Abstract
Persisters are a small fraction of quiescent bacterial cells that survive lethal antibiotics or stresses but can regrow under appropriate conditions. Persisters underlie persistent and latent infections and post-treatment relapse, posing significant challenges for the treatment of many bacterial infections. The current definition of persisters has drawbacks, and a Yin–Yang model is proposed to describe the heterogeneous nature of persisters that have to be defined in highly specific conditions. Despite their discovery more than 70 years ago, the mechanisms of persisters are poorly understood. Recent studies have identified a number of genes and pathways that shed light on the mechanisms of persister formation or survival. These include toxin–antitoxin modules, stringent response, DNA repair or protection, phosphate metabolism, alternative energy production, efflux, anti-oxidative defense and macromolecule degradation. More sensitive single-cell techniques are required for a better understanding of persister mechanisms. Studies of bacterial persisters have parallels in other microbes (fungi, parasites, viruses) and cancer stem cells in terms of mechanisms and treatment approaches. New drugs and vaccines targeting persisters are critical for improved treatment of persistent infections and perhaps cancers. Novel treatment strategies for persisters and persistent infections are discussed.
Keywords: mechanisms, persistence, persisters, treatment strategies
BACTERIAL PERSISTERS
The phenomenon of bacterial persisters was first discovered by Gladys Hobby1 in 1942, when penicillin was found to kill 99% of a streptococcal culture, leaving 1% of the bacterial population intact. This surviving 1% of the bacterial population not killed by penicillin was subsequently termed ‘persisters' by Joseph Bigger2 in 1944. The original definition of persisters by Bigger refers to a small population of dormant or non-growing bacteria that have non-heritable tolerance to penicillin but have the capacity to regrow and remain susceptible to the same antibiotic. This definition has drawbacks that recently are becoming increasingly recognized for several reasons. First, earlier studies did not appreciate the heterogeneity of persisters,2,3 and it is only recently that persisters are found to be quite heterogeneous.4,5,6,7,8,9,10 Second, Bigger's definition of persisters does not specify antibiotic exposure time and the time required to resume growth upon removal of antibiotics and culture media involved in cultivation. In fact, persisters are found to be relative,4 and the age of bacterial culture, the type of antibiotics, antibiotic concentrations, length of antibiotic exposure, medium composition and aeration during antibiotic exposure can all affect the level of persisters.4,10,11,12 This means that persisters in one condition may not be persisters in another condition. Third, the current persister definition is based on growth in fresh medium,2,13 often quantified via colony-forming unit assays in which the number of bacteria growing on agar plates or, less commonly, where growth in liquid medium is monitored. This persister definition has limitations as it excludes viable but non-culturable14 bacteria or dormant bacteria, which do not readily grow under ‘normal' culture conditions but can grow under some conditions (upon extended incubation in liquid medium15 or changing medium composition10 or addition of resuscitation factors16) and are clinically relevant as part of the persister continuum (see below). Thus, a new persister definition is required to address the above issues not covered by the current definition. The new definition of persisters can be as follows: persisters refer to genetically drug susceptible quiescent (non-growing or slow growing) organisms that survive exposure to a given cidal antibiotic or drug and have the capacity to revive (regrow or resuscitate and grow) under highly specific conditions (see above for conditions affecting persister counts). The definition of persisters may be extended or broadened to include cidal stresses in place of cidal antibiotics in which case antibiotics can be viewed as a type of the cidal stress.
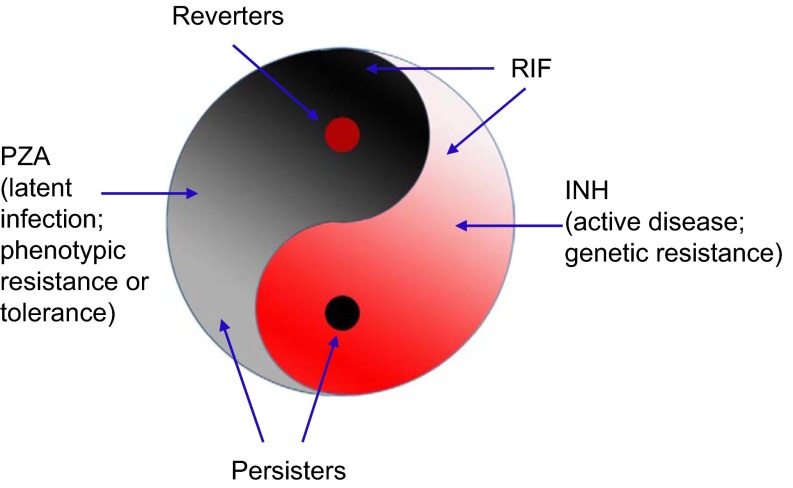
A Yin–Yang model is proposed to describe a dynamic and complex heterogeneous bacterial population consisting of growing (Yang, in red) and non-growing persister cells (Yin, in black) that are in varying growth and metabolic states in continuum5,8 and can interconvert in vitro and in vivo (Figure 1). This Yin–Yang model is compatible with the above new definition of persisters and can account for the heterogeneity of persisters. Although there may not be persisters in an actively growing log phase culture initially, when the growing population (Yang) reaches a certain age and density, a small population of non-growing or slowly growing persisters (Yin) can emerge and increase in numbers as the culture ages.12 The persister population (Yin) is heterogeneous and composed of various subpopulations with varying metabolic states in continuum in varying hierarchy, from shallow to deep persisters, which can encompass viable but non-culturable and various dormant variants with or without morphological changes as part of the persister continuum. Persisters not killed by antibiotics could revert to replicating forms (reverters) or damaged persister forms, which under appropriate conditions may have varying degrees of recovery or reversion and cause relapse or prolonged infections with symptoms. The Yin–Yang model can also be applied to genetic drug resistance (Yang resistance) in growing bacteria where bacteria grow in the presence of antibiotics due to spontaneous mutations or mobile genetic elements (plasmid or transposon), as well as phenotypic resistance17,18 or antibiotic tolerance (Yin resistance, non-inheritable), in non-growing persisters due to physiological or epigenetic changes (gene expression, protein or DNA modifications). The two types of resistances may overlap and interconvert. The Yin–Yang model can also be used to explain varying hierarchy or spectrum of latent infections (Yin) and active disease (Yang) at the host level and their respective interconversions.8 This Yin–Yang model can also be applied to other microbes besides bacteria, such as fungi, parasites, and viruses (viral infected cells), and their infections and even cancer and cancer treatments (see below).19 A list of studies and observations that support or are consistent with the Yin–Yang model is presented in Table 1.
Figure 1.
