Abstract
Given the significant spatial and temporal heterogeneity in antimicrobial resistance distribution and the factors that affect its evolution, dissemination, and persistence, it is important to highlight that antimicrobial resistance must be viewed as an ecological problem. Monitoring the resistance prevalence of indicator bacteria such as Escherichia coli and enterococci in wild animals makes it possible to show that wildlife has the potential to serve as an environmental reservoir and melting pot of bacterial resistance. These researchers address the issue of antimicrobial-resistant microorganism proliferation in the environment and the related potential human health and environmental impact.
Keywords: antimicrobial resistance, Escherichia coli, enterococci, phylogenetic groups, wild animals, virulence factors
INTRODUCTION
Microorganisms play an important role in the cycling of elements at a global scale, thus profoundly and directly affecting the environments, in which all of life evolves. While microorganisms affect the environment, the environment in turn also engenders evolutionary pressures on the microorganisms themselves (Reid and Buckley, 2011).
Recent advances in DNA sequencing, high-throughput technologies, and genetic manipulation systems have permitted empirical studies that directly characterize the molecular and genomic bases of evolution (Wagner, 2008; Conrad et al., 2011). This launched the challenge of unraveling the genotype-phenotype connection, with implications not only for the investigation of evolution, but also physiology, disease risk, development, and biodiversity (Reid and Buckley, 2011).
In microbial populations, evolutionary change is supplied by two sources of new variation: horizontal transfer of genetic material from other, sometimes distantly related species, and mutation. Though, in the short run, mutation is the primary source of new genetic variation driving evolutionary change in microbial populations. In an asexual microbial species, evolutionary changes will happen by the repeated selection of new clones carrying adaptively favorable mutations (Adams, 2004).
In fact, firstly, genomic sequencing can determine the complete set of mutations responsible for an advanced phenotype, and has led to the discovery that interactions between these mutations are very usual. Secondly, adaptive mutations commonly target regulatory mechanisms. Thirdly, principles of systems-level optimization cause the genetic changes seen in adaptive evolution, and with a systems-level understanding, these optimization principles can be harnessed for the purposes of metabolic engineering. Fourthly, mutant sub-populations of enhanced fitness invariably arise in growing populations, but their dynamics in the population are complicated due to factors such as natural selection, clonal interference, drift, and frequency-dependent selection (Barrick et al., 2009).
Antibiotic-resistant bacteria are extremely important to human health, but the wild reservoirs of resistance determinants are poorly understood. The origins of antimicrobial resistance in the wildlife is important to human health because of the increasing importance of zoonotic diseases as well as the need for predicting emerging resistant pathogens. Wild animals provide a biological mechanism for the spread of antibiotic resistance genes. Antimicrobial-resistant Escherichia coli and Enterococcusspp. isolates originating from wildlife species were reported for the first time from Japanese wild birds (Sato et al., 1978). With the new millennium several studies, in different continents, have described the occurrence of antimicrobial-resistant in these bacteria species in wildlife (Souza et al., 1999; Sherley et al., 2000; Gakuya et al., 2001; Lillehaug et al., 2005; Skurnik et al., 2006; Dolejska et al., 2007; Gaukler et al., 2009; Schierack et al., 2009; Allen et al., 2010; Silva et al., 2010; Marinho et al., 2013; Navarro-Gonzalez et al., 2013; Pesapane et al., 2013; Santos et al., 2013). On the other hand, antimicrobial resistance have been described in others important pathogens, such as Salmonella spp. (Botti et al., 2013) and methicillin-resistant Staphylococcus aureus (MRSA; Porrero et al., 2013) in wild animals.
Proximity to human activities influences the antibiotic resistance profiles of the gut bacteria of wild mammals, which live in densely populated microbial habitats in which antibiotics select for resistance. While various bacterial species are important in terms of multiresistance and nosocomial infections in human and veterinary medicine, we consider the Gram-positive vancomycin-resistant enterococci (VRE) and Extended-spectrum Beta-Lactamases producing Gram-negative bacteria like E. coli (ESBL-E. coli) as being key indicator pathogens to trace the evolution of multiresistant bacteria in the environment and wildlife.
Enterococcus spp. are Gram-positive facultative anaerobic bacteria, spherical, which occur singly, in pairs or short chains and fit within the general definition of lactic acid bacteria (Ciftci et al., 2009). Most enterococci are not virulent and are considered relatively harmless, with little potential for human infection. However, they have also been identified as nosocomial opportunistic pathogens with increased resistance to antimicrobial approved agents (Chenoweth and Schaberg, 1990). Incidence of VRE among wild animals has been reported in several countries (Mallon et al., 2002; Figueiredo et al., 2009; Ishihara et al., 2013), including in remote areas (Silva et al., 2011a).
Escherichia coli is the head of the large bacterial family, Enterobacteriaceae, the enteric bacteria, which are facultatively anaerobic Gram-negative (Sorum and Sunde, 2001). This intestinal bacterium can be easily disseminated in different ecosystems. For this reason, fecal E. coli is considered to be an important indicator for the selective pressure exerted by the use of antimicrobials on intestinal populations of bacteria (van den Bogaard and Stobberingh, 2000). The production of ESBLs by Enterobacteriaceae, specifically by E. coli, has caused a major concern in several countries, being frequently implicated in human infections. Previous reports have described ESBL-containing E. coli strains in healthy wild animals (Pinto et al., 2010; Guenther et al., 2011).
The common occurrence of antimicrobial resistance in wildlife has several implications such as: the potential to serve as an environmental reservoir and melting pot of bacterial resistance; the zoonotic potential of enteric bacteria; and the potencial problems of the medical treatment of wildlife. This review aims to summarize the current knowledge on ESBL-E. coli and VRE in wildlife, in turn underlining the need for more large scale research, in particular sentinel studies to monitor the impact of multiresistant bacteria on wildlife.
PHYLOGENETIC HISTORY AND GENETIC STRUCTURE
The combination of mutation and horizontal transfer has created the overall phylogenetic structure of E. coli and enterococci, resulted to currently recognized four main phylogenetic groups for E. coli and different species for enterococci; and their respective lineages. Thus, an appreciation of mutation and horizontal transfer as important evolutionary processes within bacteria, in general, contributes in understanding of their roles, distribution and mechanistic modes of behavior (Johnson, 2002).
A multilocus enzyme electrophoresis (MLEE)-based phenogram using 38 enzymes (Selander et al., 1986; Goullet and Picard, 1989) identified four main phylogenetic groups (A, B1, B2, and D) and two accessory groups (C and E) in E. coli (Selander et al., 1987; Herzer et al., 1990; Tenaillon et al., 2010). These phylogenetic groups were recovered using the 1,878 genes of the Escherichia spp. core genome and the 2.6 million nucleotides of the E. coli chromosomal backbone (Touchon et al., 2009), which permitted a robust phylogeny to be developed; the first split in the E. coli phylogenetic history leads to one branch including the strains of group B2 and a subgroup within D that we called group F75 and another branch containing the rest of the species (Touchon et al., 2009). The remaining strains of group D then appeared from this second branch, followed by group E (Tenaillon et al., 2010). Finally, groups A and B1 are sister groups whereas group B2 is included in an ancestral branch (Jaureguy et al., 2008; Touchon et al., 2009). The B2 group reveals the highest diversity at both the nucleotide and the gene content level (Touchon et al., 2009), supporting its early occurrence in the species lineage and suggesting that it has subspecies status (Lescat et al., 2009).
Clermont et al. (2000) described a triplex polymerase chain reaction (PCR) strategy to assign E. coli isolates quickly to one of these phylogroups. It sought three phylogenetic group markers, the chuA and yjaA genes encoding hypothetical proteins and the TSPE4.C2 DNA sequences situated within a gene encoding putative lipase esterase, and groups were assigned based on different combinations of presence and/or absence of the three amplicons (Clermont et al., 2000).
