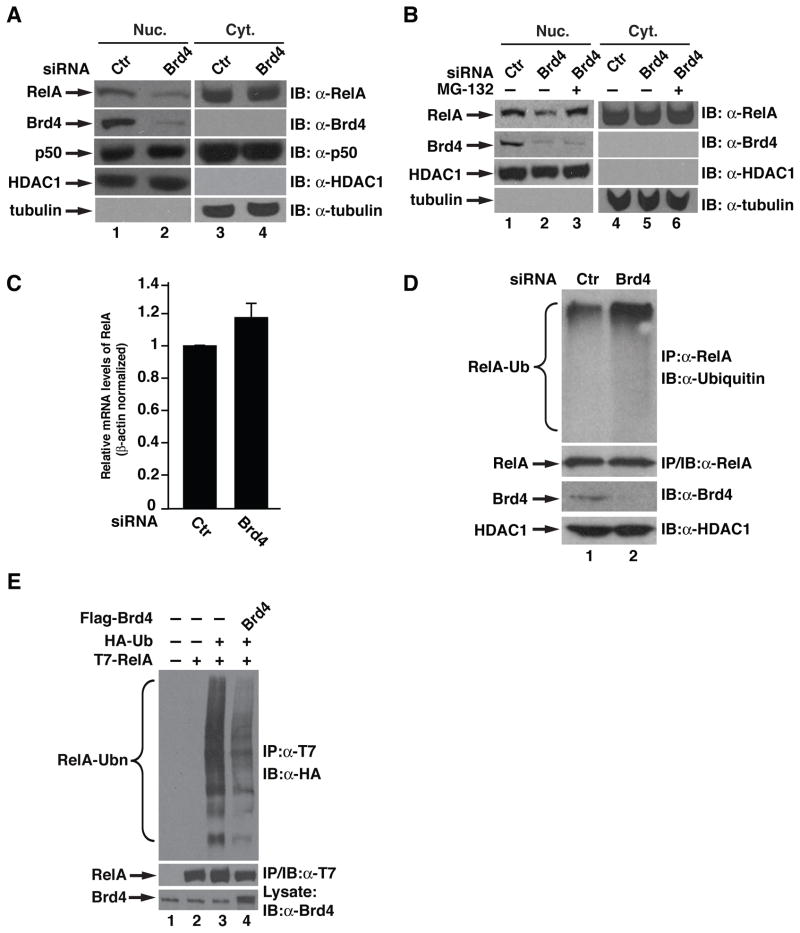
Figure 4. Brd4 stabilizes nuclear RelA by preventing its ubiquitination.
(A) A549 cells were transfected with control or Brd4 siRNA for 48 hr, the nuclear (Nuc.) and cytoplasmic (Cyt.) extracts were immunoblotted for the levels of endogenous RelA and Brd4. HDAC1 and tubulin were used as nuclear or cytoplasmic protein control, respectively. (B) A549 cells were transfected with siRNA as in (A). Forty hr after transfection, cells were treated with MG132 (10 μM) for 8 hr. Nuclear or cytoplasmic extracts were then immunoblotted for levels of RelA, Brd4, HDAC1 and tubulin. (C) A549 cells were transfected with control or Brd4 siRNA for 48 hr before total RNA was extracted. Quantitative RT-PCR was performed to analyze the expression level of RelA. Data represent the average of three independent experiments +/−SD. (D) A549 cells were transfected with control or Brd4 siRNA and nuclear RelA immunoprecipitates were immunoblotted for ubiquitination with anti-ubiquitin antibodies. To prevent the degradation of RelA, cells were treated with MG-132 (10μM) for 2 hr before lysis of cells. (E) HEK293T cells were transfected with indicated combinations of expression vectors for T7-RelA, HA-ubiquitin and Flag-Brd4. T7-RelA immunoprecipitates were immunoblotted for ubiquitination with anti-HA antibodies.
