Abstract
In response to the limited nutrients and stressful conditions of their habitats, many microorganisms including Salmonella form a biofilm by secreting a polymeric matrix to interweave individual cells and to build structural communities on an abiotic or living surface. The biofilm formation in Salmonella is tightly regulated by a regulatory network that involves multiple transcriptional regulators. As a master transcriptional regulator in biofilm formation, curli subunit gene D (csgD) functions by activating the biosynthesis of the extracellular polymeric matrix composed of exopolysaccharide cellulose, curli and biofilm-associated proteins (Baps), assisting bacterial cells in transitioning from the planktonic stage to the multicellular state. The expression of CsgD itself is affected by cell growth stage and environmental stimuli through the action of other transcriptional factors, bis-(3′–5′)-cyclic dimeric guanosine monophosphate (c-di-GMP), regulatory small RNAs (sRNAs) and other elements. The formation of biofilm confers new physiological characteristics on the bacteria within, especially resistance against unfavorable environmental conditions. Herein, we summarize the CsgD regulatory network of Salmonella biofilm formation and the new traits acquired by Salmonella when within biofilm.
Keywords: biofilm, CsgD, physiological characteristic, Salmonella
INTRODUCTION
Salmonella enterica is an intracellular facultative gram-negative pathogen that infects many hosts including humans and causes diseases ranging in severity from gastroenteritis and diarrhea to life-threatening systemic syndrome through food poisoning. When the nutrient supply is abundant and environment conditions are optimal, Salmonella reproduces exponentially and moves freely. As nutrients are consumed or when harsh conditions develop, Salmonella grows more slowly and secrets extracellular polymers to encase itself in an interwoven structure called biofilm. Biofilm formation is important for the spread of Salmonella because this biofilm is resistant to disinfectants, environmental stresses, antibiotics and the host immune system, consequently promoting bacterial dispersal and survival and enhancing its virulence.1,2,3 Thus, biofilm formation is associated with the outbreak of salmonellosis and persistent infections in patients.4,5,6,7 Although a fitness advantage is provided by biofilm for bacterial survival, biofilm formation is an energy-consuming process. Therefore, biofilm formation is tightly regulated in response to environmental changes. Previous studies identified a variety of environmental factors affecting Salmonella biofilm formation such as temperature, pH value and osmolarity.8 Recent studies revealed underlying molecular mechanisms that link biofilm formation with environmental stresses by showing that multiple transcriptional regulators including sigma factor RpoS and response regulators of two-component systems, bis-(3′–5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) and regulatory small RNAs (sRNAs) relay signals from the extracellular environment to a master regulator of biofilm formation, curli subunit gene D (csgD).9 CsgD integrates these signals and synchronizes the synthesis of several important constituents of the extracellular polymeric matrix to form biofilm.10 Once Salmonella biofilm is formed, new traits emerge in the bacteria within these multicellular aggregates such as increased resistance to unfavorable conditions, enhanced virulence and altered metabolism.