Johnson et al. (2001) found that strains from phylogenetic groups B2 and D contained more virulence factors than strains from the phylogenetic groups A and B1. Usually, the extraintestinal pathogenic strains belong to groups B2 and D (Picard et al., 1999; Johnson and Stell, 2000), the commensal strains to groups A and B1 (Bingen et al., 1998), whereas the intestinal pathogenic strains belong to groups A, B1, and D (Pupo et al., 1997).
Nowadays, these phylogenetic groups differ in their ecological niches, life-history (Gordon and Cowling, 2003) and some characteristics, such as their ability to exploit different sugar sources, their antimicrobial resistance profiles and their growth rate (Carlos et al., 2010). A recent survey (Walk et al., 2007) demonstrated that the majority of the E. coli strains that are able to persist in the environment belong to the B1 phylogenetic group (Carlos et al., 2010).
Various researchers analyzed the distribution of the main phylogenetic groups among E. coli strains isolated from human and animal faces; it was found that the relative abundance of phylogenetic groups among mammals is dependent on the host diet, body mass, and climate (Gordon and Cowling, 2003; Carlos et al., 2010). A study analyzing fecal strains isolated from birds, non-human mammals, and humans, observed the prevalence of groups D and B1 in birds, A and B1 in non-human mammals, and A and B2 in humans. These different reports concluded that one of the main forces that shapes the genetic structure of E. coli populations among the hosts is domestication (Escobar-Paramo et al., 2006). Furthermore, other study in the south of French Guiana, human strains very rarely were observed as belonging to B2 phylogroup (3.7%) whereas wild animal strains were characterized by 46.1% belonging to B2 phylogroup (Lescat et al., 2013). Moreover, feces from zoo animals were analyzed and a prevalence of group B1 in herbivorous animals and a prevalence of group A in carnivorous and omnivorous animals were found (Baldy-Chudzik et al., 2008). Furthermore, domesticated animals have a decreased proportion of B2 strains than wild animals (from 30% in wild animals to 14 and 11% in farm and zoo animals, respectively) and an increased proportion of A strains (from 14% in wild animals to 27 and 26% in farm and zoo animals, respectively; Tenaillon et al., 2010).
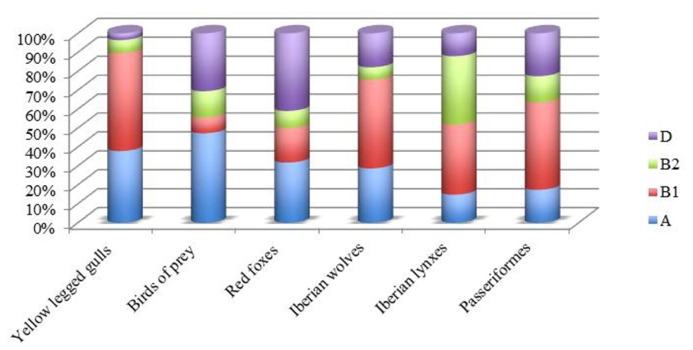
In Portugal, the prevalence of E. coli of groups A and B1 was observed in wild birds as seagulls (Radhouani et al., 2009), birds of prey (Radhouani et al., 2012b), and Passeriformes (Santos et al., 2013), but also in wild mammals as Iberian lynxes (Gonçalves et al., 2012a), red foxes (Radhouani et al., 2013), and in Iberian wolf (Gonçalves et al., 2013a; Figure 1). In addition, a report conducted by Simões et al. (2010) shows that 37% of all ESBL-E. coli isolated from seagulls belong to B2 or D phylogroup, a higher rate than previously reported (27% of all E. coli; Poeta et al., 2008).
FIGURE 1.
Phylogenetic group distributions of E. coli in wild animal from Portugal.
It is interesting to note that fecal samples from red foxes showed that the E. coli isolates from phylogenetic groups A and D were predominant. Similar results were observed in chickens and swine (Machado et al., 2008), in wild boars (Poeta et al., 2009) and in wild birds (Radhouani et al., 2010a,b) in the same geographical area.
With the exception of E. faecium and E. faecalis, the enterococci are infrequently described to be involved in human pathogenesis (Jett et al., 1994; Devriese and Pot, 1995). Though, in some countries association of strains of E. faecalis and E. faecium with human disease has reached proportions of serious concern (Jett et al., 1994; Leclercq, 1997; Fisher and Phillips, 2009). However, these enterococci species can show significant differences in the incidence of virulence factors and antimicrobial resistance. Generally, E. faecalis appears to harbor more virulence traits while E. faecium strains were generally free of virulence factors (Eaton and Gasson, 2001). In addition, considering the distribution of the antimicrobial resistance according to the species, the E. faecium possessed a higher level of resistance than E. faecalis (Gin and Zhanel, 1996; Franz et al., 2001). Actually, the majority of hospital-derived isolates of E. faecalis cluster in three clonal complexes, CC2, CC9, and CC87, and included the highest proportion of multiresistant isolates. (Ruiz-Garbajosa et al., 2006; van Schaik and Willems, 2010; Weng et al., 2013), while the CC17 is a major group of genetic lineage of E. faecium that has widely spread worldwide and it is associated with hospital outbreaks (Willems et al., 2005; Willems and van Schaik, 2009). Although E. faecalis CC2 and CC9 strains has been detected outside hospitals in farm animals and environment (Freitas et al., 2011; Novais et al., 2013), to our knowledge has not been identified in wild animals. On the other hand, E. faecium CC17 was recovered from seagulls in Portugal (Radhouani et al., 2010c).
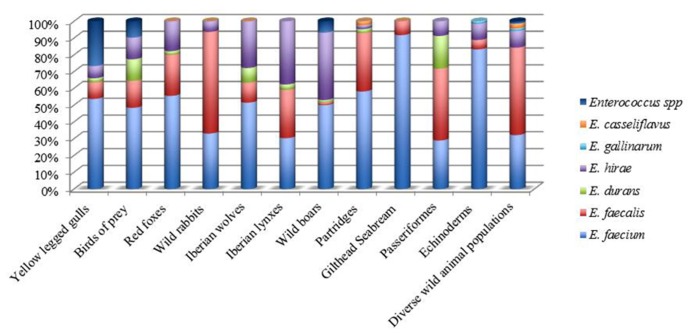
Recent comparisons of available genome sequences support the concept of a hospital-associated clade that is genetically distinct from most commensal isolates from animals and humans (Willems and van Schaik, 2009; van Schaik and Willems, 2010; Werner et al., 2011; Galloway-Pena et al., 2012). In Portugal, concerning wild animals, E. faecium were found to be the main species in seagulls (Radhouani et al., 2011b); birds of prey (Radhouani et al., 2012b), partridges (Silva et al., 2011b), Iberian wolf (Gonçalves et al., 2013a), red foxes (Radhouani et al., 2013), wild boars (Poeta et al., 2007a), gilthead seabream (Barros et al., 2011), and Echinoderms (Marinho et al., 2013), while E. faecalis was dominant in Passeriformes (Santos et al., 2013) and wild rabbits (Silva et al., 2010). On the other hand, E. hirae was the predominant specie isolated from Iberian lynx (Gonçalves et al., 2013b,c; Figure 2).
FIGURE 2.
Distribution of Enterococcus species in wild animals from Portugal.