The reduced nuclear levels of RelA in Brd4 knockdown A549 cells prompted us to investigate whether Brd4 might regulate RelA stability. We treated the Brd4 knockdown cells with MG-132, a proteasome inhibitor, and examined the levels of nuclear RelA. Treatment with MG-132 reversed the decreased levels of nuclear RelA in Brd4 knockdown cells (Figure 4B). Additionally, depletion of Brd4 had no effect on the level of RelA mRNA (Figure 4C), suggesting that Brd4 regulates RelA at the protein level rather than at the mRNA level and that Brd4 regulates the stability of nuclear RelA.
We next examined the effect of Brd4 on the ubiquitination of RelA, an event that is required for proteasome-mediated degradation. We knocked down the expression of Brd4 and examined the ubiquitination of nuclear RelA in A549 cells. Nuclear RelA was moderately ubiquitinated in cells transfected with control siRNA, reflecting a basal level turnover of nuclear RelA (Figure 4D). However, depletion of Brd4 with Brd4 siRNA enhanced the ubiquitination of nuclear RelA (Figure 4D), indicating that Brd4 regulates nuclear RelA ubiquitination. Furthermore, when exogenously expressed Brd4 was examined for its effect on the ubiquitination of nuclear RelA, we found that Brd4 suppressed the ubiquitination of RelA (Figure 4E). In contrast, depletion of endogenous Brd4 or overexpression of Brd4 had not effect on the ubiquitination of p50 (Supplementary Figures S1 and S2). Collectively, these data suggest that Brd4 stabilizes nuclear RelA by inhibiting its ubiquitination.
JQ1 induces the ubiquitination and degradation of nuclear NF-κB
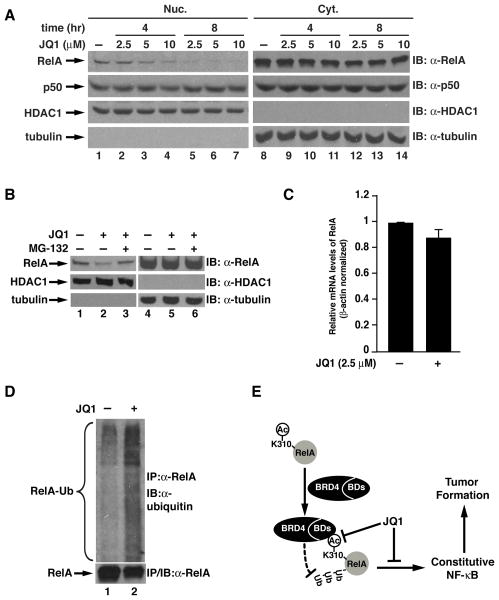
Since JQ1 blocks the interaction between RelA and Brd4 and inhibits transcription activity of NF-κB (Figures 2), we next determined whether JQ1 might have a similar effect as knockdown of Brd4. We treated A549 cells with different concentrations of JQ1 at different time points and examined the cytoplasmic and nuclear levels of RelA. The nuclear levels of RelA decreased with the increased concentration of JQ1 either at 4 hr or 8 hr (Figure 5A). Similar to the Brd4 knockdown cells, the levels of cytoplasmic RelA were not changed by JQ1 (Figure 5A). The effect of JQ1 on nuclear RelA appears to be specific since both the nuclear and the cytoplasmic levels of p50 remained unaltered (Figure 5A). Similarly, proteasome inhibitor MG-132 reversed the decreased nuclear levels of RelA (Figure 5B) and the transcription of RelA was not affected by JQ1 (Figure 5C). These data suggest that reduced levels of RelA in the nucleus is due to proteasome-mediated degradation.
Figure 5. JQ1 induces the ubiquitination and degradation of nuclear RelA.