The Yin–Yang model of persisters and latent infections.5,8,19 In a growing population of bacteria (Yang, red), there is a small population of non-growing or slowly growing persisters (Yin, black). In the persister population, there is a small number of growing bacteria (reverters). The persister population (Yin) or the growing population (Yang) is again heterogeneous and composed of various subpopulations with varying metabolic or dormant states in continuum in varying hierarchy (expressed by color from light to dark). The black spot in Yang (red) is connected to and the root of the Yin half (black), and the red spot in Yin, reverters, is connected to the Yang half (red). In the case of TB, INH kills growing bacteria (Yang) and RIF kills some growing bacteria, as well as slowly growing persisters, whereas PZA kills only persisters. Persisters not killed by antibiotics could revert to replicating forms (reverters) and cause relapse. The Yin–Yang model can be used to better describe latent infections (Yin) and active disease (Yang) at the host level and their respective interconversions.8,19 As drug treatment and immune responses inhibit or kill the growing bacteria (Yang) and some of the persisters, some persisters (Yin) still remain and the infection becomes latent (Yin), but may revert and cause relapse or sustained chronic infections with symptoms. In a hierarchical manner and among heterogeneous persister cell populations, there are a few true ‘stem' persister cells or mother cells (black spot in Yang) that have the capacity to derive other persisters (Yin) and initiate disease or cause reactivation. The Yin–Yang model proposes use of drugs targeting both replicating and non-replicating cells in combination or sequentially in a dynamic fashion and in cycles for better treatment of persistent bacterial infections. This Yin–Yang model can also be applied to other microbes, such as fungi, parasites, viruses, and their infections and even cancer and the respective treatments of infections and cancer.19
Table 1. Studies and observations that support or are consistent with the Yin–Yang model (see Figure 1).
| Setting | Organisms | References |
|---|---|---|
| Inclusion of pyrazinamide that kills persisters with other drugs that kill growing bacilli shortens TB treatment in mice and humans | M. tuberculosis | 78,106,107,108 |
| Two phases of TB therapy where the first phase involves a combination of INH, RIF, EMB and PZA followed by the second phase of only INH and RIF. INH is a drug that only kills growing bacteria and its inclusion in the second phase of treatment is to kill the ‘reverters' from the persisters not killed by the first phase treatment | M. tuberculosis | 8,109 |
| Use of isoniazid, a drug that is only active for growing bacteria, for treatment of LTBI; during LTBI, there are growing TB bacteria (reverters) that are susceptible to INH | M. tuberculosis | 8,109 |
| Spectrum or varying levels of persistence during latent TB infection and treatment | M. tuberculosis | 51,106 |
| Rapidly growing bacteria can give rise to persisters, whereas stationary phase bacteria can have cryptic growth | E. coli | 12,110 |
| Heterogeneity of persisters as demonstrated by varying antibiotic exposure times: ‘shallow' persisters and ‘deep' persisters | E. coli | 4,9 |
| Cancer stem cell drug candidates used in combination with current cancer drugs improve cancer treatment in mice | Breast cancer | 111,112,113 |
Abbreviations: EMB, ethambutol; LTBI, latent TB infection.
The Yin–Yang model simplifies and provides a unified model for the complex persister phenomenon and heterogeneity and hierarchy of persisters at the bacterial level and also persistent infections at the host level (see below). In addition, the Yin–Yang model explains the current practice of using isoniazid (INH), a drug only active against growing mycobacteria, for the treatment of latent tuberculosis (TB) infection as well as the current practice of two phase TB therapy where the second phase continues use of INH after the first phase of treatment with four drugs (INH, rifampin, pyrazinamide and ethambutol), which should have killed all growing bacteria already (Table 1). In addition, the Yin–Yang model proposes the use of multiple drugs targeting different bacterial populations, both persisters (Yin) and growing bacteria (Yang) for improved therapeutic effect. (See Figure 1 for more details.)
Persisters have been divided into two groups. Type I persisters (non-growing persisters formed in response to external triggers such as starvation) exit slowly from the stationary phase and do not grow in numbers from log phase to stationary phase. Type II persisters (slowly growing) are formed by phenotypic switching in the absence of external triggers and can switch back to normal phenotype and grow in numbers during the growth phase.20 The classification of type I and type II persisters is useful in characterizing persisters; however, it is worth noting that persisters are much more heterogeneous than the terms type I and type II suggest because either type I or type II persisters themselves again consist of different heterogeneous persisters within each category and the two types of persisters may interconvert as described in the Yin-Yang model.
Persister phenomenon is present in virtually all bacterial species, but the degree of persistence may vary among species as well as within species.21 In addition, persisters can adopt varying sizes and shapes from regular morphology to altered morphologies (granular or coccoid) as found in old cultures, biofilms and L-form bacteria.8,18,22,23 L-form bacteria are atypical, pleomorphic cell wall-deficient forms that are formed as part of the life cycle of stressed bacteria and have been implicated in persistent infections.23 Similarities between L-form bacteria, biofilm bacteria and persisters have been found22 and are discussed below (see the section on ‘MECHANISMS OF PERSISTER FORMATION AND SURVIVAL').
Persisters and multidrug tolerance
Persisters show tolerance to various bactericidal antibiotics, a property called multidrug tolerance (MDT). It was proposed that MDT in persisters is due to the prevention of ‘corruption' of drug targets by antibiotics in persister bacteria,13 but there is no evidence to support this hypothesis, and detailed mechanisms involved in MDT are not well understood. Recent studies have shown that there are multiple mechanisms of MDT. These mechanisms include reduced production in persisters of reactive oxygen species (ROS) influenced by the levels of antioxidant enzymes,24,25 inhibition of macromolecule synthesis by toxin–antitoxin (TA) modules,26 increased suppression of cellular metabolism mediated by PhoU4 and the presence of defects in trans-translation pathway that confer a broad defect in MDT.27 Decreased antibiotic uptake was recently shown to be involved in drug tolerance to fluoroquinolones, rifampin and linezolid in nutrient starved Mycobacterium tuberculosis.28 It remains to be seen whether reduced permeability to antibiotics is also found in other bacterial species as a mechanism for MDT in persisters. Although antibiotic tolerance in persisters is thought to be phenotypic, it is possible that under some conditions, antibiotic tolerant persisters may acquire mutations and develop genetic resistance. Similarly, a genetically antibiotic resistant mutant (Yang resistance) could also develop persisters with tolerance (Yin resistance); thus, genetic resistance and tolerance may interconvert and overlap.8
Stress and persisters
Because persisters are tolerant to not only antibiotics but also other stresses, susceptibility to stresses of mutants is often tested as part of the persister phenotypes in evaluating persister-defective mutants. For example, phoU and sucB mutants with defects in persisters are highly susceptible to not only antibiotics but also a variety of stresses.4,9 On the other hand, stresses can slow and inhibit bacterial growth, resulting in lower metabolic status and facilitates persister formation. Nutrient (amino acid or carbon) depletion has been shown to induce drug tolerant persisters.29 The carbon starvation mediated persister formation is mediated through activation of the ppGpp-SpoT metabolic TA module, which then leads to inhibition of DNA-negative supercoiling, a process that is affected by FIS, IHF, HU and SeqA DNA-binding proteins that participate in ppGpp-dependent persister formation through modulating DNA negative supercoiling.30,31 However, the persisters induced by transient nutrient depletion seem to lack the sustainable, multidrug-tolerant phenotype of persisters in the stationary-phase population.29 Heat, acidic pH and oxidative stresses have been shown to induce persister formation.32,33 Notably, bacterial persisters can tolerate antibiotics by reducing production of hydroxyl radicals.24 Although defects in the stringent response genes relA and spoT are known to cause decreased antibiotic tolerance,31 this phenotype was recently shown to be mediated through reduced production of the antioxidant defense enzymes superoxide dismutase and catalase.25,34 Furthermore, inactivation of enzymes involved in hydrogen sulfide (H2S) production in various bacteria rendered the bacteria highly sensitive to a variety of antibiotics due to loss of H2S antagonism of the reactive oxygen species induced by antibiotics.35 Like H2S, NO has also been shown to induce antibiotic tolerance through antioxidative defense.36 Low concentrations of antibiotics, such as ciprofloxacin, which presumably causes reactive oxygen production and reduced membrane potential via toxin TisB, could induce persister formation.37 More recently, antibiotics, such as the RNA synthesis inhibitor rifampin, protein synthesis inhibitor tetracycline and energy inhibitor CCCP, were shown to induce persister formation and enrich the proportion of persisters in cultures.38 Arrested protein synthesis caused by the above diverse stresses seems to be involved in persister formation.38