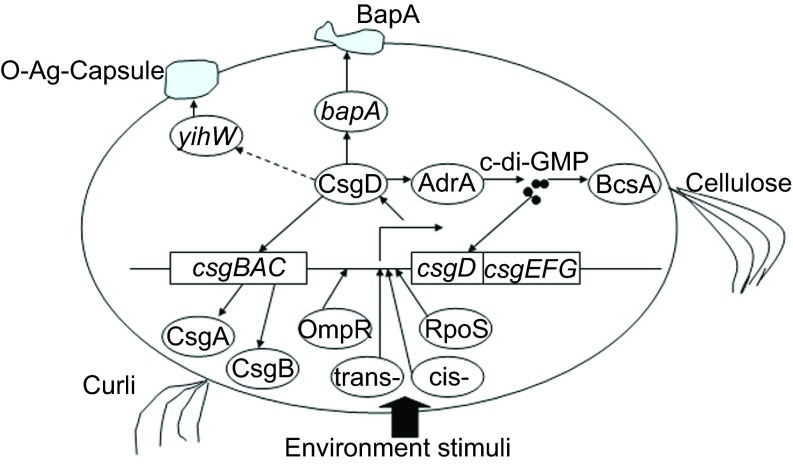
CsgD REGULATES Salmonella BIOFILM FORMATION
The main constituents of the extracellular matrix of Salmonella biofilm are cellulose, curli fimbriae, biofilm-associated protein (Bap),10 O-antigen-capsule (O-Ag-capsule), lipopolysaccharides and extracellular DNA. Cellulose, aβ-1-4-𝒟-glucose polymer, is an important biofilm-associated extracellular polymeric substance (EPS) that interweaves with other extracellular components to form three-dimensional structures and to promote cell–cell interactions and protect cells from unfavorable environments. Its synthesis requires genes encoded in the bcsABZC–bcsEFG operons. Curli fimbria, an amyloid-like proteinaceous substance, is another major component of the extracellular matrix in biofilm and is encoded by the csgBAC–csgDEFG operons.11 BapA is a large proline–threonine-rich protein that is essential for bacterial aggregation and subsequent pellicle formation at the air/liquid interface in Luria-Bertani medium.12 BapA is encoded by bapA and is secreted to the cell surface by a genetically linked BapBCD type I secretion system. The O-Ag-capsule is crucial for S. Typhimurium and S. Typhi gallstone biofilm formation.13 The yihU–yshA and yihVW operons are responsible for O-Ag-capsule assembly and translocation.
With the decline of nutrient availability, Salmonella cells begin their transition from an exponential growth phase to a stationary phase and begin to form biofilm. The synthesis of the extracellular matrix components of Salmonella biofilm is regulated by a highly complex regulatory network that has many components including sigma factors, transcriptional factors, several nucleotide messengers and sRNAs. As a master transcriptional regulator, CsgD is major control and integration unit for Salmonella biofilm formation due to its regulation of the expression of Salmonella biofilm-associated extracellular matrix substances including curli and cellulose.14 Curli production is directly activated by the binding of the transcription factor CsgD to the promoter of the csgBAC operon,15 which encodes the major curli subunit CsgA as well as the nucleator protein CsgB. Recently, Zakikhany et al.16 revealed the mechanism in Escherichia. coli by which unphosphorylated CsgD directly binds to an 11 bp variant and thus activates csgBAC transcription. CsgD also regulates cellulose synthesis at a post-transcriptional level by controlling the activity of several cellulose synthases including BcsB through the synthesis of c-di-GMP by AdrA. Bearing a GGDEF domain, AdrA synthesizes c-di-GMP as diguanylate cyclase, and its transcription is under the control of CsgD. The c-di-GMP binds to the PilZ domain in cellulose synthases and increases their enzyme activity via the allosteric change of the protein conformation.17
CsgD is considered to be a regulator for BapA expression in Salmonella because a special binding motif, which is very similar to the CsgD-binding sequence in the adrA promoter, is present in the bapA promoter. Thus, CsgD may influence bapA transcription or expression.10,18 However, in one controversial report, Zakikhany et al. did not find any evidence that CsgD regulates bapA transcription or expression using a combined bioinformatics and global transcription approach.16
As a component of EPS in S. enteritidis biofilm, the O-Ag-capsule is assembled and translocated by proteins encoded by the divergently oriented operons yihU–yshA and yihVW. A gene expression assay revealed that CsgD regulates the yih operons, although the detailed mechanism of this is unknown.19
Although CsgD is essential for biofilm formation, an individual bacterium may not express CsgD at levels similar to that in the multicellular community. Grantcharova et al.20 identified the bistable characteristic of CsgD expression: a subpopulation of bacteria in biofilm has a high level of CsgD expression, whereas in another subpopulation of bacteria, the expression of CsgD is low. The bacteria with upregulated CsgD expression are responsible for generating the extracellular matrix and for forming the biofilm structure that encases the low CsgD-expressing bacteria. Because bistable CsgD expression saves energy in biofilm production, it may confer additional advantages to the entire bacterial community while maintaining the developmental potential of the population.