(A) JQ1 reduces the nuclear levels of RelA in a dose- and time-dependent manner. A549 cells were treated with indicated dose of JQ1 for 4 or 8 hr. The nuclear and cytoplasmic extracts were immunoblotted for the levels of endogenous RelA and Brd4 as in Figure 4A. (B) MG-132 reverses the JQ1-induced down-regulation of nuclear RelA. A549 cells were treated with 2.5 μM JQ1 for 8 hr, followed by treatment with or without MG-132 (10 μM) for 4 hr. The nuclear and cytoplasmic extracts were immunoblotted for the indicated proteins. (C). JQ1 does not affect the transcription of RelA. A549 cells were treated with 2.5 μM JQ1 for 8 hr before total RNA was extracted. Complementary DNA was synthesized and quantitative real-time PCR was performed. Levels of RelA mRNA were normalized with the expression of actin. Data represent the average of three independent experiments +/−SD. (D) JQ1 induces the ubiquitination of nuclear RelA. A549 cells were treated with 10 μM JQ1 for 4 hr followed by treatment with MG-132 (10 μM) for 2 hr to prevent the degradation of RelA. Nuclear extracts were isolated and subjected to immunoprecipitation with anti-RelA antibodies. RelA immunoprecipitates were then immunoblotted with anti-ubiquitin antibodies for the ubiquitination of endogenous nuclear RelA. (E) Schematic model for the binding of Brd4 to acetylated lysine-310 of RelA to prevent the ubiquitination and degradation of nuclear RelA and the potential role of this interaction in the maintenance of constitutively active NF-κB and in tumor formation.
Furtehrmore, when we examined the ubiquitination of nuclear RelA in the presence and absence of JQ1, we observed that JQ1 treatment enhanced the ubiquitination of nuclear RelA (Figure 5D), indicating that JQ1 promotes RelA ubiquitination. Consistent with the finding that nuclear p50 levels were not affected by JQ1 (Figure 5A), ubiquitination of p50 was barely affected by JQ1 (Supplementay Figure S3). Collectively, these data demonstrate that JQ1 induces the ubiquitination and degradation of nuclear RelA.
Discussion
Constitutively active NF-κB is observed in a variety of cancers and is responsible for many features of cancer cells (8–10). While signals triggering the activation of NF-κB in cancer have been intensively studied and elucidated, much less is known about how the activated NF-κB is maintained in cancer cells. In this study, we have unveiled a mechanism by which bromodomain-containing factor Brd4 plays an important role in this process. Brd4 stabilizes nuclear RelA in A549 lung cancer cells by preventing RelA ubiquitination and degradation through a specific recognition of the acetylated lysine-310 of RelA via its two bromodomains (Figure 5E). More importantly, inhibiting the specific interaction using small molecule JQ1 suppresses the transcriptional activation of NF-κB and inhibits the tumorigenesis of A549 lung cancer cells (Figure 5E). These studies underscore the potential of targeting posttranslationally modified NF-κB and its associated epigenetic regulators in the prevention and treatment of cancer.
Emerging evidence suggests that reversible acetylation of the RelA subunit of NF-κB plays an important role in controlling the functions of NF-κB under physiological and pathological conditions (11, 15). Acetylation of lysine-310 of RelA facilitates the expression of NF-κB-mediated inflammatory genes and is involved in bacterial and viral infection-induced inflammation (15, 36, 37). Additionally, hyperacetylated RelA at lysine-310 is responsible for Stat3-mediated sustained NF-κB activation in lung cancer (19). Targeting the acetylation of NF-κB has therefore been proposed as a potential approach for the treatment of inflammatory diseases and cancer (15, 38, 39). Our current structural studies identified a specific interaction between the acetylated lysine-310 and the highly conserved asparagines in the two bromodomains of Brd4 (Figures 1A &1B). Mutation of the asparagines inhibited Brd4’s interaction with acetylated RelA and impaired the coactivation function of Brd4 for NF-κB (Figures 1C, 1D, and 1E). Consistently, WT but not the RelA binding-defective mutant Brd4 rescued down-regulated TNF-α-induced transcriptional activation of NF-κB in Brd4 knockdown cells (Figure 1F and 1G).