Persister assays and models
The current persister assays consist of exposing bacterial cultures or cells to bactericidal antibiotics (cell wall inhibitors, aminoglycosides or quinolones) for a short period of time (usually 2–6 h) and then scoring the number of surviving bacteria by colony-forming unit assay.39,40 Some studies added antibiotics directly to stationary phase cultures, which has more persisters not killed, whereas others resuspended or diluted stationary phase cultures in fresh medium containing antibiotics,41 which typically leads to fewer persisters due to elevated metabolic activity of stationary phase bacteria being resuspended in fresh medium. These different conditions affect persister counts. In addition, the type of antibiotics, antibiotic exposure time, antibiotic concentrations, age of cultures, aeration and culture media all affect persister numbers.4,9,41 The recovery time after antibiotic exposure may vary among persister cells.2,10 An automated method, ScanLag, was recently developed to detect delayed growth of persisters and is useful for measuring the slow recovery of persisters.42 There is a tendency in the field toward frequently using short antibiotic exposure times of no more than 6–8 h in persister assays. It must be emphasized that while a short exposure time to antibiotics is sufficient for demonstrating the presence of persisters, it may not be sufficient to demonstrate persister defects in some mutants that are obvious only after prolonged antibiotic exposure.4,9 In fact, the original studies by Hobby and Bigger used penicillin exposure times of 24 h or 48 h and even up to 3–11 days.1,2 If one understands the enormous heterogeneity of persisters, as expressed in ‘shallow' and ‘deep' persisters9 and best captured in the Yin–Yang model,5,8 one may not need to be so dogmatic about sticking to short antibiotic exposure times in persister assays. In addition, although the original persister phenomenon was demonstrated with bactericidal antibiotics, stress conditions have also been used as an equivalent to antibiotics in persister studies.4,9,43,44 It is likely that there is overlap between antibiotic persisters and stress persisters despite individuality or heterogeneity and specificity of persisters to particular conditions. This can be addressed using single-cell techniques such as utilizing a microfluidic device (see below). In addition, one has to determine which persister model among different models to use and whether one persister model is more relevant than others in persister studies. Finally, it must be realized that in vitro persisters are not the same as in vivo persisters due to differences in the environments that the bacteria reside in and the presence or absence of antibiotic exposure. Thus, a drug that can kill all in vitro persisters is not guaranteed to do so in vivo. Nevertheless, the in vitro persisters may share some common features of in vivo persisters and in vitro persister models should still have value in persister studies as surrogates of in vivo persisters. Even in vivo, persisters are not all the same and are subject to hierarchy and heterogeneity of persisters as expressed in the Yin–Yang model (Figure 1).
Persisters and single-cell analysis
Although persister cells were found to be dormant or non-growing at the population level in the 1940s, the presence of single persister bacteria tolerant to antibiotics was demonstrated convincingly only recently, using a single-cell microfluidic device.20 There is recent interest in the use of single-cell techniques for the study of persisters.20,45 The single-cell techniques are powerful for demonstrating tolerance to cidal antibiotics in a single persister,20 yet so far no transcriptomic or proteomic data are available for single persister cells due to the lack of sensitivity of the current methods. With increasing appreciation of the heterogeneity of persisters,5,7,8 the single-cell technique also faces some challenges as to which persister to study and whether the persister cell obtained in one in vitro system would be representative of other persisters in the population in vitro and persister cells in vivo. Recently, microfluidics studies revealed that Mycobacterium smegmatis cells expressing lower levels of KatG expression were tolerant to INH and grew in the presence of INH.45 INH is a prodrug that needs to be activated by the KatG enzyme, mutations of which cause INH resistance.46 It was proposed that stochastic expression of KatG leading to various bacterial populations expressing different amounts of KatG can lead to different INH tolerant persister populations.45 Although this in vitro model explains varying susceptibility or tolerance to INH as a function of the level of KatG expression in an artificial system, this may not be used as an argument against persisters being non-growing or dormant. It remains to be determined if this is a relevant persister mechanism for generation of real INH persisters in vivo or even in vitro in stationary phase cultures.
PERSISTERS, LATENT INFECTIONS AND PATHOGENESIS
Persisters pose significant challenges for the treatment of many chronic and persistent bacterial infections such as TB,8 Lyme disease47 and urinary tract infections (Table 2). Persisters underlie latent infections, chronic and recurrent infections, biofilm infections and lengthy therapy of certain bacterial infections, such as TB, and post-treatment persistence and relapse.8,13,18,48,49 While the most attention has been given to genetic drug resistance either in bacteria, viruses or even cancer, persistence or tolerance to antibiotics (Yin resistance) is equally important to, if not more important than, genetic drug resistance (Yang resistance) because prolonged and repeated treatment of persistent infections may lead to genetic drug resistance, which could occur during TB treatment.
Table 2. Diseases with known bacterial persistence problems.
| Disease | Pathogen | Treatment |
|---|---|---|
| Tuberculosis | M. tuberculosis | Isoniazid, rifampin, pyrazinamide, ethambutol |
| Syphilis | Treponema pallidum | Penicillins, doxycycline, macrolide |
| Lyme disease | Borrelia burgdorferi | Doxycycline, amoxicillin |
| Urinary tract infections | E. coli, Enterococcus, Pseudomonas aeruginosa, Chlamydia, Mycoplasma genitalium | Trimethoprim, amoxicillin, nitrofurantoin, quinolones, doxycycline, macrolide |
| Peptic ulcer | Helicobacter pylori | Amoxicillin, clarithromycin, metronidazole, omeprazole, doxycycline, bismuth |
| Bacteremia/sepsis | Staphylococcus aureus, Group B Streptococcus | Various antibiotics |
| Endocarditis | Streptococcus, Staphylococcus, Enterococcus | Penicillins, vancomycin |
| Otitis media | S. pneumoniae, Haemophilus influenzae, Moraxella catarrhalis | Amoxicillin, azithromycin |
| Brucellosis | Brucella abortus | Doxycycline, rifampin |
| Biofilm infections, periodontitis, prosthetic device infections | Various pathogens | Refractory to antibiotic treatment |
Persistent and latent infections are more complex than previously thought and are found to be of varying hierarchy50 and in continuous spectrum51 and can be expressed in the Yin–Yang model (Figure 1).8 Persistent or latent infections can be pre-antibiotic persistent or post-antibiotic persistent. Pre-antibiotic persistence that is formed under the pressure of the host immune responses refers to initial latent infection before the development of active disease and antibiotic treatment, whereas post-antibiotic persistence refers to the presence of persisters that survive antibiotic treatment and can relapse after treatment. Pre-antibiotic persistence may not be the same as post-antibiotic persistence, which may be more similar to ‘deep' persistence. In addition, microbial variants with increased persistence or antibiotic tolerance may develop during treatment as observed in chronic Pseudomonas aeruginosa infection in cystic fibrosis patients,52 but the molecular basis involved is unclear.
Persistence seems to be a widespread phenomenon. However, different bacterial species seem to have different capacities for persistence in vitro and in vivo such that bacterial infections have varying degrees of difficulty to treat or cure (Table 2). For example, Streptococcus pneumoniae seems to have poor ability to form persisters such that its cure by a single antibiotic can be achieved readily in a week or two. In addition, immune clearance of a small number of residual S. pneumonia seems effective, so there is usually no relapse after antibiotic treatment. In contrast, some bacterial species, such as M. tuberculosis, cause a chronic persistent infection that takes at least 6 months to cure while the immune system seems to be less adequate to clear residual persisters left over from chemotherapy. More recently, Borrelia burgdorferi has been demonstrated to have a persistence problem despite antibiotic treatment using mouse and monkey models,53,54 which may provide some explanation for persisting chronic Lyme disease observed in some patients.47 In addition to bacterial factors that vary in persistence, the host susceptibilities that vary among individuals play a role in the degree of persistence during infection as well. These variations at the levels of bacterial persistence and host defense mechanisms can have implications in treatment of bacterial infections and might explain why some individuals develop chronic disease and relapse after treatment, whereas others seem to have a stable cure. A variety of conditions, such as host immune and hormonal factors, physical and psychological stresses, and co-infections, such as HIV, measles and mixed bacterial infections, might cause relapse or reactivation of latent infections.