THE INTERACTION BETWEEN CsgD AND OTHER REGULATORS IN BIOFILM FORMATION
It has long been known that environmental factors (temperature, nutrients and starvation, ethanol, oxygen tension, osmolarity, iron and pH) have important effects on biofilm EPS production and their composition. These environmental factors affect Salmonella biofilm formation by acting on CsgD expression through RNA polymerase sigma factor RpoS, multiple transcriptional factors and small nucleotides.21 The intergenic region between csgBAC and csgDEFG operons is 582 bp, providing the binding sites with multiple transcriptional factors and making it a hub for integrating different environmental signals.9 When Salmonella cells enter the stationary growth phase, sigma factor RpoS becomes dominant in competition with sigma factors. RpoS joins RNA polymerase to form holoenzyme, binding to the promoter sequences of a set of stationary phase and stress-induced genes and initiating mRNA transcription, which is assisted by other transcriptional regulators and accessory proteins.22 RpoS-containing RNA polymerase has a low activity until it binds to accessory protein Crl, which also accumulates in the stationary phase. Crl is more stable at low temperatures (28 °C) than at high temperatures (37 °C). Biofilm forms at 28 °C, indicating that Crl serves as a temperature sensor in biofilm formation.23
Multiple transcriptional factors bind to the promoter sequence of csgD and regulate its transcription, including OmpR, integrating host factor (IHF), the histone-like nucleoid-structuring protein and the MerR-like regulator (MlrA). OmpR, a response regulator in the EnvZ/OmpR two-component regulatory system, is phosphorylated in response to environmental stimuli such as osmolarity and pH and binds to the csgD promoter to enhance its expression. OmpR-binding sites in the csgD promoter were identified using a combination of genetic studies and in vitro experimentation.24 Similar to the response to osmolarity, EnvZ/OmpR is involved in the ethanol-inducing upregulation of csgD.2 IHF is a key architectural protein involved in a variety of cellular processes and binds to consensus sequences in the csgD promoter to facilitate the activation or repression of csgD transcription.25 Mutants in IHF subunits showed altered and reduced biofilm morphotypes on Congo Red agar plates. Because the same phenotypes were observed at different temperatures, IHF appears to be uninvolved in temperature-dependent biofilm morphotype regulation. Histone-like nucleoid-structuring protein is a DNA structural protein that plays a significant role in integrating a wide range of environmental stimuli such as osmolarity and temperature.26 Histone-like nucleoid-structuring protein was demonstrated to bind AT-rich sites in the intergenic csgBAC–csgDEFG region and has indirect effects on csgD expression.9 MlrA, the expression of which is regulated by RpoS, binds upstream of the csgD promoter and induces the transcription of the csgD gene. MlrA specifically acts on the transcription of operon csgDEFG primarily in response to metal ions.27 Recent evidence in E. coli found that MlrA directly binds to a 33 bp palindromic sequence between IHF- and OmpR-binding sites in the upstream sequence of the csgD promoter.28 These three positive regulators of csgD expression (IHF, OmpR and MlrA) are found to bind to the same site of csgD promoter and function independently without strong cooperation.28
c-di-GMP is a newly identified message molecule that naturally occurs in bacteria and is responsible for regulating many biological functions including biofilm formation, motility, virulence, the cell cycle, differentiation and other processes.29 Due to the importance and prevalence of c-di-GMP, the interactions between the second message and biofilm have drawn significant attention. It was demonstrated that increased c-di-GMP enhances csgD expression. We have previously mentioned that AdrA, the expression of which is promoted by CsgD, was the first identified diguanylate cyclase involving in the production of c-di-GMP. The produced c-di-GMP also provides positive feedback for csgD expression in some strains. However, phosphodiesterase degrades c-di-GMP, thereby inhibiting csgD expression.