The specific interaction between acetylated RelA and Brd4 could be blocked by the small molecule JQ1, which likely displaces the acetylated lysine-310 RelA from the binding pocket through its competitive binding to the same asparagines within the bromodomains (Figure 2). Importantly, JQ1 not only suppressed the transcriptional activation of NF-κB (Figure 2) but also inhibited the tumorigenic potential of A549 lung cancer cells (Figure 3). Therefore, in addition to directly targeting the acetylation of RelA, modulating the binding of effector proteins to acetylated RelA could also be an effective approach for the prevention and treatment of NF-κB-driven diseases. It has to be noted that JQ1 is an inhibitor targeting all BET family proteins, which include Brd2, Brd3, Brd4, and a testis specific Brdt (40), raising the possibility that the inhibitory effect of JQ1 on NF-κB might be derived from its inhibition on other BET family proteins. However, both Brd2 and Brd3 don’t seem to be involved in the transcriptional activation of NF-κB since co-expression of Brd2 or Brd3 failed to coactivate transcriptional activity of NF-κB and depletion of Brd2 or Brd3 by siRNA did not inhibit TNF-α-induced NF-κB target gene expression (Supplementary Figures S5, S6 and S7). Therefore, the inhibitory effect of JQ1 on NF-κB might reflect its specific inhibition on Brd4 by blocking its interaction with acetylated lysine-310. Further supporting this, we found that RelA-deficient mouse embryonic fibroblasts (MEFs) reconstituted with WT-RelA were much more sensitive to JQ1-mediated cell growth inhibition compared to cells reconstituted with either vector or RelA-K310R (Supplementary S4).
BET family protein inhibitors, including JQ1 and I-BET, have recently emerged as promising therapeutic molecules for the treatment of cancers and inflammatory diseases (30, 32, 41). These small molecules bind to bromodomains of BET family proteins and compete with acetylated-lysine histone peptides (30, 32). The anti-tumor or anti-inflammatory response activities of these small molecules are ascribed partially to their abilities to displace the binding of Brd4 from chromatin and suppress the expression of genes involved in these processes. Oncogene Myc has been identified as the primary target of JQ1 in acute myeloid leukemia and multiple myeloma (29, 31, 34). JQ1 down-regulates the expression of Myc by dislocating Brd4 from the promoter of Myc (29, 31, 34). Our studies demonstrate that RelA is also a direct target of JQ1; JQ1 down-regulated the nuclear levels of RelA (Figure 5A). However, different from its effect on Myc, JQ1 targets RelA at the protein level rather than at the transcription level since the transcription of RelA was barely affected by JQ1 (Figure 5C). Consistently, JQ1 induced the degradation of RelA, which was prevented by treatment with proteasome inhibitor MG-132 (Figure 5B). It appears that JQ1 might modulate the properties of different proteins via distinct mechanisms, depending on the functional consequences of the binding of Brd4 to the acetylated target proteins. Due to their structural similarities, other BET bromodomain inhibitors might also target RelA and control the functions of NF-κB through a similar mechanism. A recent study showed that BET-specific bromodomain inhibitor MS417 inhibited NF-κB activity and NF-κB-mediated inflammatory response (42). It seems likely that the anti-inflammatory effect of I-BET might also partially result from suppression of NF-κB activity (32).
Our results demonstrate that depletion of Brd4 or treatment of cells with JQ1 induced the ubiquitination and degradation of nuclear RelA, but not the cytoplasmic RelA (Figure 4 and Figure 5). In contrast, the levels of p50 were not affected by the depletion of Brd4 or JQ1 treatment (Figures 4A and 5A). Consistently, the ubiquitination of p50 was not altered (Supplementary Figures S1–S3). These data suggest that Brd4 or JQ1 specifically targets RelA. However, it remains unclear how Brd4 prevents the ubiquitination and degradation of nuclear RelA. It has been shown that acetylation of RelA at lysine-310 prevents Set9-mediated methylation of RelA (43), which triggers the ubiquitination and degradation of chromatin-associated NF-κB (44). Deacetylation of lysine-310 enhances the methylation and the subsequent ubiquitination and degradation of RelA (43). It is possible that binding of acetylated lysine-310 to the binding pocket of bromodomains of Brd4 might block the access of the lysine-310 deacetylases, thus prolonging the acetylation signal and the activity of NF-κB. Brd4 has been shown to stabilize papillomavirus E2 protein through its direct interaction with E2 and the blockage of the recruitment of the ubiquitin E3 ligase complex (45–47). Alternatively, Brd4 might utilize a similar mechanism to stabilize NF-κB. Binding of Brd4 to acetylated RelA might directly impair the recruitment of the ubiquitination machinery to NF-κB, which is subject to ubiquitination and degradation after activation (48, 49).