It is possible that not all bacterial cells of a given pathogenic species can cause successful infections. We hypothesize that ‘seeding' with persisters or mother cells (dormant cells where heterogeneous persisters are derived) may be critical for successful establishment of infection and disease. In addition to the metabolic status of the bacterial cells that enter the host, the heterogeneity of host phagocytes might also influence the outcome of infection. Thus, interactions of the heterogeneous nature of populations of bacteria, such as M. tuberculosis, and of the macrophages that ingest them might cause a diverse range of possible outcomes. These outcomes include unsuccessful infection, successful infection with a transient immune response (lost after some time due to bacterial clearance), successful infection with a stable prolonged immune response and successful infection with an immune response and disease pathology. This hypothesis needs to be addressed with animal models in future studies. At the level of granuloma lesions, there might be a varying degree of heterogeneous granulomatous tissue correlating to the degree of inflammation, ranging from quiescent granulomas with low inflammation to more active and dynamic granulomas with more active inflammation even in the same lungs, and over time. Recently, it has been shown that there are varying degrees of latent TB infection, ranging from nearly active TB to a latent state with a remote chance of reactivation.51
At the host level, it is possible that infection of host stem cells (or stem-like cells or progenitor cells, including quiescent resting memory cells) by pathogens, such as the intracellular bacteria M. tuberculosis and Brucella abortus, and viruses, such as HIV55 and HBV,56 might contribute to increased persistence problems and protracted or chronic disease courses due to the longevity of the stem cells. It is of interest to note that infection with M. leprae,57 which causes chronic leprosy, could reprogram the host cell into a stem cell-like phenotype that survives a long time, though it may not be easy to distinguish if the infected cell is stem cell-like before or after infection. More recently, it was shown that M. tuberculosis could reside in bone marrow CD271+/CD45− mesenchymal stem cells, which could provide a niche for dormant infection.58 It remains to be seen if the chronicity of infections by certain pathogens, such as mycobacteria, could involve host stem cells as a niche for perpetuation of the infection.
MECHANISMS OF PERSISTER FORMATION AND SURVIVAL
Mechanisms of persister formation are not well understood as persisters are elusive, small in number, heterogeneous, and transient and can change with environment, which poses significant challenges to their study. Epigenetic factors can promote bacterial persister formation through bistable gene expression,59 mediated through stochastic or induced expression of persister related genes,60 or through changes in DNA modifications or signaling protein modifications. Thus, permutations at the levels of expression of multiple persister genes (Table 3), regulatory RNA, modifications of DNA and post-translational modifications of proteins could produce enormous diversity and heterogeneity of persisters as expressed in the Yin–Yang model (Figure 1). Although senescence or aging has been proposed as a persister mechanism,61 aging itself can hardly be a mechanism of persisters as aging must in turn be acting through certain cellular processes, which could involve persister mechanisms. Although various persister genes have been identified (Table 3), what and how cells sense to form persisters remain unclear.
Table 3. Persister mechanisms in bacteria.
| Persister pathways | Genes involved | Mechanisms/features | References |
|---|---|---|---|
| Toxin–antitoxin modules | hipBA, relBE, mazEF, tisAB, mqsR, hhA, hokA, cspD, pasT | Toxin–antitoxin modules inhibit protein or nucleic acid synthesis; Lon protease can degrade the antitoxin to regulate persister formation | 3,12,37,40,69,114,115 |
| Alternative energy production | sucB, ubiF, glpD, plsB, tgs1 | Provision of energy under stress conditions | 9,63,116 |
| Stringent response | relA, dksA | ppGpp synthesized by RelA inhibits RNA synthesis | 31,34,68,117 |
| SOS response/DNA repair and protection of DNA | lexA, recA, recB, xerC, xerD, dps | Repair of DNA damage caused by ROS | 29,118,119,120 |
| Antioxidant defense H2S, NO | Superoxide dismutase, catalase | Removal of ROS and hydroxyl radical | 24,33,35,36 |
| Enhanced efflux or transporter activity | Various | Removal of toxic substances or antibiotic buildup, underlying tolerance to antibiotics and stresses | 33,121,122 |
| Phosphate metabolism | phoU | PhoU is a negative regulator of phosphate uptake, mutant has dramatic defect in persister phenotype; shutdown of metabolic activity | 4,67,123 |
| Trans-translation | ssrA, smpB, rpsA | Degradation of toxic proteins and mRNA and recycling of ribosomes | 27,81 |
| Signaling pathways | comE/comC; tnaA, oxyR, flu, pspBC | Quorum sensing peptide or homoserine lactone or indole, acting through TA or antioxidant defense OxyR and phage-shock pathways | 32,124,125 |
The approaches used to identify persister genes are worth mentioning. Although persisters are caused by epigenetic changes, mutagenesis has been traditionally used to isolate genes involved in persister formation and has led to identification of a range of persister related genes, such as hipA, relA, phoU, sucB and ubiF, just as sporulation is an epigenetic trait that has become reasonably well understood using the mutagenesis approach. The mutagenesis approach has been used to identify persister-related genes whose mutations caused either reduced persistence or increased persistence. Mutations that cause decreased persistence include relA,31 phoU,4 sucB and ubiF.9 Mutations that cause increased persistence map to the following genes: hipA encoding toxin,3 metG encoding methionyl-tRNA synthetase, tktA encoding transketolase A and glpD encoding glycerol-3-phosphate dehydrogenase.62 In an overexpression study, glpD and glpABC encoding glycerol-3-phosphate dehydrogenase and plsB encoding glycerol-3-phosphate acyltransferase were found to confer increased persistence.63 It is intriguing that glpD had opposite phenotypes in the two different studies.62,63 However, the mutagenesis approach is only useful for identifying non-essential dominant genes that have a major effect on the phenotype and is less useful for identifying a phenotype that is determined by multiple genes of minor effect. The fact that certain mutagenesis screens to identify persister genes did not provide much insight into persister mechanisms 11,13 does not invalidate this approach to studying persisters. Factors that might have contributed to failure to identify persister genes by the transposon mutant approach might include screening a partial mutant library, short antibiotic exposure and aeration during antibiotic exposure. The duration of antibiotic exposure in the mutant screen is critical. A short exposure of 6 h with ofloxacin was used to screen the Escherichia coli KEIO mutant library and identified many genes involved in stress responses and global regulation with minor or ‘shallow' persister phenotypes.39 It is unrealistic to expect a complete loss of persisters (a ‘persisterless' phenotype) by a mutant in a screen with a brief antibiotic exposure of a few hours, especially when using stationary phase cultures. A longer antibiotic exposure or higher antibiotic concentrations may be needed for identification of true or ‘deep' persister genes and, indeed, has led to the discovery of phoU,4 sucB and ubiF9 as persister genes. It is likely that different persister genes will be identified at different antibiotic exposure times. However, a potential limitation of the use of a deletion mutant library for persister mutant screens is that compensatory mutations could mask the persister defective phenotype, which may lead to an inability to identify critical persister genes. Although microarray analysis has been used for profiling persister related genes,64,65 the data were obtained mostly on heterogeneous populations, which could mask the signals in individual or single persister cells. In addition, the genes involved in persistence are likely to vary according to the specific environment or models used in the study. These are the challenges facing studies aimed to identify persister genes.