sRNAs are involved in the regulation of biofilm formation.30 Most sRNAs are 80–200 nt long and directly bind to their target mRNAs in the 5′-UTR or in the open reading frame to execute their regulation activity by changing the exposure of the SD sequence of mRNAs to ribosomes or altering the stability of mRNAs.31 Chaperone protein Hfq protects sRNAs from degradation and facilitates their binding to the target mRNAs. Recently, Hfq-dependent sRNAs (ArcZ and SdsR) were found to positively affect csgD expression and biofilm formation.32 The mRNA of csgD forms a relatively long and stable stem-loop structure in its 5′-UTR. In E. coli, csgD mRNA is the target of McaS, RprA or OmrA/OmrBsRNAs. McaS sRNA binds csgD mRNA, which results in csgD mRNA degradation. RprA and OmrA/OmrB sRNAs interfere with csgD mRNA translation.33
Lytic transglycosylases were found to have effects on the production and regulation of Salmonella biofilm through the csgD pathway. Lytic transglycosylases are a group of ubiquitous bacterial enzymes involved in the turnover of the bacterial cell wall constituent peptidoglycan. Among these, two lytic transglycosylases (MltE and MltC) are able to uniquely affect biofilm production by mediating csgD expression.34 The mRNA level of csgD in Salmonella ΔmltE and ΔmltC double mutant was found to be approximately 47% of that of the wild type, indicating its role in the regulation of csgD expression. Although RpoS and OmpR were indicated to mediate several factors to affect Salmonella biofilm production,15 RpoS and OmpR were not found to participate in the process of MltE- and MltC-mediated regulation of csgD expression. However, a high concentration of the second message c-di-GMP inside the cells can reverse the change of adar (for red, dry and rough) morphotypes and csgD production in the ΔmltE and ΔmltC double mutant, suggesting that c-di-GMP signaling acts downstream of MltE and MltC.
Taken together, CsgD is a major biofilm formation regulator that controls the production of the extracellular matrix of biofilm. Meanwhile, environment factors regulate csgD expression through the action of sigma factors, transcription factors, c-di-GMP and sRNAs, thereby determining biofilm formation (Figure 1).
Figure 1.
Regulation of Salmonella biofilm formation by CsgD.
BIOFILM FORMATION REGULATED BY CsgD COINCIDES WITH A METABOLIC CHANGE
A number of studies demonstrated that the switch from the planktonic state to the biofilm mode in bacteria can cause a global shift in metabolism.15,35 White et al.36 used metabolomics to compare Salmonella cells in rdar colony biofilms to isogenic csgD deletion mutants and found that there are 25 detected metabolites at significantly different concentrations between the two populations. In wild-type colonies, many end products and important intermediates of gluconeogenesis are detected at higher levels than in colonies formed by csgD deletion mutants. Three of the major osmoprotectants used by S. Typhimurium trehalose, as well as glycine betaine (betaine) and glutamate, were found at higher concentrations in wild-type colonies. Other metabolites more abundant in wild-type colonies included nicotinamide adenine dinucleotide, octanoic acid, pyroglutamate and glutathione. However, the upper tricarboxylic acid cycle (cycle) intermediates, adenosine monophosphate and polyamine compounds were detected in csgD deletion mutant colonies. Thus, the hypothesis that the accumulation of tricarboxylic acid cycle intermediates and polyamine compounds would block the gluconeogenesis in csgD mutant cells is reasonable. Adenylate kinase catalyzes the reaction of ATP+adenosine monophosphate into two ADP molecules and is known to buffer ATP levels during periods of rapid ATP consumption.37 Adenylate kinase expression was upregulated in wild-type cultures compared with csgD mutant cultures, suggesting that cells living in biofilm have increased ATP requirements. These results could explain the increased adenosine monophosphate levels detected in csgD mutant colonies.35 speA, speB, speC and cadBA, the main genes encoding polyamine biosynthesis enzymes, had similar magnitudes of expression in wild-type and csgD mutant cells. These results suggest that the accumulated polyamines in csgD mutant cells are caused by the reduced carbon flux of gluconeogenesis and other biosynthetic pathways.