Along with seagulls and birds of prey in Portugal, migratory Canada geese report showed also the occurrence of E. faecium and E. faecalis species. Furthermore, different reports found that all enterococci isolates detected in wild mammals (Mallon et al., 2002) and also in glaucous gulls (Drobni et al., 2009) were E. faecium species. Another study performed in Brazil, showed the occurrence of E. faecalis in non-human primate (capuchin monkeys and common marmoset) fecal samples (Xavier et al., 2010). In American bison fecal samples, E. casseliflavus was the predominant species recovered with 62.4%, followed by E. faecalis (16%; Anderson et al., 2008).
With the arrival of next-generation sequencing (Mardis, 2008), it was quickly possible to investigate hundreds of strains to get a better knowledge at the whole-genome level the evolutionary processes acting in populations (Liti et al., 2009; MacLean et al., 2009; Schacherer et al., 2009), opening the era of “population genomics” (Tenaillon et al., 2010). Certainly further studies using new high throughput technologies are mandatory to completely understand the evolution of predominant clones and species in different hosts and environments (Santagati et al., 2012).
DIVERSITY AND COMPLEXITY OF BACTERIAL NICHES
VIRULENCE FACTORS
Large-scale epidemiological studies provide insight into the diversity and complexity of E. coli and enterococci niches (Tenaillon et al., 2010; Arias and Murray, 2012). Thus, genome plasticity has contributed to the emergence of new virulence traits: some clusters of genes or genomic islands, including pathogenicity islands (PAIs) should be discovered only in a subset of strains and favored in some specific environments. Furthermore, several alternative combinations of genes could promote similar adaptations to a given environment (Tenaillon et al., 2010). Gene gain and loss make fundamental contributions to new habitat adaptation and the emergence of new lineages (Dagan and Martin, 2007). Strains related to hospital infections were found to have significantly larger overall average genome size than strains from non-hospitalized humans or animals groups (Lebreton et al., 2013). The prevalence of virulence factors genes is variable among commensal populations. On a global scale, the human microbiota is characterized by a higher prevalence of virulence genes than the microbiota of other organisms (Skurnik et al., 2008). In animals, the presence of virulence genes increases with body mass, which reveals the gut complexity of larger animal (Skurnik et al., 2008). Thus, virulence factors and their change in prevalence among hosts may reflect some local adaptation to commensal habitats rather than virulence per se (Tenaillon et al., 2010).
INTRA-SPECIES INTERACTIONS
Interactions between community members are required for community development and maintenance; and can also drive some diversification. Many genes that are carried by mobile elements code for traits that are expressed outside of the cell. Such traits are involved in bacterial sociality, such as the production of public goods, which benefit a cell’s neighbors, or the production of bacteriocins, which harm a cell’s neighbors. To out-compete other clones, the production of colicins can represent a useful strategy in a structured environment (Chao and Levin, 1981). Colicins are the most expansively studied bacteriocins produced by E. coli. This could permit unadapted strains to colonize the gut and, hence, allow numerous clones to coexist in the long term. It may also promote diversification in a clone, as some strains may try to benefit from the production of the colicin but avoid paying the associated cost. However, the secretion of bacteriocins can be defined as a “spiteful” behavior in which are costly for the producer and cause harm to other members of the population (Rankin et al., 2011).
Horizontal gene transfer is a key step in the evolution of bacterial pathogens. Chromosomal structures such as PAIs have been shown to extensively contribute to the evolution of bacterial pathogens by providing dynamic changes of the bacterial genome composition leading to a bacterial evolution in quantum leaps. Different mechanisms have been proposed for transfer of PAIs across the species border including phage transfer, mobilization by conjugative transposons and plasmids (Schubert et al., 2009). The fact that these microorganisms are less virulent than others is not reassuring, since the acquirement of virulence genes by bacteria is possible (Leclercq, 1997). Further research is required to characterize molecular and cellular interactions between the host and enterococci which lead to intra-species genetic transfer and virulence factors in enterococci species (Giridhara Upadhyaya et al., 2009).
ANTIMICROBIAL RESISTANCE
Antimicrobial resistance is a worldwide problem in human and veterinary medicine. Commonly, it is usual that the major risk factor for the increase of this situation is an extensive use of antimicrobials that leads to the dissemination of resistant bacteria and resistance genes in animals and humans (van den Bogaard and Stobberingh, 2000). The appearance of multiresistant bacteria of human and veterinary origin is probably accompanied by co-contamination of the environment apparently leading to a great health concern (Grobbel et al., 2007). As in the hospital environment, the agricultural use of antimicrobials agents selects for antibiotic resistance. These antimicrobial drugs from both hospital and agricultural sources can persist in soil and aquatic environments, and the selective pressure imposed by these compounds may affect the treatment of human diseases (Thiele-Bruhn, 2003; Segura et al., 2009). In addition, the prophylactic use of antibiotics in fish farms has led to a rise in the number of resistant bacteria (Cabello, 2006). However, the use of antibiotics by humans is not the only selective pressure for antibiotic resistance in natural microbial communities: compounds and conditions that occur in these communities may provide additional selection pressures. In fact, most antimicrobial agents are produced by strains of fungi and bacteria that occur naturally in all environments, including soil (Martin and Liras, 1989).
Bacteria may also acquire resistance determinants without direct exposure to an antimicrobial through horizontally mobile elements including conjugative plasmids, integrons, and transposons (Middleton and Ambrose, 2005). These mobile elements can simply transfer antimicrobial resistance genes from one bacterium to another (Coque et al., 2008). Antimicrobial agents exert a selective pressure not only on pathogenic, but also on commensal bacteria of the intestinal tract of humans and animals (van den Bogaard and Stobberingh, 2000). In a broader view, increasing evidence suggests that components such as integrons and their gene cassettes played significant roles in genome evolution and fluidity within the bacterial kingdom (Duriez et al., 2001; Davies and Davies, 2010).
In E. coli, the phylogenetic group A strains (Mammeri et al., 2009) and some group D strains (Deschamps et al., 2009) are especially permissive to the development of resistance to third-generation cephalosporins. Conversely, phylogenetic group B2 strains are less resistant than the remaining strains (Picard and Goullet, 1989; Johnson et al., 1991, 1994), regardless of the molecular mechanism implicated in the acquisition of resistance, and have a lower prevalence of integrons in commensal E. coli strains from both human (Skurnik et al., 2005) and animal hosts (Skurnik et al., 2006). This could reveal the relative decrease of phylogenetic group B2 strains in domesticated animals in which antimicrobials are used considerably (Tenaillon et al., 2010).
The class of beta-lactam antibiotics is among the most important groups of antimicrobial agents in human and veterinary medicine. Besides the first widely used antimicrobial substance penicillin, other members of this family have gained a similar importance over the last decades, namely the first- to fourth- generation cephalosporins and the beta-lactamase-inhibitors. All beta-lactams interfere with the final stage of peptidoglycan synthesis through acting on penicillin-binding proteins, thereby preventing the bacterial cell wall from forming. The most common resistance mechanism of Enterobacteriaceae spp. against beta-lactams is the inactivation of the antibiotic by breaking up the nitrogen-carbonyl bond in the beta-lactam ring (Walsh, 2003). The emergence and wide dissemination of ESBLs among clinical E. coli isolates in hospitals, has caused a major concern in several countries, being frequently implicated in human infections. These infections have a great impact on public health due to an increased incidence of treatment failure and severity of disease. ESBLs mainly include TEM, SHV, and CTX-M enzymes. Among them, the highest number of variants described in the last years corresponds to the CTX-M family (Naas et al., 2007). The presence of CTX-M enzymes render E. coli resistant to a variety of beta-lactams, and are transferred via plasmids that can also include resistance genes to several unrelated classes of antimicrobial agents (Canton et al., 2012).