Although different bacterial species may differ in terms of their ability to form persisters, they share many common features and mechanisms. It is increasingly clear that multiple mechanisms of varying hierarchy and importance are involved in persister formation in different models of persistence (Table 3). Our comparative analyses of the pathways involved in persister formation and survival between E. coli and M. tuberculosis8 indicate that while persister genes and pathways may vary, the overall persister mechanisms and pathways in different bacterial species are largely conserved (convergent evolution) (Table 3). In addition, the genes and pathways in persisters and biofilm bacteria and L-form bacteria have been found to overlap and share significant similarities,22 which include SOS response and DNA repair, iron homeostasis, signaling, efflux/transporter, envelope/membrane stress, energy production, phosphate metabolism, sulfur metabolism, signaling, phage shock proteins and protein degradation (protease and trans-translation). These findings suggest that biofilm bacteria, L-form bacteria and persisters are related entities that share common mechanisms.
Given the recent advances in understanding persister mechanisms, it remains to be seen whether the in vitro identified persister mechanisms (Table 3) are operative and valid for in vivo persisters. Some persister genes, such as phoU and relA, that have been shown to be a persister gene in vitro4,31 are also involved in virulence66 and persistence in vivo.67,68 Deletion of TA module PasTI, but not other TA modules, such as HipBA and HigBA, in pathogenic E. coli, was shown to have reduced persister formation and decreased virulence in mice.69
PERSISTERS, L-FORM AND BIOFILM BACTERIA, AND CANCER STEM CELLS
There are significant parallels between bacterial persisters and cancer stem cells. In cancer, there is a situation analogous and equivalent to bacterial persisters, termed ‘cancer stem cells'. Cancer stem cells are defined as ‘a small subset of cancer cells within a cancer that constitutes a reservoir of self-sustaining cells with the exclusive ability to self-renew and to cause the heterogeneous lineages of cancer cells that comprise the tumor.70 It was proposed that cancer stem cells resemble bacterial persister cells in 2007 (http://forms.asm.org/microbe/index.asp?bid551533),71 based on the common pathways between bacterial persisters, biofilm and L-form bacteria (cell wall-defective variants formed under cell membrane stress)22,23,72 and cancer stem cells.19,73 In E. coli, L-form bacteria, which can be considered as a type of deep or true persisters (mother cells), occur at the frequency of 104–105 cells,22 which is about two orders of magnitude less frequent than persisters. Like persister cells, cancer stem cells are also quite heterogeneous and resist chemotherapy drugs and stresses and cause relapse and metastasis.19,74 There is significant recent interest in the analogy between bacterial persisters and cancer stem cells.75,76,77 The above analyses19,22,73 revealed that although the genes involved in the common pathways between bacterial persisters, L-form and biofilm bacteria, and cancer stem cells do not show significant homology, they have similar functions. Such parallels in bacterial persisters and cancer stem cells may not only help to shed light on their mechanisms via convergent evolution but also may allow common treatment strategies to be developed for more effective treatment of persistent infections and cancer in the future (see below).
TAMING PERSISTERS: TREATMENT STRATEGIES
While different bacterial infections seem to have different capacities for persistence and varying degrees of difficulty for treatment, their cure relies on the combined action of antibiotics and the host immune system. In addition, the type of drugs and the status of the target cells affect treatment outcome. Here it may be instructive to examine in some detail the interesting example of the unique TB persister drug PZA, which may shed light on the treatment of persistent bacterial infections in general and even cancers. PZA plays a key role in shortening TB therapy from 9–12 months to 6 months by killing a subpopulation of persisters not killed by other TB drugs (Figure 1).78 PZA is an unconventional and paradoxical drug that acts only on non-growing persisters at acidic pH.78,79 Unlike common antibiotics that act on growing bacteria, PZA is completely dissimilar in that it has no activity against growing M. tuberculosis bacteria.78 In contrast to common antibiotics that inhibit cell wall, protein, and nucleic acid synthesis and are active only against growing bacteria, PZA inhibits energy production80 and the trans-translation process, which recycles ribosomes and degrades toxic protein buildup under stress,81 and perhaps coenzyme A synthesis82, which is required for survival of M. tuberculosis persisters. It is these unique properties of PZA that are critical for killing persisters and shortening TB therapy. It is of interest to note that PZA also inhibits the quiescent malaria parasite in the mouse model83 and is also active against E. coli ampicillin tolerant persisters.84 Although there is considerable recent interest in developing antibiotics targeting persisters,13,85,86 PZA is the only prototype persister drug so far that has been shown to improve the treatment of a persistent infection. Nevertheless, PZA validates an important principle that drugs targeting dormant persisters, when used in combination with drugs that target growing organisms, are critical for shortening the treatment. The story of PZA has important implications for developing future antibiotics and cancer drugs that target persisters and cancer stem cells to improve treatment of both persistent infections and cancers and perhaps even latent viral infections, such as HIV and HBV, which hide in quiescent stem-like cells, and also persistent parasites or fungi.
In addition to the insights from the above example, several approaches should be explored to better control persisters. One approach would be to directly target persisters with drugs, but unfortunately all current antibiotics, except the TB drug PZA, are predominantly active against growing bacteria. Current antibiotics generally have no activity against persisters because these types of cells were not used during the screening. There is currently increasing interest in developing new drugs active against bacterial persisters.7,8,49,87 Some candidate compounds that are active against persisters8 have been identified and, if they pass the safety and efficacy phase, are expected to be used together with current antibiotics or drugs for improved treatment based on the common principle of targeting both growing bacteria or cells and non-growing persisters.5,19 This is exemplified in the case of INH (which kills growing bacteria) and PZA (which kills persisters) for TB treatment (Figure 1). However, it is preferable that the drugs in combination interfere with different pathways in the cells and kill different cell populations to optimize the potential for killing of persisters.
A second approach would be to ‘wake up' or alter the metabolic status of persisters,8,18 so they respond to antibiotic treatment. Although resuscitation factors have been found for bacteria,16,88 they have not been used therapeutically in animal models to demonstrate feasibility. Recently, metabolites, such as glucose, glycerol and relatively less efficient carbon sources (mannose, fructose, sorbitol, pyruvate, lactate and acetate), and nucleotides, such as thymidine, uridine and inosine, have been shown to potentiate activity of aminoglycoside activity for persisters in vitro.89 Such an approach needs to be validated in animal or human studies in the future.
A third approach would be to enhance the activity of current antibiotics by certain agents to kill some persister cells.90,91,92 For example, aspirin, ibuprofen and iron have been shown to enhance the activity of the persister drug PZA against M. tuberculosis.90,91 In addition, sugar mannitol can enhance the killing activity of persisters by aminoglycoside antibiotics through stimulating the proton motive force needed for increased uptake of the antibiotic in the mouse model of urinary tract infection.92 However, it is unclear whether mannitol works through its diuretic effect to wash off the bacteria more effectively by increasing the amount of urine and/or through its effect on enhancing the uptake of aminoglycoside. In addition, this is a highly specific case, and the sugar only increases the activity of aminoglycosides but not other antibiotics. Furthermore, it remains to be seen whether the use of mannitol is effective in patients. A related approach to enhancing the effectiveness of the existing antibiotics in killing persisters is to increase ROS production.93 Recently, it has been shown that silver, which produces ROS, enhanced the activity of vancomycin, improving the treatment of bacterial infections in mice.94 In addition, 3-[4-(4-methoxyphenyl)piperazin-1-yl]piperidin-4-yl biphenyl-4-carboxylate (C10)95 and (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5H)-one (BF8)96 were found to convert antibiotic tolerant persisters to an antibiotic sensitive phenotype. It remains to be determined how the compounds work and whether they can be used to resuscitate persisters for improved treatment of persistent infections.