CsgD REGULATING BIOFILM RESISTANCE TO STRESS FACTORS
Salmonella is profoundly adapted to live in non-host environments, with biofilm formation acting as a primary contributor. Salmonella biofilm has been shown to confer resistance to desiccation, disinfectants and antibiotics and has drawn significant attention in the food and medical fields. White et al.38 demonstrated that S. Typhimurium cells in biofilm formed in ATM have a survival rate of 10% after 9 months of storage and 68% after 3 months of storage. Several studies have shown that Salmonella biofilms on the surface of various materials are significantly more resistant to the sanitizers chlorine, hypochlorite, iodine, glutaraldehyde and cationic tensides.39 Further studies showed that curli pili, cellulose and other CsgD-regulated compounds play a key role in enhanced the long-term survival and persistence of Salmonella in biofilm under stressful conditions. After the knockout of the csgD, csgA and bcsA genes, the biofilm's resistance to stress factors was significantly reduced.38 Similar to the csgD mutant, O-Ag-capsule mutants (yihP, yihQ and yihO) have reduced survival ability against desiccation stress, demonstrating the role of the O-Ag-capsule in desiccation tolerance.19
Doctors usually treat bacterial infections with antibiotics that have been validated via drug susceptibility tests. However, the antibiotic sensitivity of bacteria in the planktonic state is different from that in a biofilm state, which can also cause infections in patients. This may explain why the therapeutic effect in vivo is inconsistent with drug susceptibility tests in vitro. Studies were performed to compare the effects of several antibiotics (enrofloxacin, gentamicin, erythromycin, tilmicosin, ampicillin, oxytetracycline and trimethoprim-sulfadoxine) on planktonic cells and the established biofilm of clinical Salmonella Typhimurium and Salmonella Bredeney isolates on polystyrene plates.40 With the exceptions of erythromycin and tilmicosin, planktonic cells were found to be sensitive to all antibiotics, whereas Salmonella in biofilm was only sensitive to enrofloxacin and ampicillin. Furthermore, Tabak et al.41 discovered that Salmonella biofilms are up to 2000-fold more resistant to ciprofloxacin compared with planktonic cells. This is of great significance as ciprofloxacin together with a third-generation cephalosporin is commonly used to treat non-typhoid Salmonella infections. Recently, Papavasileiou et al.43 investigated 194 strains of Salmonella enterica isolated from patients for their ability to form biofilm on silicon disks and compared the biofilm status of strains with their corresponding planktonic form regarding their susceptibility to nine antibiotic types. The cells in biofilm showed increased antimicrobial resistance to all antimicrobial agents compared with the planktonic bacteria, with the highest resistance rates for gentamicin (90%) and ampicillin (84%).
At present, research into bacterial biofilm resistance mechanisms is very active. There are several types of resistance mechanisms: (i) bacteria in a biofilm state have lowered metabolic and growth rates, especially those deep within the biofilm, which decreases their sensitivity to drugs;44 (ii) EPS acting as an adsorbent prevents antibiotic contact with bacteria;45 (iii) the physiological characteristics of biofilm-state bacteria are different from those in a planktonic state;46 and (iv) biofilm bacteria often express special protective factors such as multidrug efflux pumps and stress response regulon.47 Additionally, c-di-GMP may participate in antibiotic resistance in bacteria within biofilm. Chua et al.48 found that P. aeruginosa cells with a low level of c-di-GMP are more resistant to colistin than cells with high c-di-GMP. This finding contradicts the current view that dispersed cells are more susceptible to antibiotics than their sessile counterparts.
CsgD MEDIATING BIOFILM FORMATION INFLUENCES BACTERIAL INVASION AND VIRULENCE
Crull et al.49 created a mouse tumor model by inoculating Salmonella through the tail vein after injecting CT26 cells into the mouse body. The surface of the tumor was covered by Salmonella Typhimurium biofilm, but the bacteria were not able to invade into the tumor cells, which contradicts the in vitro observation that Salmonella Typhimurium can invade CT26 cells. To further investigate this finding, the researchers used the Salmonella csgD knockout mutant to repeat the experiments in a mouse tumor model and found that the mutant was detected intracellularly. csgD is a major control and integration unit for Salmonella biofilm formation and regulates the expression of some important Salmonella biofilm-associated matrix compounds. A csgD mutant lacked any multicellular behavior, as visualized via a morphotype on Congo red agar plates.50 These tests confirmed the important role of csgD in the process of bacteria invasion in vivo, but the detailed mechanism remains to be explored.