Taken together, our findings indicate that Brd4 plays an essential role in maintaining constitutively active NF-κB in cancer cells. Depletion of Brd4 or treatment of cancer cells by JQ1 down-regulates the nuclear levels and activity of NF-κB (Figures 3, 4 &5). Furthermore, JQ1 suppresses the proliferation and transformation potential of A549 lung cancer cells (Figure 3). JQ1 also inhibits the proliferation of a variety of cancer cells with constitutively active NF-κB (Supplementary S8). As such, regulation of the interaction between RelA and Brd4 by JQ1 or other BETi might be a potential approach for the treatment of cancer. In many cancers, chemotherapy and radiotherapy induce constitutive activation of NF-κB, resulting in the resistance of tumors to treatment. Blocking the interaction between RelA and Brd4 by small molecules might also reduce the resistance to chemotherapy and radiotherapy and increase the effectiveness of these treatments. Therefore, identification of Brd4 as a novel regulator of constitutively active NF-κB in cancer not only provides new insights into the tumor-promoting function of NF-κB in cancer, but also provides potential approaches for the prevention and treatment of NF-κB-driven cancer by targeting epigenetic regulator Brd4.
Materials and methods
Protein expression, crystallization, and structure determination
The expression and purification of recombinant bromodomains of Brd4 from Escherichia coli has been described previously (21). Information for crystallization and structure determination is provided in Supplementary Information.
Cell lines, recombinant proteins, and plasmids
Human A549 lung carcinoma cells and HEK293T cells were purchased from ATCC and cultured in DMEM supplemented with 10% fetal bovine serum (FBS). The GST fusion proteins of GST-BD1/BD2 were prepared as previously described (21). Expression vectors for Brd4 point mutation constructs were generated using Quickchange site-directed mutagenesis (Stratagene, San Diego, CA, USA) and all mutations were confirmed by sequencing.
Antibodies
Antibodies against RelA, p50, HDAC1, Flag, HA, GST, ubiquitin and tubulin were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). Anti-Brd4 antibodies were from Bethyl Laboratories (Montgomery, TX, USA). Anti-T7 antibodies were from Covance (Emeryville, CA, USA). Anti-acetylated lysine-310 antibodies have been previously described (50).
In vitro protein-protein interaction
Purified recombinant proteins of GST-bromodomains of Brd4 were incubated with biotin labeled acetylated lysine-310 RelA peptide bound to Streptavidin agarose beads. JQ1 small molecules were added to the binding buffer to a final concentration of 2 μM. Binding of RelA to bromodomains of Brd4 was detected by immunoblotting with anti-GST antibodies.
Immunoprecipitation and immunoblotting analysis, luciferase reporter assay and quantitative real-time PCR analysis
Immunoprecipitation, immunoblotting analysis, luciferase reporter assay, and quantitative real-time PCR analysis were performed as previously described (21).
In vivo ubiquitination assays
HEK293T cells transfected with expression vectors for T7-RelA or T7-p50, HA-ubiquitin or Brd4 for 48 hr were lysed with a lysis buffer (50 mM Tris, pH 8.0; 120 mM NaCl; 0.5% NP40; and 2% (w/v) sodium dodecyl sulfate (SDS) followed by sonication. Before immunoprecipitation with the anti-T7 antibody-conjugated agaroses, the concentration of SDS in the binding buffer was diluted to 0.1% with dilution buffer (50 mM Hepes, pH8.0; 250 mM NaCl; 1% NP40; 1 mM EDTA; 1 mM PMSF and 1X protease inhibitor cocktails). The immunoprecipitates were subjected to immunoblotting with anti-HA antibodies. For ubiquitination of endogenous RelA or p50 in A549 cells, cells were treated with MG-132 (10μM) for 2 hr before lysis of cells. Nuclear RelA or p50 were isolated and subjected to immunoprecipitation with anti-RelA or p50 antibodies followed by immunoblotting with anti-ubiquitin antibodies.
Proliferation assay and soft-agar colony formation assay
Proliferation assay and soft-agar assays were performed as previously described (51). Colony growth was scored after 4 weeks of cell incubation at the normal condition with or without JQ1 treatment. All the proliferation and colony formation assays presented in this study were repeated in at least 3 independent experiments.