The fourth approach would be to harness the host immune system to control persisters and cancer stem cells through enhancing innate and acquired immunity in the form of immune-modulating cytokines or immunotherapeutic vaccines that encompass antigens from both growing cells and non-growing cells (persisters and cancer stem cells). For example, inclusion of antigens from both growing bacteria (Antigen 85 and ESAT-6) and dormancy antigen Rv2660c or HspX from M. tuberculosis could enhance vaccine efficacy in prophylactic and therapeutic vaccines in animal models.97 Combined immunotherapy with chemotherapy for persisters should also be explored for improved treatment.98,99
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Despite significant progress in our understanding of persisters in recent years, much remains to be learned about the biology of persisters. Classical genetic approaches have identified multiple genes and pathways that are involved in persister formation or survival. However, there are some limitations with the classical genetic mutant approach due to problems of compensatory mutations and with the reductionist approach of looking at one gene at a time. With the application of the ‘omics' (transcriptome, proteome, metabolome, epigenome) and next-generation sequencing techniques including Tn-seq,100 new knowledge about persisters will undoubtedly be gained in the near future. Networks and systems biology approaches remain to be applied to the study of persister mechanisms. It is not enough to say that the whole is more than its parts in terms of persister mechanisms. More importantly, how different components interact in a dynamic manner in the context of systems biology to cause complex persister phenotypes needs to be addressed in the future. An even more crucial challenge is to understand what these complex data mean and whether useful intervention or treatment strategies can be derived from them. In addition, significant technical hurdles exist when applying the ‘omics' tools to single or rare true persister cells or mother cells due to lack of sufficiently sensitive techniques. For example, current single-cell techniques cannot yet identify the transcriptomic or proteomic profiles of an individual persister cell. In addition, future studies may need to explore new dimensions of persister mechanisms, including studying possible roles of bio-electromagnetic fields and information flow (‘Qi' or flow of energy or life force) in cellular circuits that maintain viability of persisters in the context of thermodynamics as in dissipative systems. It is in this context that the relationships between stress, cell death, aging, persistence and longevity and the nature of life need to be investigated in the future.
While most persister mechanistic studies have been performed mainly with E. coli, it is important to study the persister mechanisms in other bacterial pathogens. As an evolutionarily useful strategy to survive harmful stresses in the environment, the persister phenomenon occurs not only in bacteria but also in all life forms (kingdoms). For example, persister phenomenon has been found in fungi,101 parasites,102 cancer stem cells19 and viral infected host cells.55 It would be of interest to compare and contrast the common mechanisms among bacteria, fungi, parasites102 and viral (HIV,55 HBV, HPV) infected host cells and cancer stem cells.19
It would be of interest to study the latent forms of the disease (i.e., latent infections) rather than just the advanced and complicated forms of the disease. Future studies by ecological approaches need to examine the microenvironment of persisters and assess the environmental factors, as well as host factors (including role of host microbiota), that affect reactivation, progression and outcome of the disease. In addition, it would be of interest to develop more sensitive diagnostic tools to detect dormant persister organisms in clinical specimens and in affected tissues. Moreover, it will be necessary to identify immune mechanisms that control latent infections. Such information will be useful for developing interventions based on altering the microenvironment needed for survival of persisters and developing immunotherapeutic vaccines for their effective control. It is important to understand why some individuals are not cured while others are cured. Future investigations are needed to understand why some individuals seem to have chronic persistent and recurrent infections, whereas other individuals are cured by standard treatment in the context of varying degrees of host susceptibilities (defined in a broad sense not necessarily restricted by genetic factors) and bacterial persistence.
It is important to establish more relevant models of persisters or persistence for mechanistic studies that are representative of in vivo situations, as well as developing drugs that kill in vivo persisters and improve treatment. It would be quite challenging to develop persister drugs as one ponders which model to use for drug screens, considering the diverse and variable nature of persisters as expressed in the Yin–Yang model (Figure 1).8 The above problems with bacterial persisters, also apply to cancer stem cells,19,103 and will be a major stumbling block for both fields and a major topic of interest for the future. The current in vitro models of persisters or cancer stem cells may have significant limitations and it remains to be seen if the data obtained in vitro can be validated in vivo in animal models or patients.
There are currently significant debates, as well as interest, about persister mechanisms and drugs. To capture the current status of the field, it may be fitting to end the article with the parable about the blind men and the elephant. The elephant, which is analogous to persisters or cancer stem cells, is described as a snake, a spear, a fan, a tree, a wall and a rope by blind men touching different parts of the elephant, which represent different models and pathways of persisters or cancer stem cells and are only partially right. This partial knowledge, which largely results from the limitations of current methodologies, is not perfect and is an intermediate state of knowledge that is useful and acceptable with reservation. The ultimate test of this partial knowledge will be whether we can devise useful drugs and therapeutic strategies targeting persisters for improved treatment in the future. There is a convergence of interest in both the persister field and the cancer stem cell field to develop new drugs targeting the quiescent forms (‘Yin') (i.e., persisters)8,49,85,87 of cancer stem cells for improved treatment.19,104,105 The identified pathways in bacterial persisters could serve as potential targets for development of new persister drugs. From the prototype persister drug PZA, one may see the future of antibiotic and even cancer drug development. Future studies are needed to test whether drugs analogous to PZA that target persisters and cancer stem cells can improve treatment of persistent infections and cancers.
Acknowledgments
The support from NIH AI099512, Lyme Research Alliance and lymedisease.org is gratefully acknowledged. I thank Peng Cui (Huashan Hospital, Fudan University, China) for help with drawing Figure 1.