Further studies indicate that c-di-GMP can control the bacterial invasion ability through csgD.51 Intracellular c-di-GMP was regulated by a series of proteases containing GGDEF or EAL domains, which involve the catalytic activity of guanylate cyclase and phosphodiesterase, respectively. To date, more than 20 protease types with diguanylate cyclase or phosphodiesterase activity have been discovered in Salmonella. Using gene mutant and gene overexpression methods, Ahmad et al.52 found that STM3611 and STM4264 proteins significantly promote the activity of Salmonella in invading human colon cancer cells (HT-29). Both proteins contain EAL domain and exert phosphodiesterase catalytic activity to degrade the c-di-GMP. To confirm the involvement of csgD in the process of the high level of intracellular c-di-GMP inhibiting bacterial invasion, the Ahmad team performed further studies on csgD and bcsA. The STM4264 mutant of Salmonella Typhimurium had reduced invasion due to the high level of c-di-GMP. However, the STM4264 mutant with knocked-out csgD or bcsA restored the ability to invade HT-29. CsgD and BcsA appear to play a role downstream of STM4264. This finding suggests that the c-di-GMP degraded by STM4264 restrains the invasive ability of S. Typhimurium through CsgD and BcsA. In the background of the STM3611 mutant, knocking out csgD or bcsA has little effect on Salmonella invasion.49 Other genes may be involved in the pathway of this mutant. csgD expression inhibits the functions of effecter proteins SopE2 and SipA of Salmonella T3SS-1 (type III secretion system-1).30 In the STM3611 mutant or STM4264 mutant, the production of effecter protein SipA was increased when the gene csgD was knocked out. The increased SipA in Salmonella with csgD and the STM3611 double mutant obviously failed to reverse the invasive ability of Salmonella.
Biofilm formation has been considered to be related with chronic infections. It has been found that the expression of biofilm-related genes is apparently able to increase the expression of bacterial virulence genes in E. coli.53 In some bacteria, the expression of some virulence genes under biofilm conditions is higher than that in a planktonic state.34 In Salmonella, there are many essential virulence genes such as the sipA gene in S. Typhimurium, which encodes an outer-membrane protein of the Salmonella pathogenicity island-2 type III secretion system essential for host infection.54 It has been demonstrated that knocking out the sipA gene can impair the ability of bacteria to form biofilm, which indicates that virulence is closely related with the ability of bacteria to form biofilm.55 A further study found that in biofilm conditions, both wild-type S. Typhimurium and the sipA mutant S. Typhimurium are more adhesive than in planktonic conditions in vitro; moreover, in a biofilm state, Salmonella has more proliferative capacity in macrophages than in a planktonic state.55 The strong evidence presented above shows that biofilm formation promotes bacterial virulence, but the mechanism of the interaction between virulence and the formation of biofilm remains to be elucidated. The studies on biofilm affecting virulence primarily focus on the molecular level and have been performed in vitro; this phenomenon should be further investigated in vivo. The csgD gene could become a novel target for the future control of Salmonella infection.
Biofilm formation is an efficient mechanism that bacteria utilize to evade recognition and elimination by the host immune system. Biofilm may play a key role in bacteria adaptation to adverse environments in a way that makes them both more persistent and less invasive.56,57 Biofilm increases the resistance of bacteria and makes them less conspicuous to the immune system due to hidden antigens and repressed key ligands.58 Additionally, it has been reported that bacterial cells isolated from chronically infected patients lack flagella, pili and motility. In in vitro experiments, P. aeruginosa living in mature biofilm lost flagella, pili and motility.59,60 These adaptations contribute to the change of virulence of bacteria living in biofilm.
CONCLUSION
Taken together, biofilm formation in Salmonella is controlled by a regulatory network composed of multiple transcriptional factors, secondary messengers, and sRNAs. The formation of biofilm offers a fitness advantage to Salmonella and the ability to survive in unfavorable conditions. Biofilm formation is related with Salmonella spread and its pathogenesis. Thus, further studies are needed to clearly understand the relationship between biofilm and microbial physiological functions in Salmonella in an effort to develop new tools and strategies to prevent and treat biofilm-related diseases.