Carbapenems such as imipenem or meropenem possess the most consistent activity against ESBL-producing Enterobacteriaceae strains. Both antibiotics are considered the agents of choice in the treatment of infections due to ESBL-producing organisms (Nordmann et al., 2011) However, Enterobacteriaceae-producing carbapenemases have rapidly emerged and disseminated worldwide, including in the wild (Fischer et al., 2013). The carbapenemases, such as the New Delhi metallo-beta-lactamase 1 (NDM-1) hydrolyze all β-lactam antibiotics, including carbapenems, and their high potential for rapid, wide dissemination constitutes a major clinical and public health threat (Nordmann et al., 2009; Nordmann et al., 2011).
The increase in the occurrence of nosocomial infections caused by enterococci in particular E. faecium, is at least partly due to the wide variety of intrinsic and acquired resistances to glycopeptides and aminoglycosides, among others, posing a challenge to therapeutic options. Moreover, infections caused by other Enterococcus species (E. faecalis, E. durans, E. mundtii, E. avium, E. raffinosus, E. gallinarum, and E. casseliflavus) occasionally occur and warrant attention (Murray, 1990; Prakash et al., 2005). Enterococci are expert in acquiring and transferring elements that confer resistance to antimicrobials and they are also known to be intrinsically resistant to numerous antimicrobials. As a result, therapeutic alternatives for treatment of enterococcal infections are increasingly limited (Murray, 1990). Evolution of enterococci toward resistance to multiple antimicrobials is also a major cause of concern.
The acquisition of vancomycin resistance by enterococci has seriously affected the treatment and infection control of these organisms. Different types of glycopeptide resistance and their biochemical mechanisms have been described in enterococci: acquired type (vanA, vanB, vanD, vanE, vanG, and vanL); and low-level intrinsic type (vanC, associated with the E. gallinarum, E. casseliflavus, and E. flavescens species). VanA-type resistance, which was the first to be elucidated and which is the most common, is characterized by high levels of resistance to glycopeptides, vancomycin, and teicoplanin and is mediated by transposon Tn1546 or closely related elements that are chromosomally or plasmid located (Arthur et al., 1996). The development of newer antimicrobial drugs, such as linezolid and daptomycin with activity against many VRE strains (Lee et al., 2007). has improved this situation; however resistance to these agents has already been described (Miller et al., 2013; Patel et al., 2013).
WILDLIFE AS RESERVOIRS OF ANTIBIOTIC RESISTANCE
An important difficulty in evaluating the causal relationship between antimicrobial use and resistance is the confounding influence of geography: the co-localization of resistant bacterial species with antimicrobial use does not essentially involve causation and could represent the presence of environmental conditions and factors that have independently contributed to the incidence of resistance (Singer et al., 2006).
The collection of all antimicrobial resistance genes and their precursors in pathogenic and non-pathogenic bacteria and also in antimicrobial producing-organisms is referred as the antimicrobial resistome, a concept that has been advanced to serve as a framework for understanding the ecology of resistance on a global scale (Wright, 2010).
Usually, wildlife is not exposed to clinical antimicrobial agents but can acquire antimicrobial-resistant bacteria through contact with humans, animals and the environment, where water polluted with feces appears to be the most significant vector of contamination. The incidence of commensal and pathogenic bacteria in fecal contaminations can be expected to be a connection between the environment and settings with regular or even constant antimicrobial pressure (aquaculture, livestock farming, human, and veterinary clinical settings), resulting in a constant release of antimicrobial-resistant human and animal bacteria into the environment through wastewater or manure (Martinez, 2009). Additionally, the detection of antimicrobial-resistant bacteria in aquatic environments affected by human and animal wastewater and soil provides evidence for this hypothesis (Kummerer and Henninger, 2003). In this context the common use of antimicrobials in aquaculture is also of utmost importance due to possible direct influences on wild animals (Smith, 2008). As intestinal bacteria like E. coli and enterococci can be easily disseminated in different ecosystems through water, they are intensively used as indicator species for fecal pollution (Guenther et al., 2011). In this sense, it is essential to interpret the evolutionary and ecological forces that influence in the population structure of the commensal strains to fully understand the antimicrobial resistance and virulence of pathogenic strains. Certainly, the selective pressures in the habitats of commensal strains may coincidentally promote the emergence of antimicrobial resistance and virulence factors, rendering commensal strains reservoirs of virulent and resistant strains (Tenaillon et al., 2010).
Despite the commensal character of E. coli and enterococci, they are commonly involved in animal and human infections that implicate the use of antimicrobials, which increases public health preoccupations to the list of implications that arise from the spread of ESBL-E. coli and VRE into wildlife. Furthermore, the increasing frequency of community-acquired ESBL-E. coli and VRE infections and the occurrence in livestock farming has been observed recently, suggesting a successful transmission as well as persistence of ESBL-E. coli and VRE strains outside hospital settings. An additional parallel global phenomenon is the spread of ESBL-E. coli and VRE into the environment beyond human and domesticated animal populations, and this appears to be directly induced by antimicrobial practice (Guenther et al., 2011). This might be a significant cause of the community-onset of ESBL-E. coli and VRE infections but can result (i) in an involvement of wildlife in ESBL-E. coli and VRE spread and transmission into fragile environmental niches, (ii) in subsequent colonization of wild animal populations which can turn into an infectious source or even a reservoir of ESBL-E. coli and VRE, (iii) in new putative infection cycles between wildlife, domesticated animals and humans, and (iv) in difficulties of wildlife medical treatment (Guenther et al., 2011).
Monitoring the prevalence of resistance in indicator bacteria such as fecal E. coli and enterococci in different populations (animals, patients and healthy humans) makes it possible to compare the prevalence of resistance and to detect transfer of resistant bacteria or resistance genes from animals to humans and vice versa (Martel et al., 2001). Just recently, there has been increasing interest in resistant bacteria and resistance genes isolated from wild animals (Allen et al., 2011).
The degree of colonization varies a lot between different animal species (Gordon and Cowling, 2003). Therefore, ESBL-E. coli and VRE prevalence is clearly influenced by sampling schemes, by geographic regions, by host spectrum of these bacteria and by the degree of synanthropic behavior shown by host species (Allen et al., 2010). It is important also to take in consideration the limitations that occur interpreting these results. For instance, ESBL-E. coli prevalence in different Portuguese geographical areas ranged from 0.5% in birds of the remote Azores islands in the Atlantic Ocean (Silva et al., 2011a) to 32% for birds of the Portugal’s Northern Portuguese coast (Simões et al., 2010). A lower prevalence of ESBL-E. coli was also observed (0.8%) in glaucous-winged gulls of Kamchatka peninsula in Russia (Hernandez et al., 2010). These findings suggest that wild animals living in urban areas are more susceptible to carry ESBL-E. coli than those living in remote areas.
Due to their diversity in ecological niches and their ease in picking up human and environmental bacteria, wild birds might act as mirrors of human activities. Within the heterogeneous class of birds, two groups seem to be in the focus of ESBL-E. coli and VRE carriage in wildlife: birds of prey (Costa et al., 2006; Pinto et al., 2010; Radhouani et al., 2010a,b; Silva et al., 2011a) and waterfowl/water related species (Poeta et al., 2008; Bonnedahl et al., 2009; Dolejska et al., 2009; Guenther et al., 2010a; Hernandez et al., 2010; Literak et al., 2010a; Radhouani et al., 2010c; Simões et al., 2010; Garmyn et al., 2011). However, recent studies in Passeriformes have also described a significant incidence of VRE (Silva et al., 2011a, 2012; Oravcova et al., 2013a,b).