References
- Hobby GL, Meyer K, Chaffee E. Observations on the mechanism of action of penicillin. Proc Soc Exp Biol NY. 1942;50:281–285. [Google Scholar]
- Bigger JW. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. [Google Scholar]
- Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Y. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob Agents Chemother. 2007;51:2092–2099. doi: 10.1128/AAC.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Advances in the treatment of tuberculosis. Clin Pharmacol Ther. 2007;82:595–600. doi: 10.1038/sj.clpt.6100362. [DOI] [PubMed] [Google Scholar]
- Gefen O, Balaban NQ. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol Rev. 2009;33:704–717. doi: 10.1111/j.1574-6976.2008.00156.x. [DOI] [PubMed] [Google Scholar]
- Allison KR, Brynildsen MP, Collins JJ. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr Opin Microbiol. 2011;14:593–598. doi: 10.1016/j.mib.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yew WW, Barer MR. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother. 2012;56:2223–2230. doi: 10.1128/AAC.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Sim S, Shi W, Du L, Xing D, Zhang Y. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol Lett. 2010;303:33–40. doi: 10.1111/j.1574-6968.2009.01857.x. [DOI] [PubMed] [Google Scholar]
- Joers A, Kaldalu N, Tenson T. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J Bacteriol. 2010;192:3379–3384. doi: 10.1128/JB.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Coates AR. Transposon mutagenesis identifies genes which control antimicrobial drug tolerance in stationary-phase Escherichia coli. FEMS Microbiol Lett. 2005;243:117–124. doi: 10.1016/j.femsle.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microbial Ecology. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- Hobby GL, Auerbach O, Lenert TF, Small MJ, Comer JV. The late emergence of M. tuberculosis in liquid cultures of pulmonary lesions resected from humans. Am Rev Tuberc. 1954;70:191–218. doi: 10.1164/art.1954.70.2.191. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Woods A, Cotter RJ, Sun Z. Resuscitation of dormant Mycobacterium tuberculosis by phospholipids or specific peptides. Biochem Biophys Res Commun. 2001;284:542–547. doi: 10.1006/bbrc.2001.4993. [DOI] [PubMed] [Google Scholar]
- Greenwood D. Phenotypic resistance to antimicrobial agents. J Antimicrob Chemother. 1985;15:653–655. doi: 10.1093/jac/15.6.653. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Persistent and dormant tubercle bacilli and latent tuberculosis. Front Biosci. 2004;9:1136–1156. doi: 10.2741/1291. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhang Y. Cancer stem cells: models, mechanisms and implications for improved treatment. Cell Cycle. 2008;7:1360–1370. doi: 10.4161/cc.7.10.5953. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Stewart B, Rozen DE. Genetic variation for antibiotic persistence in Escherichia coli. Evolution. 2012;66:933–939. doi: 10.1111/j.1558-5646.2011.01467.x. [DOI] [PubMed] [Google Scholar]
- Glover WA, Yang Y, Zhang Y. Insights into the molecular basis of L-form formation and survival in Escherichia coli. PLoS ONE. 2009;4:e7316. doi: 10.1371/journal.pone.0007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingue GJ. Demystifying pleomorphic forms in persistence and expression of disease: are they bacteria, and is peptidoglycan the solution. Discov Med. 2010;10:234–246. [PubMed] [Google Scholar]
- Kim JS, Heo P, Yang TJ, et al. Bacterial persisters tolerate antibiotics by not producing hydroxyl radicals. Biochem Biophys Res Commun. 2011;413:105–110. doi: 10.1016/j.bbrc.2011.08.063. [DOI] [PubMed] [Google Scholar]
- Bink A, Vandenbosch D, Coenye T, Nelis H, Cammue BP, Thevissen K. Superoxide dismutases are involved in Candida albicans biofilm persistence against miconazole. Antimicrob Agents Chemother. 2011;55:4033–4037. doi: 10.1128/AAC.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Maisonneuve E. Bacterial persistence and toxin–antitoxin loci. Annu Rev Microbiol. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L, Shi W, Xie J, Zhang Y. Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J Antimicrob Chemother. 2013;68:2477–2481. doi: 10.1093/jac/dkt231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarathy J, Dartois V, Dick T, Gengenbacher M. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung DK, Chan EW, Chin ML, Chan RC. Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Antimicrob Agents Chemother. 2010;54:1082–1093. doi: 10.1128/AAC.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol Cell. 2013;50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- Leung V, Levesque CM. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol. 2012;194:2265–2274. doi: 10.1128/JB.06707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Vulic M, Keren I, Lewis K. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother. 2012;56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan BW, Valenta JA, Benedik MJ, Wood TK. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother. 2013;57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S, Lewis K, Vulic M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother. 2008;52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Wang X, O'Connor HF, Benedik MJ, Wood TK. Bacterial persistence increases as environmental fitness decreases. Microb Biotechnol. 2012;5:509–522. doi: 10.1111/j.1751-7915.2011.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luidalepp H, Joers A, Kaldalu N, Tenson T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J Bacteriol. 2011;193:3598–3605. doi: 10.1128/JB.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Reisman I, Gefen O, Fridman O, et al. Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nat Methods. 2010;7:737–739. doi: 10.1038/nmeth.1485. [DOI] [PubMed] [Google Scholar]
- Scherrer R, Moyed HS. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J Bacteriol. 1988;170:3321–3326. doi: 10.1128/jb.170.8.3321-3326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl S, Gabay C, Kishony R, Oppenheim A, Balaban NQ. Nongenetic individuality in the host–phage interaction. PLoS Biol. 2008;6:e120. doi: 10.1371/journal.pbio.0060120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamoto Y, Dhar N, Chait R, et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- Stricker RB, Johnson L. The pain of chronic Lyme disease: moving the discourse backward. FASEB J. 2011;25:4085–4087. doi: 10.1096/fj.11-1203LTR. [DOI] [PubMed] [Google Scholar]
- McDermott W. Microbial persistence. Yale J Biol Med. 1958;30:257–291. [PMC free article] [PubMed] [Google Scholar]
- Fauvart M, de Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- McDermott W. Microbial persistence. Harvey Lecture. 1969;63:1–31. [PubMed] [Google Scholar]
- Barry CE, 3rd, Boshoff HI, Dartois V, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy LR, Burns JL, Lory S, Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, Hodzic E, Imai DM, Feng S, Yang X, Luft BJ. Ineffectiveness of tigecycline against persistent Borrelia burgdorferi. Antimicrob Agents Chemother. 2010;54:643–651. doi: 10.1128/AAC.00788-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embers ME, Barthold SW, Borda JT, et al. Persistence of Borrelia burgdorferi in rhesus macaques following antibiotic treatment of disseminated infection. PLoS ONE. 2012;7:e29914. doi: 10.1371/journal.pone.0029914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Zoulim F. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J Hepatol. 2005;42:302–308. doi: 10.1016/j.jhep.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Masaki T, Qu J, Cholewa-Waclaw J, Burr K, Raaum R, Rambukkana A. Reprogramming adult Schwann cells to stem cell-like cells by leprosy bacilli promotes dissemination of infection. Cell. 2013;152:51–67. doi: 10.1016/j.cell.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Kashino SS, Pulu I, et al. CD271+ bone marrow mesenchymal stem cells may provide a niche for dormant Mycobacterium tuberculosis. Sci Transl Med. 2013;5:170ra13. doi: 10.1126/scitranslmed.3004912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, et al. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci USA. 2010;107:12541–12546. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper I, Gilbert P, Ayati BP, Dockery J, Stewart PS. Senescence can explain microbial persistence. Microbiology. 2007;153:3623–3630. doi: 10.1099/mic.0.2007/006734-0. [DOI] [PubMed] [Google Scholar]
- Girgis HS, Harris K, Tavazoie S. Large mutational target size for rapid emergence of bacterial persistence. Proc Natl Acad Sci USA. 2012;109:12740–12745. doi: 10.1073/pnas.1205124109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AL, Vulic M, Lewis K. GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol. 2006;188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts J, Lukey P, Robb L, McAdam R, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Buckles EL, Wang X, Lockatell CV, Johnson DE, Donnenberg MS. PhoU enhances the ability of extraintestinal pathogenic Escherichia coli strain CFT073 to colonize the murine urinary tract. Microbiology. 2006;152:153–160. doi: 10.1099/mic.0.28281-0. [DOI] [PubMed] [Google Scholar]