In vivo tumorigenicity assays
Five-week old female severe combined immunodeficient (SCID) mice (Harlan Laboratories, Indianapolis, IN, USA) were subcutaneously implanted with A549 cells (5X106) for two weeks. Mice bearing A549 xenografts were treated with vehicle or JQ1 daily for 32 days with the dosage of 50mg/kg (mpk). The recipient mice were monitored daily by palpation. Mice were killed and dissected for tumor evaluation when tumor volume reached 2,000 mm3.
Materials and methods
Protein crystallization and structure determination
For crystallization, an additional size exclusion chromatography (Superdex 75 16/60; GE Healthcare, Pittsburgh, PA, USA) was added at the end of purification. Peptide complexes were formed by incubation of the recombinant bromodomains with a 5-fold stoichiometric excess of each peptide for 30 min on ice prior to initiation of crystallization. Crystals of the bromodomain-peptide complexes were grown using the hanging drop method. Briefly, 1 μl of the respective bromodomain-peptide complex was mixed with an equal volume of precipitant solution (for the first bromodomain BD1: 0.2M NaNO3, 20% PEG3350, and for the second bromodomain BD2: 2.5 M (NH4)2SO4, 0.1 M Tris PH 8.5). Crystals appeared overnight and reached the maximal size in about 3 days. Crystals of each bromodomain-peptide complex was briefly immersed in the corresponding mother liquor supplemented with 20% glycerol prior to vitrification by direct immersion in liquid nitrogen. Selenomethionine labeled BD2 was grown as described (REF) and crystals of SeMet BD2 were grown and manipulated under similar conditions.
All diffraction data were collected at an insertion device synchrotron source (Sector 21 ID-D, Advanced Photon Source, Argonne National Labs, IL) using a Mar 300 CCD detector. All data were indexed and scaled using the HKL2000 package. Initial crystallographic phases were determined by single wavelength anomalous diffraction methods using crystals of SeMet BD2. The structures of BD1 and of each of the peptide complexes were determined by molecular replacement using the refined coordinates of SeMet BD2 as a search probe. For data from crystals of SeMet BD2, a six-fold redundant data set was collected to 2.0 Å resolution with an overall Rmerge=0.067 and I/σ(I)=5 in the highest resolution shell. The heavy atom substructure was determined using HySS and refinement of heavy atom parameters using Phaser, as implemented in the PHENIX software package, yielded a Fig. 1 of Merit of 0.513. The resultant experimental map was of exceptional quality allowing the entire polypeptide chain to be automatically traced using ARP/wARP. Further manual fitting using XtalView (50) was interspersed with rounds of refinement using REFMAC5. Cross-validation, using 5% of the data for the calculation of the free R factor was utilized throughout model building process in order to monitor building bias.
For the peptide complex structure, although clear and continuous density could be observed for the peptide using initial phases from molecular replacement, the peptides were only built into the model after the free R factor dropped below 30%. The stereochemistry of the models was routinely monitored throughout the course of refinement using PROCHECK. Relevant data collection and refinement parameters are provided in Table 1 in supplementary information. The refined coordinates have been deposited in the Protein Data Bank.
Supplementary Material
Acknowledgments
We thank members in the Chen lab for discussion and A. Lamb for assistance in the preparation of the manuscript. This work is supported in part by funds provided by UIUC (to L.F.C.) and NIH grants DK-085158 (to L.F.C.)