Although wild birds, such as birds of prey, have only rare contact with antimicrobial agents, in disagreement with the existence of direct selective pressure, they can be contaminated or colonized by resistant bacteria. Water contact and acquisition via food seem to be major aspects of transmission of resistant bacteria of human or veterinary origin to wild animals (Cole et al., 2005). In the other hand, wild birds such as seagulls are often opportunistic marine feeders along the shoreline or offshore, but also eat the food sources provided by humans, especially garbage. Migrating birds that travel long distance seem to act as transporters, or as reservoirs, of resistant bacteria and may consequently have a significant epidemiological role in the dissemination of resistance, as well as being mirrors of the spectrum of pathogenic microorganisms present in humans (Radhouani et al., 2010c; Silva et al., 2011a). Reports on marine fish showed the presence of ESBL-E. coli (Sousa et al., 2011) and VRE (Barros et al., 2012) in gilthead seabream, indicating a dissemination of ESBL-E. coli and VRE into the Atlantic ocean. Moreover, it has previously been demonstrated that seagulls shared strains with isolates cultured from wastewater treatment plants and landfills (Nelson et al., 2008). This highlights the possibility of bacterial exchange between human sewage and birds.
Another important host of these bacteria appears to be in wild rodents. Although these animals have previous been in the focus of research on ESBL in wildlife in different continents (Gilliver et al., 1999; Kozak et al., 2009; Guenther et al., 2010b; Literak et al., 2010b; Allen et al., 2011), they have only been detected in urban rats (Guenther et al., 2010a; Ho et al., 2011). On the other hand, VRE have been earlier described in wild rodents (Mallon et al., 2002). This synantropic species can easily pickup human waste and frequently interacts with human feces in the sewage system in urban environments and can therefore easily acquire multiresistant bacteria. Remarkably, wild boars have also been described as hosts of these bacteria in Europe, which might expose their omnivorous feeding behavior (Poeta et al., 2007b, 2009; Literak et al., 2010b). Recent studies revealed the presence of ESBL-producing E. coli (Gonçalves et al., 2012a,b) and VRE isolates (Gonçalves et al., 2011) in Iberian wolf and/or Iberian lynx. The incidence of ESBL-E. coli (Radhouani et al., 2012a) and VRE (Radhouani et al., 2011a) in red foxes may be due their diet as these wild animals usually hunt wild rabbits, small rodents and birds. It is important to point out that some studies reported the presence of antimicrobial-resistant isolates in wild rabbits (Figueiredo et al., 2009; Silva et al., 2010) and wild rodents (Kozak et al., 2009; Guenther et al., 2010b). Foxes are on top of the food chain, perhaps accumulating multiresistant bacteria from their prey (Grobbel et al., 2012). All these evidences may contribute in the acquisition and spread of antimicrobial-resistant bacteria even in the absence of direct antimicrobial pressure.
These wild animals act as reservoirs of resistance genes and they could spread resistant bacteria throughout the wild environment. These researchers address the issue of antimicrobial-resistant microorganism proliferation in the environment and the related potential human health and environmental impact.
The level of resistant bacteria detected in wild animals seems to relate well with the degree of association with human activity (Skurnik et al., 2006). In fact, human density, natural preservation state, livestock or the reserve of an area may be significant criteria for the proliferation of antimicrobial-resistant bacteria (Allen et al., 2010). Nevertheless, several studies report the occurrence of multidrug-resistant bacteria in remote places or preservation areas therefore underlining the complexity of the spread of antimicrobial resistance in wild animals. These discoveries propose, on one hand, an influence of migratory behavior of wild birds into remote areas, or on the other hand the omnipresence of human influence in various ecological niches of the planet via human feces (Guenther et al., 2011). Different reports showed that areas with high livestock and human density and an assumable frequent interaction of wildlife with human influenced habitats of any kind (livestock farms, landfills, sewage systems, or wastewater treatment facilities) result in a higher risk for wildlife to acquire antimicrobial-resistant bacteria (Allen et al., 2010).
CONCLUSION
The strength of trillions upon trillions of microorganisms, combined with the ancient force of evolution by constant, insistent variation, will inevitably overpower the drugs. Their spectrum is selected to involve pathogenic bacteria and antimicrobials constantly select naturally resistant bacteria (American Academy of Microbiology, 2009). As bacteria quickly evolve to acquire resistance to the available antimicrobials, it is a constant race for scientists to develop effective strategies to combat infection and to reveal new therapeutic targets (Davies and Davies, 2010).
Moreover, antimicrobial resistance evolving and spreading among bacterial pathogens is a public health problem of increasing magnitude. Since the beginning of the antimicrobial era, the selective pressure caused by the use of antimicrobials in clinical, veterinary, husbandry, and agricultural practices is considered the major factor responsible for the occurrence and spread of antimicrobial-resistant bacteria. The evolution of antimicrobial resistance in bacteria is related to the evolution of antimicrobial production. Though, resistance has also been discovered in the absence of antimicrobial exposure, as in bacteria from wildlife, raising an interest about the mechanisms of emergence and persistence of resistant strains under similar conditions, and the implications for resistance control strategies (Pallecchi et al., 2008). Monitoring antimicrobial resistance in wildlife from remote areas could also be a useful tool to evaluate the impact of anthropic pressure (Thaller et al., 2010).
Singer et al. (2006) support that in ecological studies of antimicrobial resistance, there has possibly been too much focus on resistant organisms and not enough on resistance genes. Due to the capability of bacteria to transfer resistance genes, even among distantly related bacteria, analyses of antimicrobial resistance emergence, dissemination and persistence might be better conducted at the gene level (Singer et al., 2006).
Until now, genomics-based investigation into E. coli and enterococci has focused on the identification of genes directly implicated in virulence. However, the fundamental physiology and response mechanisms to environmental conditions of E. coli and enterococci remained relatively poorly understood. This is a serious oversight because during infection the microbial fitness is an important reason in the success of any microbial pathogen. Further genome-wide reports aiming to define genes that are important during infection and colonization or exposure to antimicrobials will deliver significant data on relevant aspects of E. coli and enterococcal biology. This knowledge can consequently be useful for the development of novel treatment approaches to combat microbial infections.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Hajer Radhouani and Susana Correia were supported by grants of FCT (Fundação para a Ciência e a Tecnologia) and POPH/FSE (Programa Operacional Potencial Humano/Fundo Social Europeu) with references SFRH/BD/60846/2009 and SFRH/BD/75160/2010, respectively.