- Shi W, Zhang Y. PhoY2 but not PhoY1 is the PhoU homologue involved in persisters in Mycobacterium tuberculosis. J Antimicrob Chemother. 2010;65:1237–1242. doi: 10.1093/jac/dkq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JL, Kraus CN, Boshoff HI, et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci USA. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JP, Mulvey MA. Toxin–antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog. 2012;8:e1002954. doi: 10.1371/journal.ppat.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Holzman D.Genetic switch plays key role, converting ordinary to “persister” bacteria Washington, DC; Microbe Magazine; 2007. Available at http://forms.asm.org/microbe/index.asp?bid551533 (accessed 24 March 2013). [Google Scholar]
- Allan EJ, Hoischen C, Gumpert J. Bacterial L-forms. Adv Appl Microbiol. 2009;68:1–39. doi: 10.1016/S0065-2164(09)01201-5. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wulfkuhle J, Zhang H, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras A. Cancer stem cells in tumor heterogeneity. Adv Cancer Res. 2011;112:255–281. doi: 10.1016/B978-0-12-387688-1.00009-0. [DOI] [PubMed] [Google Scholar]
- Glickman MS, Sawyers CL. Converting cancer therapies into cures: lessons from infectious diseases. Cell. 2012;148:1089–1098. doi: 10.1016/j.cell.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson CC, Intapa C, Jabra-Rizk MA. “Persisters”: survival at the cellular level. PLoS Pathog. 2011;7:e1002121. doi: 10.1371/journal.ppat.1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Jacob E, Coffey DS, Levine H. Bacterial survival strategies suggest rethinking cancer cooperativity. Trends Microbiol. 2012;20:403–410. doi: 10.1016/j.tim.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: a review. Int J Tuberc Lung Dis. 2003;7:6–21. [PubMed] [Google Scholar]
- Zhang Y, Permar S, Sun Z. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol. 2002;51:42–49. doi: 10.1099/0022-1317-51-1-42. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother. 2003;52:790–795. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- Shi W, Zhang X, Jiang X, et al. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;333:1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen J, Shi W, Liu W, Zhang WH, Zhang Y. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect. 2013;2:e34. doi: 10.1038/emi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deye GA, Gettayacamin M, Hansukjariya P, et al. Use of a rhesus Plasmodium cynomolgi model to screen for anti-hypnozoite activity of pharmaceutical substances. Am J Trop Med Hyg. 2012;86:931–935. doi: 10.4269/ajtmh.2012.11-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MM, Zhang Y. Effects of weak acids, UV and proton motive force inhibitors on pyrazinamide activity against Mycobacterium tuberculosis in vitro. J Antimicrob Chemother. 2006;58:936–941. doi: 10.1093/jac/dkl358. [DOI] [PubMed] [Google Scholar]
- Coates AR, Hu Y. Targeting non-multiplying organisms as a way to develop novel antimicrobials. Trends Pharmacol Sci. 2008;29:143–150. doi: 10.1016/j.tips.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Nathan C. Fresh approaches to anti-infective therapies. Sci Transl Med. 2012;4:140sr2. doi: 10.1126/scitranslmed.3003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- Mukamolova GV, Turapov OA, Young DI, Kaprelyants AS, Kell DB, Young M. A family of autocrine growth factors in Mycobacterium tuberculosis. Mol Microbiol. 2002;46:623–635. doi: 10.1046/j.1365-2958.2002.03184.x. [DOI] [PubMed] [Google Scholar]
- Orman MA, Brynildsen MP. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob Agents Chemother. 2013;57:4398–4409. doi: 10.1128/AAC.00372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoskovi A, Wade MM, Sun Z, Zhang Y. Iron enhances the antituberculous activity of pyrazinamide. J Antimicrob Chemother. 2004;53:192–196. doi: 10.1093/jac/dkh042. [DOI] [PubMed] [Google Scholar]
- Byrne ST, Denkin SM, Zhang Y. Aspirin and ibuprofen enhance pyrazinamide treatment of murine tuberculosis. J Antimicrob Chemother. 2007;59:313–316. doi: 10.1093/jac/dkl486. [DOI] [PubMed] [Google Scholar]
- Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci USA. 2012;109:12147–12152. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morones-Ramirez JR, Winkler JA, Spina CS, Collins JJ. Silver enhances antibiotic activity against gram-negative bacteria. Sci Transl Med. 2013;5:190ra81. doi: 10.1126/scitranslmed.3006276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Heo P, Yang TJ, et al. Selective killing of bacterial persisters by a single chemical compound without affecting normal antibiotic-sensitive cells. Antimicrob Agents Chemother. 2011;55:5380–5383. doi: 10.1128/AAC.00708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Bahar AA, Syed H, Ren D. Reverting antibiotic tolerance of Pseudomonas aeruginosa PAO1 persister cells by (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5H)-one. PLoS ONE. 2012;7:e45778. doi: 10.1371/journal.pone.0045778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard C, Hoang T, Dietrich J, et al. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat Med. 2011;17:189–194. doi: 10.1038/nm.2285. [DOI] [PubMed] [Google Scholar]
- Wang CC, Zhu B, Fan X, Gicquel B, Zhang Y. Systems approach to tuberculosis vaccine development. Respirology. 2013;18:412–420. doi: 10.1111/resp.12052. [DOI] [PubMed] [Google Scholar]
- Lowrie DB, Tascon RE, Bonato VL, et al. Therapy of tuberculosis in mice by DNA vaccination. Nature. 1999;400:269–271. doi: 10.1038/22326. [DOI] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Kyle DE, Gatton ML. Artemisinin resistance in Plasmodium falciparum: A process linked to dormancy. Int J Parasitol Drugs Drug Resist. 2012;2:249–255. doi: 10.1016/j.ijpddr.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality. Nat Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JB, Zhang Y. Preclinical development of cancer stem cell drugs. Expert Opin Drug Discov. 2009;4:741–752. doi: 10.1517/17460440903002059. [DOI] [PubMed] [Google Scholar]
- McCune RM, Jr, McDermott W, Tompsett R. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J Exp Med. 1956;104:763–802. doi: 10.1084/jem.104.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries K, Verhasselt P, Guillemont J, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- Fox W, Ellard GA, Mitchison DA. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council tuberculosis units, 1946–1986, with relevant subsequent publications. Int J Tuberc Lung Dis. 1999;3:S231–S279. [PubMed] [Google Scholar]
- The World Health Organization Treatment of tuberculosis: guidelines4th ed. Geneva; WHO; 2010 [PubMed] [Google Scholar]
- Ryan FJ. Bacterial mutation in a stationary phase and the question of cell turnover. J Gen Microbiol. 1959;21:530–549. doi: 10.1099/00221287-21-3-530. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhang H, Gu P, Bai J, Margolick JB, Zhang Y. NF-kappaB pathway inhibitors preferentially inhibit breast cancer stem-like cells. Breast Cancer Res Treat. 2008;111:419–427. doi: 10.1007/s10549-007-9798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhang H, Gu P, Margolick JB, Yin D, Zhang Y. Cancer stem/progenitor cell active compound 8-quinolinol in combination with paclitaxel achieves an improved cure of breast cancer in the mouse model. Breast Cancer Res Treat. 2009;115:269–277. doi: 10.1007/s10549-008-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Wang F, Chen G, et al. Effective elimination of cancer stem cells by a novel drug combination strategy. Stem Cells. 2013;31:23–34. doi: 10.1002/stem.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Kawata K, Taniuchi A, Kakinuma K, May T, Okabe S. RelE-mediated dormancy is enhanced at high cell density in Escherichia coli. J Bacteriol. 2012;194:1169–1176. doi: 10.1128/JB.06628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wood TK. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2009;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb C, Lee CM, Dubey VS, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS ONE. 2009;4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka K, Ghosh B, Dasgupta A, Basu J, Kundu M, Bose I. Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS ONE. 2008;3:e1771. doi: 10.1371/journal.pone.0001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbia EA, Roveta S, Schito AM, Gualco L, Marchese A. Antibiotic persistence: the role of spontaneous DNA repair response. Microb Drug Resist. 2001;7:335–342. doi: 10.1089/10766290152773347. [DOI] [PubMed] [Google Scholar]
- Dorr T, Lewis K, Vulic M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5:e1000760. doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almiron M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KN, Takaki K, Connolly LE, et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Mao Y, Yu J, et al. PhoY2 of mycobacteria is required for metabolic homeostasis and stress response. J Bacteriol. 2013;195:243–252. doi: 10.1128/JB.01556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moker N, Dean CR, Tao J. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J Bacteriol. 2010;192:1946–1955. doi: 10.1128/JB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat Chem Biol. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]