Footnotes
Conflict of interest
The authors declare no conflict of interest
References
- 1.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109 (Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16(2):225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–83. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nature reviews Cancer. 2002;2(4):301–10. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nature reviews Immunology. 2005;5(10):749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 6.Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–30. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 7.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nature reviews Drug discovery. 2009;8(1):33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer cell. 2004;6(3):203–8. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi MM, Sung B, Yadav VR, Kannappan R, Aggarwal BB. NF-kappaB addiction and its role in cancer: ‘one size does not fit all’. Oncogene. 2011;30(14):1615–30. doi: 10.1038/onc.2010.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 11.Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nature reviews Cancer. 2012;12(2):121–32. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 12.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9(10):815–20. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 14.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25(51):6717–30. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 15.Huang B, Yang XD, Lamb A, Chen LF. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal. 2010;22(9):1282–90. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen LF, Greene WC. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol. 2004;5(5):392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 17.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 2002;21(23):6539–48. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23(12):80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer cell. 2009;15(4):283–93. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai Y, Rahmani M, Dent P, Grant S. Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-kappaB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol Cell Biol. 2005;25(13):5429–44. doi: 10.1128/MCB.25.13.5429-5444.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29(5):1375–87. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22(5):151–3. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 23.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141–5. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 24.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 biology reports. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117(3):349–60. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 26.Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, et al. A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol. 2002;22(18):6509–20. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, et al. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci U S A. 1998;95(15):8538–43. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.French CA. Pathogenesis of NUT midline carcinoma. Annual review of pathology. 2012;7:247–65. doi: 10.1146/annurev-pathol-011811-132438. [DOI] [PubMed] [Google Scholar]
- 29.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2011;468(7327):1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119–23. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung CW. Small molecule bromodomain inhibitors: extending the druggable genome. Prog Med Chem. 2012;51:1–55. doi: 10.1016/B978-0-12-396493-9.00001-7. [DOI] [PubMed] [Google Scholar]
- 34.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9(2):240–6. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 36.Choi KC, Lee YH, Jung MG, Kwon SH, Kim MJ, Jun WJ, et al. Gallic acid suppresses lipopolysaccharide-induced nuclear factor-kappaB signaling by preventing RelA acetylation in A549 lung cancer cells. Molecular cancer research : MCR. 2009;7(12):2011–21. doi: 10.1158/1541-7786.MCR-09-0239. [DOI] [PubMed] [Google Scholar]
- 37.Ishinaga H, Jono H, Lim JH, Kweon SM, Xu H, Ha UH, et al. TGF-beta induces p65 acetylation to enhance bacteria-induced NF-kappaB activation. EMBO J. 2007;26(4):1150–62. doi: 10.1038/sj.emboj.7601546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adcock IM, Tsaprouni L, Bhavsar P, Ito K. Epigenetic regulation of airway inflammation. Curr Opin Immunol. 2007;19(6):694–700. doi: 10.1016/j.coi.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 39.Gupta SC, Sundaram C, Reuter S, Aggarwal BB. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta. 2010;1799(10–12):775–87. doi: 10.1016/j.bbagrm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nature reviews Cancer. 2012;12(7):465–77. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliver SS, Denu JM. Disrupting the reader of histone language. Angew Chem Int Ed Engl. 2011;50(26):5801–3. doi: 10.1002/anie.201101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang G, Liu R, Zhong Y, Plotnikov AN, Zhang W, Zeng L, et al. Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem. 2012 doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang XD, Tajkhorshid E, Chen LF. Functional interplay between acetylation and methylation of the RelA subunit of NF-kappaB. Mol Cell Biol. 2010;30(9):2170–80. doi: 10.1128/MCB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28(8):1055–66. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng G, Schweiger MR, Martinez-Noel G, Zheng L, Smith JA, Harper JW, et al. Brd4 regulation of papillomavirus protein E2 stability. J Virol. 2009;83(17):8683–92. doi: 10.1128/JVI.00674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee AY, Chiang CM. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J Biol Chem. 2009;284(5):2778–86. doi: 10.1074/jbc.M805835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jang MK, Kwon D, McBride AA. Papillomavirus E2 proteins and the host BRD4 protein associate with transcriptionally active cellular chromatin. J Virol. 2009;83(6):2592–600. doi: 10.1128/JVI.02275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200(1):107–13. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sen R, Smale ST. Selectivity of the NF-{kappa}B response. Cold Spring Harb Perspect Biol. 2010;2(4):a000257. doi: 10.1101/cshperspect.a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, et al. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25(18):7966–75. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang B, Qu Z, Ong CW, Tsang YH, Xiao G, Shapiro D, et al. RUNX3 acts as a tumor suppressor in breast cancer by targeting estrogen receptor alpha. Oncogene. 2012;31(4):527–34. doi: 10.1038/onc.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.