REFERENCES
- Adams J. (2004). Microbial evolution in laboratory environments. Res. Microbiol. 155 311–318 10.1016/j.resmic.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Allen H. K., Donato J., Wang H. H., Cloud-Hansen K. A., Davies J., Handelsman J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8 251–259 10.1038/nrmicro2312 [DOI] [PubMed] [Google Scholar]
- Allen S. E., Boerlin P., Janecko N., Lumsden J. S., Barker I. K., Pearl D. L., et al. (2011). Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in southern Ontario, Canada. Appl. Environ. Microbiol. 77 882–888 10.1128/AEM.01111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Academy of Microbiology. (2009). Antibiotic Resistance: An Ecological Perspective on an Old Problem. Washington, DC: American Academy of Microbiology [Google Scholar]
- Anderson J. F., Parrish T. D., Akhtar M., Zurek L., Hirt H. (2008). Antibiotic resistance of enterococci in American bison (Bison bison) from a nature preserve compared to that of enterococci in pastured cattle. Appl. Environ. Microbiol. 74 1726–1730 10.1128/AEM.02164-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C. A., Murray B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10 266–278 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Reynolds P., Courvalin P. (1996). Glycopeptide resistance in enterococci. Trends Microbiol. 4 401–407 10.1016/0966-842X(96)10063-9 [DOI] [PubMed] [Google Scholar]
- Baldy-Chudzik K., Mackiewicz P., Stosik M. (2008). Phylogenetic background, virulence gene profiles, and genomic diversity in commensal Escherichia coli isolated from ten mammal species living in one zoo. Vet. Microbiol. 131 173–184 10.1016/j.vetmic.2008.02.019 [DOI] [PubMed] [Google Scholar]
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., Schneider D., et al. (2009). Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461 1243–1247 10.1038/nature08480 [DOI] [PubMed] [Google Scholar]
- Barros J., Andrade M., Radhouani H., Lopez M., Igrejas G., Poeta P., et al. (2012). Detection of vanA-containing Enterococcus species in faecal microbiota of gilthead seabream (Sparus aurata). Microbes Environ. 27 509–511 10.1264/jsme2.ME11346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros J., Igrejas G., Andrade M., Radhouani H., Lopez M., Torres C., et al. (2011). Gilthead seabream (Sparus aurata) carrying antibiotic resistant enterococci. A potential bioindicator of marine contamination? Mar. Pollut. Bull. 62 1245–1248 10.1016/j.marpolbul.2011.03.021 [DOI] [PubMed] [Google Scholar]
- Bingen E., Picard B., Brahimi N., Mathy S., Desjardins P., Elion J., et al. (1998). Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177 642–650 10.1086/514217 [DOI] [PubMed] [Google Scholar]
- Bonnedahl J., Drobni M., Gauthier-Clerc M., Hernandez J., Granholm S., Kayser Y., et al. (2009). Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS ONE 4:e5958 10.1371/journal.pone.0005958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botti V., Navillod F. V., Domenis L., Orusa R., Pepe E., Robetto S., et al. (2013). Salmonella spp. and antibiotic-resistant strains in wild mammals and birds in north-western Italy from 2002 to 2010. Vet. Ital. 49 195–202 [PubMed] [Google Scholar]
- Cabello F. C. (2006). Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8 1137–1144 10.1111/j.1462-2920.2006.01054.x [DOI] [PubMed] [Google Scholar]
- Canton R., Akova M., Carmeli Y., Giske C. G., Glupczynski Y., Gniadkowski M., et al. (2012). Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18 413–431 10.1111/j.1469-0691.2012.03821.x [DOI] [PubMed] [Google Scholar]
- Carlos C., Pires M. M., Stoppe N. C., Hachich E. M., Sato M. I., Gomes T. A., et al. (2010). Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 10:161 10.1186/1471-2180-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L., Levin B. R. (1981). Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. U.S.A. 78 6324–6328 10.1073/pnas.78.10.6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth C., Schaberg D. (1990). The epidemiology of enterococci. Eur. J. Clin. Microbiol. Infect. Dis. 9 80–89 [DOI] [PubMed] [Google Scholar]
- Ciftci A., Findik A., Ica T., Bas B., Onuk E. E., Gungordu S. (2009). Slime production and antibiotic resistance of Enterococcus faecalis isolated from arthritis in chickens. J. Vet. Med. Sci. 71 849–853 10.1292/jvms.71.849 [DOI] [PubMed] [Google Scholar]
- Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66 4555–4558 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D., Drum D. J., Stalknecht D. E., White D. G., Lee M. D., Ayers S., et al. (2005). Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 11 935–938 10.3201/eid1106.040717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T. M., Lewis N. E., Palsson B. O. (2011). Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 7 509 10.1038/msb.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque T. M., Novais A., Carattoli A., Poirel L., Pitout J., Peixe L., et al. (2008). Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14 195–200 10.3201/eid1402.070350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa D., Poeta P., Saenz Y., Vinue L., Rojo-Bezares B., Jouini A., et al. (2006). Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 58 1311–1312 10.1093/jac/dkl415 [DOI] [PubMed] [Google Scholar]
- Dagan T., Martin W. (2007). Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc. Natl. Acad. Sci. U.S.A. 104 870–875 10.1073/pnas.0606318104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Davies D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74 417–433 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps C., Clermont O., Hipeaux M. C., Arlet G., Denamur E., Branger C. (2009). Multiple acquisitions of CTX-M plasmids in the rare D2 genotype of Escherichia coli provide evidence for convergent evolution. Microbiology 155 1656–1668 10.1099/mic.0.023234-0 [DOI] [PubMed] [Google Scholar]
- Devriese L. A., Pot B. (1995). “The genus Enterococcus,” in The Lactic Acid Bacteria: The Genera of Lactic Acid Bacteria Vol. 2 eds Wood B. J. B., Holzapfel W. H. (London: Blackie Academic; ) 327–367 10.1007/978-1-4615-5817-0_10 [DOI] [Google Scholar]
- Dolejska M., Bierosova B., Kohoutova L., Literak I., Cizek A. (2009). Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106 1941–1950 10.1111/j.1365-2672.2009.04155.x [DOI] [PubMed] [Google Scholar]
- Dolejska M., Cizek A., Literak I. (2007). High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from Black-headed Gulls in the Czech Republic. J. Appl. Microbiol. 103 11–19 10.1111/j.1365-2672.2006.03241.x [DOI] [PubMed] [Google Scholar]
- Drobni M., Bonnedahl J., Hernandez J., Haemig P., Olsen B. (2009). Vancomycin-resistant enterococci, Point Barrow, Alaska, USA. Emerg. Infect. Dis. 15 838–839 10.3201/eid1505.081219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duriez P., Clermont O., Bonacorsi S., Bingen E., Chaventre A., Elion J., et al. (2001). Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147 1671–1676 [DOI] [PubMed] [Google Scholar]
- Eaton T. J., Gasson M. J. (2001). Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67 1628–1635 10.1128/AEM.67.4.1628-1635.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Paramo P., Le Menac’h A., Le Gall T., Amorin C., Gouriou S., Picard B., et al. (2006). Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 8 1975–1984 10.1111/j.1462-2920.2006.01077.x [DOI] [PubMed] [Google Scholar]
- Figueiredo N., Radhouani H., Goncalves A., Rodrigues J., Carvalho C., Igrejas G., et al. (2009). Genetic characterization of vancomycin-resistant enterococci isolates from wild rabbits. J. Basic Microbiol. 49 491–494 10.1002/jobm.200800387 [DOI] [PubMed] [Google Scholar]
- Fischer J., Schmoger S., Jahn S., Helmuth R., Guerra B. (2013). NDM-1 carbapenemase-producing Salmonella enterica subsp. enterica serovar Corvallis isolated from a wild bird in Germany. J. Antimicrob. Chemother. 68 2954–2956 10.1093/jac/dkt260 [DOI] [PubMed] [Google Scholar]