References
- Berger CN, Sodha SV, Shaw RK, et al. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ Microbiol. 2010;12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- Gerstel U, Romling U. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ Microbiol. 2001;3:638–648. doi: 10.1046/j.1462-2920.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- Solano C, García B, Valle J, et al. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- Bae YM, Baek SY, Lee SY. Resistance of pathogenic bacteria on the surface of stainless steel depending on attachment form and efficacy of chemical sanitizers. Int J Food Microbiol. 2012;153:465–473. doi: 10.1016/j.ijfoodmicro.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Bowen A, Fry A, Richards G, Beuchat L. Infections associated with cantaloupe consumption: a public health concern. Epidemiol Infect. 2006;134:675–685. doi: 10.1017/S0950268805005480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Reeve KE, Gunn JS. Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J Bacteriol. 2010;192:2981–2990. doi: 10.1128/JB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhao P, Cannon JL, Doyle MP. Inactivation of Salmonella in biofilms and on chicken cages and preharvest poultry by levulinic acid and sodium dodecyl sulfate. J Food Prot. 2011;74:2024–2030. doi: 10.4315/0362-028X.JFP-11-197. [DOI] [PubMed] [Google Scholar]
- Romling U, Bokranz W, Rabsch W, Zogaj X, Nimtz M, Tschäpe H. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int J Med Microbiol. 2003;293:273–285. doi: 10.1078/1438-4221-00268. [DOI] [PubMed] [Google Scholar]
- Gerstel U, Park C, Romling U. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol. 2003;49:639–654. doi: 10.1046/j.1365-2958.2003.03594.x. [DOI] [PubMed] [Google Scholar]
- Latasa C, Roux A, Toledo-Arana A, et al. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol. 2005;58:1322–1339. doi: 10.1111/j.1365-2958.2005.04907.x. [DOI] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penadés JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Gibson DL, Kay WW, Gunn JS. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect Immun. 2008;76:5341–5349. doi: 10.1128/IAI.00786-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel U, Romling U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol. 2003;154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Hamilton S, Bongaerts RJ, Mulholland F, et al. The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms. BMC Genom. 2009;10:599. doi: 10.1186/1471-2164-10-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakikhany K, Harrington CR, Nimtz M, Hinton JC, Römling U. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2010;77:771–786. doi: 10.1111/j.1365-2958.2010.07247.x. [DOI] [PubMed] [Google Scholar]
- Zorraquino V, García B, Latasa C, et al. Coordinated cyclic-di-GMP repression of Salmonella motility through YcgR and cellulose. J Bacteriol. 2013;195:417–428. doi: 10.1128/JB.01789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latasa C, Solano C, Penadés JR, Lasa I. Biofilm-associated proteins. C R Biol. 2006;329:849–857. doi: 10.1016/j.crvi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Gibson DL, White AP, Snyder SD, et al. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J Bacteriol. 2006;188:7722–7730. doi: 10.1128/JB.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantcharova N, Peters V, Monteiro C, Zakikhany K, Römling U. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar Typhimurium. J Bacteriol. 2010;192:456–466. doi: 10.1128/JB.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AP, Gibson DL, Grassl GA, et al. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect Immun. 2008;76:1048–1058. doi: 10.1128/IAI.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Stationary phase gene regulation: what makes an Escherichia coli promoter sigmaS-selective. Curr Opin Microbiol. 2002;5:591–595. doi: 10.1016/s1369-5274(02)00372-7. [DOI] [PubMed] [Google Scholar]
- Steenackers H, Hermans K, Vanderleyden J, Sigrid CJ, de Keersmaecker Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res Int. 2012;45:502–531. [Google Scholar]
