Abstract
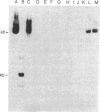
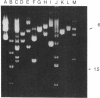
The nontranscribed spacer regions (NTS) that adjoin the coding portion of mouse ribosomal DNA are protected in nucleoli against exhaustive DNase I digestion. Since these sequences are degraded by the enzyme after they are extracted by phenol, the protection is suggested to result from the binding of specific proteins. The nucleolar structure would thus be organized to protect NTS sequences and expose the coding sequences for transcription. We show here that these protected sequences include tracts of poly(dG-dT).poly(dA-dC). We also report that these sequences are localized in regions flanking the rRNA transcription unit. These sequences can potentially form Z-DNA. The organized DNase I-resistant NTS structure in which they participate could be involved in structuring the nucleolus or in regulating transcription because poly(dG-dT).poly(dA-dC) sequences and portions of spacer rDNA can serve as transcriptional enhancer elements.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Seperack P., Banerji J., Lang R. B., Miesfeld R., Marcu K. B. Mouse rDNA nontranscribed spacer sequences are found flanking immunoglobulin CH genes and elsewhere throughout the genome. Cell. 1980 Nov;22(1 Pt 1):179–185. doi: 10.1016/0092-8674(80)90166-x. [DOI] [PubMed] [Google Scholar]
- Bolla R. I., Braaten D. C., Shiomi Y., Hebert M. B., Schlessinger D. Localization of specific rDNA spacer sequences to the mouse L-cell nucleolar matrix. Mol Cell Biol. 1985 Jun;5(6):1287–1294. doi: 10.1128/mcb.5.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman L. H., Rabin B., Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981 Oct 10;9(19):4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzoni I., Baldari C. T., Amaldi F., Buongiorno-Nardelli M. Replication of ribosomal DNA in Xenopus laevis. Eur J Biochem. 1981 Sep 1;118(3):585–590. doi: 10.1111/j.1432-1033.1981.tb05559.x. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Seidman M., Howard B. H., Gorman C. M. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984 Dec;4(12):2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami R., Urano Y., Mishima Y., Muramatsu M., Moriwaki K., Yoshikura H. Novel repetitive sequence families showing size and frequency polymorphism in the genomes of mice. J Mol Biol. 1983 Apr 5;165(2):209–228. doi: 10.1016/s0022-2836(83)80254-x. [DOI] [PubMed] [Google Scholar]
- Kominami R., Urano Y., Mishima Y., Muramatsu M. Organization of ribosomal RNA gene repeats of the mouse. Nucleic Acids Res. 1981 Jul 24;9(14):3219–3233. doi: 10.1093/nar/9.14.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M., Arnheim N. Nucleotide sequence of the genetically labile repeated elements 5' to the origin of mouse rRNA transcription. Nucleic Acids Res. 1983 Jan 11;11(1):211–224. doi: 10.1093/nar/11.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell. 1984 May;37(1):285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Krystal M., Arnheim N. A member of a new repeated sequence family which is conserved throughout eucaryotic evolution is found between the human delta and beta globin genes. Nucleic Acids Res. 1981 Nov 25;9(22):5931–5947. doi: 10.1093/nar/9.22.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirre C., Stahl A. Ultrastructural organization, sites of transcription and distribution of fibrillar centres in the nucleolus of the mouse oocyte. J Cell Sci. 1981 Apr;48:105–126. doi: 10.1242/jcs.48.1.105. [DOI] [PubMed] [Google Scholar]
- Mroczka D. L., Cassidy B., Busch H., Rothblum L. I. Characterization of rat ribosomal DNA. The highly repetitive sequences that flank the ribosomal RNA transcription unit are homologous and contain RNA polymerase III transcription initiation sites. J Mol Biol. 1984 Mar 25;174(1):141–162. doi: 10.1016/0022-2836(84)90369-3. [DOI] [PubMed] [Google Scholar]
- Möller A., Nordheim A., Kozlowski S. A., Patel D. J., Rich A. Bromination stabilizes poly(dG-dC) in the Z-DNA form under low-salt conditions. Biochemistry. 1984 Jan 3;23(1):54–62. doi: 10.1021/bi00296a009. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. Negatively supercoiled simian virus 40 DNA contains Z-DNA segments within transcriptional enhancer sequences. Nature. 1983 Jun 23;303(5919):674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]