- Fisher K., Phillips C. (2009). The ecology, epidemiology and virulence of Enterococcus. Microbiology 155 1749–1757 10.1099/mic.0.026385-0 [DOI] [PubMed] [Google Scholar]
- Franz C. M., Muscholl-Silberhorn A. B., Yousif N. M., Vancanneyt M., Swings J., Holzapfel W. H. (2001). Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 67 4385–4389 10.1128/AEM.67.9.4385-4389.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas A. R., Coque T. M., Novais C., Hammerum A. M., Lester C. H., Zervos M. J., et al. (2011). Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. J. Clin. Microbiol. 49 925–931 10.1128/JCM.01750-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakuya F. M., Kyule M. N., Gathura P. B., Kariuki S. (2001). Antimicrobial susceptibility and plasmids from Escherichia coli isolated from rats. East Afr. Med. J. 78 518–522 10.4314/eamj.v78i10.8960 [DOI] [PubMed] [Google Scholar]
- Galloway-Pena J., Roh J. H., Latorre M., Qin X., Murray B. E. (2012). Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS ONE 7:e30187 10.1371/journal.pone.0030187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmyn A., Haesebrouck F., Hellebuyck T., Smet A., Pasmans F., Butaye P., et al. (2011). Presence of extended-spectrum beta-lactamase-producing Escherichia coli in wild geese. J. Antimicrob. Chemother. 66 1643–1644 10.1093/jac/dkr148 [DOI] [PubMed] [Google Scholar]
- Gaukler S. M., Linz G. M., Sherwood J. S., Dyer N. W., Bleier W. J., Wannemuehler Y. M., et al. (2009). Escherichia coli, Salmonella, and Mycobacterium avium subsp. paratuberculosis in wild European starlings at a Kansas cattle feedlot. Avian Dis. 53 544–551 10.1637/8920-050809-Reg.1 [DOI] [PubMed] [Google Scholar]
- Gilliver M. A., Bennett M., Begon M., Hazel S. M., Hart C. A. (1999). Antibiotic resistance found in wild rodents. Nature 401 233–234 10.1038/45724 [DOI] [PubMed] [Google Scholar]
- Gin A. S., Zhanel G. G. (1996). Vancomycin-resistant enterococci. Ann. Pharmacother. 30 615–624 [DOI] [PubMed] [Google Scholar]
- Giridhara Upadhyaya P. M., Ravikumar K. L., Umapathy B. L. (2009). Review of virulence factors of Enterococcus: an emerging nosocomial pathogen. Indian J. Med. Microbiol. 27 301–305 10.4103/0255-0857.55437 [DOI] [PubMed] [Google Scholar]
- Gonçalves A., Igrejas G., Radhouani H., Correia S., Pacheco R., Santos T., et al. (2013a). Antimicrobial resistance in faecal enterococci and Escherichia coli isolates recovered from Iberian wolf. Lett. Appl. Microbiol. 56 268–274 10.1111/lam.12044 [DOI] [PubMed] [Google Scholar]
- Gonçalves A., Igrejas G., Radhouani H., Santos T., Monteiro R., Pacheco R., et al. (2013b). Detection of antibiotic resistant enterococci and Escherichia coli in free range Iberian Lynx (Lynx pardinus). Sci. Total Environ. 456–457. 115–119 10.1016/j.scitotenv.2013.03.073 [DOI] [PubMed] [Google Scholar]
- Gonçalves A., Igrejas G., Radhouani H., Santos T., Monteiro R., Marinho C., et al. (2013c). Iberian Lynx (Lynx pardinus) from the captive breeding program as reservoir of antimicrobial resistant enterococci and Escherichia coli isolates. J. Integr. OMICS 3 2 10.5584/jiomics.v3i2.142 [DOI] [Google Scholar]
- Gonçalves A., Igrejas G., Radhouani H., Estepa V., Alcaide E., Zorrilla I., et al. (2012a). Detection of extended-spectrum beta-lactamase-producing Escherichia coli isolates in faecal samples of Iberian lynx. Lett. Appl. Microbiol. 54 73–77 10.1111/j.1472-765X.2011.03173.x [DOI] [PubMed] [Google Scholar]
- Gonçalves A., Igrejas G., Radhouani H., Estepa V., Pacheco R., Monteiro R., et al. (2012b). Iberian wolf as a reservoir of extended-spectrum beta-lactamase-producing Escherichia coli of the TEM, SHV, and CTX-M groups. Microb. Drug Resist. 18 215–219 10.1089/mdr.2011.0145 [DOI] [PubMed] [Google Scholar]
- Gonçalves A., Igrejas G., Radhouani H., Lopez M., Guerra A., Petrucci-Fonseca F., et al. (2011). Detection of vancomycin-resistant enterococci from faecal samples of Iberian wolf and Iberian lynx, including Enterococcus faecium strains of CC17 and the new singleton ST573. Sci. Total Environ. 410–411 266–268 10.1016/j.scitotenv.2011.09.074 [DOI] [PubMed] [Google Scholar]
- Gordon D. M., Cowling A. (2003). The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149 3575–3586 10.1099/mic.0.26486-0 [DOI] [PubMed] [Google Scholar]
- Goullet P., Picard B. (1989). Comparative electrophoretic polymorphism of esterases and other enzymes in Escherichia coli. J. Gen. Microbiol. 135 135–143 [DOI] [PubMed] [Google Scholar]
- Grobbel M., Lubke-Becker A., Alesik E., Schwarz S., Wallmann J., Werckenthin C., et al. (2007). Antimicrobial susceptibility of Escherichia coli from swine, horses, dogs and cats as determined in the BfT-GermVet monitoring program 2004-2006. Berl. Munch. Tierarztl. Wochenschr. 120 391–401 [PubMed] [Google Scholar]
- Grobbel M., Wittstatt U., Guenther S., Ewers C. (2012). Urban red foxes (Vulpes vulpes) and their possible role in the transmission of 3rd generation beta-lactam resistant E. coli to the environment,” in Proceedings of 3rd ASM Conference on Antimicrobial resistance in Zoonotic Bacteria and Foodborne Pathogens in Animals, Humans, and the Environment Aix-en-Provence; France [Google Scholar]
- Guenther S., Ewers C., Wieler L. H. (2011). Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2:246 10.3389/fmicb.2011.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther S., Grobbel M., Beutlich J., Guerra B., Ulrich R. G., Wieler L. H., et al. (2010a). Detection of pandemic B2-O25-ST131 Escherichia coli harbouring the CTX-M-9 extended-spectrum beta-lactamase type in a feral urban brown rat (Rattus norvegicus). J. Antimicrob. Chemother. 65 582–584 10.1093/jac/dkp496 [DOI] [PubMed] [Google Scholar]
- Guenther S., Grobbel M., Heidemanns K., Schlegel M., Ulrich R. G., Ewers C., et al. (2010b). First insights into antimicrobial resistance among faecal Escherichia coli isolates from small wild mammals in rural areas. Sci. Total Environ. 408 3519–3522 10.1016/j.scitotenv.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Hernandez J., Bonnedahl J., Eliasson I., Wallensten A., Comstedt P., Johansson A., et al. (2010). Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring blaCTX-M-15, found in Glaucous-winged gull at remote Commander Islands, Russia. Environ. Microbiol. Rep. 2 329–332 10.1111/j.1758-2229.2010.00142.x [DOI] [PubMed] [Google Scholar]
- Herzer P. J., Inouye S., Inouye M., Whittam T. S. (1990). Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172 6175–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P. L., Chow K. H., Lai E. L., Lo W. U., Yeung M. K., Chan J., et al. (2011). Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to “critically important” antibiotics among food animals in Hong Kong, 2008-10. J. Antimicrob. Chemother. 66 765–768 10.1093/jac/dkq539 [DOI] [PubMed] [Google Scholar]
- Ishihara S., Bitner J. J., Farley G. H., Gillock E. T. (2013). Vancomycin-resistant gram-positive cocci isolated from the saliva of wild songbirds. Curr. Microbiol. 66 337–343 10.1007/s00284-012-0278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaureguy F., Landraud L., Passet V., Diancourt L., Frapy E., Guigon G., et al. (2008). Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560 10.1186/1471-2164-9-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jett B. D., Huycke M. M., Gilmore M. S. (1994). Virulence of enterococci. Clin. Microbiol. Rev. 7 462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R. (2002). Escherichia Coli: Virulence Mechanisms of a Versatile Pathogen. London: Academic Press [Google Scholar]