- Gerstel U, Kolb A, Romling U. Regulatory components at the csgD promoter—additional roles for OmpR and integration host factor and role of the 5′ untranslated region. FEMS Microbiol Lett. 2006;261:109–117. doi: 10.1111/j.1574-6968.2006.00332.x. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology. 2010;156 Pt 8:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- Sambanthamoorthy K, Luo C, Pattabiraman N, et al. Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob Agents Chemother. 2012;56:5202–5211. doi: 10.1128/AAC.01396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int J Mol Sci. 2013;14:4560–4579. doi: 10.3390/ijms14034560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Reimegård J, Sterk M, Grantcharova N, Römling U, Wagner EG. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, McNally RR, Sundin GW. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J Bacteriol. 2013;195:1706–1717. doi: 10.1128/JB.02056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- Monteiro C, Fang X, Ahmad I, Gomelsky M, Römling U. Regulation of biofilm components in Salmonella enterica serovar Typhimurium by lytic transglycosylases involved in cell wall turnover. J Bacteriol. 2011;193:6443–6451. doi: 10.1128/JB.00425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AP, Weljie AM, Apel D, et al. A global metabolic shift is linked to Salmonella multicellular development. PLoS ONE. 2010;5:e11814. doi: 10.1371/journal.pone.0011814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht JM, Bremer E. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system proU. FEMS Microbiol Rev. 1994;14:3–20. doi: 10.1111/j.1574-6976.1994.tb00067.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez JA, Csonka LN. Isolation and characterization of adenylate kinase (adk) mutations in Salmonella typhimurium which block the ability of glycine betaine to function as an osmoprotectant. J Bacteriol. 1995;177:390–400. doi: 10.1128/jb.177.2.390-400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AP, Gibson DL, Kim W, Kay WW, Surette MG. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J Bacteriol. 2006;188:3219–3227. doi: 10.1128/JB.188.9.3219-3227.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestby LK, Nesse LL, Storheim SE, Kotlarz K, Langsrud S. Evaluation of efficacy of disinfectants against Salmonella from the feed industry. J Appl Microbiol. 2009;106:1005–1012. doi: 10.1111/j.1365-2672.2008.04067.x. [DOI] [PubMed] [Google Scholar]
- Olson ME, Ceri H, Morck DW, Buret AG, Read RR. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res. 2002;66:86–92. [PMC free article] [PubMed] [Google Scholar]
- Tabak M, Scher K, Chikindas ML, Yaron S. The synergistic activity of triclosan and ciprofloxacin on biofilms of Salmonella Typhimurium. FEMS Microbiol Lett. 2009;301:69–76. doi: 10.1111/j.1574-6968.2009.01804.x. [DOI] [PubMed] [Google Scholar]
- Parry CM, Threlfall EJ. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr Opin Infect Dis. 2008;21:531–538. doi: 10.1097/QCO.0b013e32830f453a. [DOI] [PubMed] [Google Scholar]
- Papavasileiou K, Papavasileiou E, Tseleni-Kotsovili A, et al. Comparative antimicrobial susceptibility of biofilm versus planktonic forms of Salmonella enterica strains isolated from children with gastroenteritis. Eur J Clin Microbiol Infect Dis. 2010;29:1401–1405. doi: 10.1007/s10096-010-1015-y. [DOI] [PubMed] [Google Scholar]
- Brown MR, Allison DG, Gilbert P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect. J Antimicrob Chemother. 1988;22:777–780. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P, Allison DG, McBain AJ. Biofilms in vitro and in vivo: do singular mechanisms imply cross-resistance. J Appl Microbiol. 2002;92 Suppl:98S–110S. [PubMed] [Google Scholar]
- Chua SL, Tan SY, Rybtke MT, et al. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:2066–2075. doi: 10.1128/AAC.02499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crull K, Rohde M, Westphal K, et al. Biofilm formation by Salmonella enterica serovar Typhimurium colonizing solid tumours. Cell Microbiol. 2011;13:1223–1233. doi: 10.1111/j.1462-5822.2011.01612.x. [DOI] [PubMed] [Google Scholar]
- Romling U, Rohde M, Olsén A, Normark S, Reinköster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I, Lamprokostopoulou A, Le Guyon S, et al. Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS ONE. 2011;6:e28351. doi: 10.1371/journal.pone.0028351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Safadi R, Abu-Ali GS, Sloup RE, et al. Correlation between in vivo biofilm formation and virulence gene expression in Escherichia coli O104:H4. PLoS ONE. 2012;7:e41628. doi: 10.1371/journal.pone.0041628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Su J, Bai Y, et al. Characterization of the expression of Salmonella Type III secretion system factor PrgI, SipA, SipB, SopE2, SpaO, and SptP in cultures and in mice. BMC Microbiol. 2009;9:73. doi: 10.1186/1471-2180-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Peng D, Jiao X, Zhang X, Geng S, Liu X. Roles of the spiA gene from Salmonella enteritidis in biofilm formation and virulence. Microbiology. 2011;157 Pt 6:1798–1805. doi: 10.1099/mic.0.046185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]