- Johnson J. R., Delavari P., Kuskowski M., Stell A. L. (2001). Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183 78–88 10.1086/317656 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Goullet P., Picard B., Moseley S. L., Roberts P. L., Stamm W. E. (1991). Association of carboxylesterase B electrophoretic pattern with presence and expression of urovirulence factor determinants and antimicrobial resistance among strains of Escherichia coli that cause urosepsis. Infect. Immun. 59 2311–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Orskov I., Orskov F., Goullet P., Picard B., Moseley S. L., et al. (1994). O, K, and H antigen predict virulence factors, carboxylesterase B pattern, antimicrobial resistance, and host compromise among Escherichia coli strains causing urosepsis. J. Infect. Dis. 169 119–126 10.1093/infdis/169.1.119 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Stell A. L. (2000). Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181 261–272 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- Kozak G. K., Boerlin P., Janecko N., Reid-Smith R. J., Jardine C. (2009). Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75 559–566 10.1128/AEM.01821-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerer K., Henninger A. (2003). Promoting resistance by the emission of antibiotics from hospitals and households into effluent. Clin. Microbiol. Infect. 9 1203–1214 10.1111/j.1469-0691.2003.00739.x [DOI] [PubMed] [Google Scholar]
- Lebreton F., Van Schaik W., Mcguire A. M., Godfrey P., Griggs A., Mazumdar V., et al. (2013). Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 4 pii: e00534-13 10.1128/mBio.00534-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq R. (1997). Enterococci acquire new kinds of resistance. Clin. Infect. Dis. 24(Suppl. 1) S80–S84 10.1093/clinids/24.Supplement_1.S80 [DOI] [PubMed] [Google Scholar]
- Lee D. K., Kim Y., Park K. S., Yang J. W., Kim K., Ha N. J. (2007). Antimicrobial activity of mupirocin, daptomycin, linezolid, quinupristin/dalfopristin and tigecycline against vancomycin-resistant enterococci (VRE) from clinical isolates in Korea (1998 and 2005). J. Biochem. Mol. Biol. 40 881–887 10.5483/BMBRep.2007.40.6.881 [DOI] [PubMed] [Google Scholar]
- Lescat M., Hoede C., Clermont O., Garry L., Darlu P., Tuffery P., et al. (2009). aes, the gene encoding the esterase B in Escherichia coli, is a powerful phylogenetic marker of the species. BMC Microbiol. 9:273 10.1186/1471-2180-9-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescat M., Clermont O., Woerther P. L., Glodt J., Dion S., Skurnik D., et al. (2013). Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependant population structure. Environ. Microbiol. Rep. 5 49–57 10.1111/j.1758-2229.2012.00374.x [DOI] [PubMed] [Google Scholar]
- Lillehaug A., Bergsjo B., Schau J., Bruheim T., Vikoren T., Handeland K. (2005). Campylobacter spp., Salmonella spp., verocytotoxic Escherichia coli, and antibiotic resistance in indicator organisms in wild cervids. Acta Vet. Scand. 46 23–32 10.1186/1751-0147-46-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Literak I., Dolejska M., Janoszowska D., Hrusakova J., Meissner W., Rzyska H., et al. (2010a). Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 76 8126–8134 10.1128/AEM.01446-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Literak I., Dolejska M., Radimersky T., Klimes J., Friedman M., Aarestrup F. M., et al. (2010b). Antimicrobial-resistant faecal Escherichia coli in wild mammals in central Europe: multiresistant Escherichia coli producing extended-spectrum beta-lactamases in wild boars. J. Appl. Microbiol. 108 1702–1711 10.1111/j.1365-2672.2009.04572.x [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., James S. A., et al. (2009). Population genomics of domestic and wild yeasts. Nature 458 337–341 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado E., Coque T. M., Canton R., Sousa J. C., Peixe L. (2008). Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 62 296–302 10.1093/jac/dkn179 [DOI] [PubMed] [Google Scholar]
- MacLean D., Jones J. D., Studholme D. J. (2009). Application of “next-generation” sequencing technologies to microbial genetics. Nat. Rev. Microbiol. 7 287–296 10.1038/nrmicro2088 [DOI] [PubMed] [Google Scholar]
- Mallon D. J. P., Corkill J. E., Hazel S. M., Wilson J. S., French N. P., Bennett M., et al. (2002). Excretion of vancomycin-resistant enterococci by wild mammals. Emerg. Infect. Dis. 8 636–638 10.3201/eid0806.010247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammeri H., Galleni M., Nordmann P. (2009). Role of the Ser-287-Asn replacement in the hydrolysis spectrum extension of AmpC beta-lactamases in Escherichia coli. Antimicrob. Agents Chemother. 53 323–326 10.1128/AAC.00608-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E. R. (2008). The impact of next-generation sequencing technology on genetics. Trends Genet. 24 133–141 10.1016/j.tig.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Marinho C., Silva N., Pombo S., Santos T., Monteiro R., Goncalves A., et al. (2013). Echinoderms from Azores islands: an unexpected source of antibiotic resistant Enterococcus spp. and Escherichia coli isolates. Mar. Pollut. Bull. 69 122–127 10.1016/j.marpolbul.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Martel J. L., Tardy F., Sanders P., Boisseau J. (2001). New trends in regulatory rules and surveillance of antimicrobial resistance in bacteria of animal origin. Vet. Res. 32 381–392 10.1051/vetres:2001131 [DOI] [PubMed] [Google Scholar]
- Martin M. F., Liras P. (1989). Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43 173–206 10.1146/annurev.mi.43.100189.001133 [DOI] [PubMed] [Google Scholar]
- Martinez J. L. (2009). The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 276 2521–2530 10.1098/rspb.2009.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton J. H., Ambrose A. (2005). Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J. Wildl. Dis. 41 334–341 10.7589/0090-3558-41.2.334 [DOI] [PubMed] [Google Scholar]
- Miller C., Kong J., Tran T. T., Arias C. A., Saxer G., Shamoo Y. (2013). Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob. Agents Chemother. 57 5373–5383 10.1128/AAC.01473-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E. (1990). The life and times of the Enterococcus. Clin. Microbiol. Rev. 3 46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Oxacelay C., Nordmann P. (2007). Identification of CTX-M-type extended-spectrum-beta-lactamase genes using real-time PCR and pyrosequencing. Antimicrob. Agents Chemother. 51 223–230 10.1128/AAC.00611-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Gonzalez N., Casas-Diaz E., Porrero C. M., Mateos A., Dominguez L., Lavin S., et al. (2013). Food-borne zoonotic pathogens and antimicrobial resistance of indicator bacteria in urban wild boars in Barcelona, Spain. Vet. Microbiol. 167 686–689 10.1016/j.vetmic.2013.07.037 [DOI] [PubMed] [Google Scholar]
- Nelson M., Jones S. H., Edwards C., Ellis J. C. (2008). Characterization of Escherichia coli populations from gulls, landfill trash, and wastewater using ribotyping. Dis. Aquat. Organ. 81 53–63 10.3354/dao01937 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Cuzon G., Naas T. (2009). The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9 228–236 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Naas T., Poirel L. (2011). Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17 1791–1798 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais C., Freitas A. R., Silveira E., Antunes P., Silva R., Coque T. M., et al. (2013). Spread of multidrug-resistant Enterococcus to animals and humans: an underestimated role for the pig farm environment. J. Antimicrob. Chemother. 68 2746–2754 10.1093/jac/dkt289 [DOI] [PubMed] [Google Scholar]
- Oravcova V., Ghosh A., Zurek L., Bardon J., Guenther S., Cizek A., et al. (2013a). Vancomycin-resistant enterococci in rooks (Corvus frugilegus) wintering throughout Europe. Environ. Microbiol. 15 548–556 10.1111/1462-2920.12002 [DOI] [PubMed] [Google Scholar]
- Oravcova V., Zurek L., Townsend A., Clark A. B., Ellis J. C., Cizek A., et al. (2013b). American crows as carriers of vancomycin-resistant enterococci with vanA gene. Environ. Microbiol. 10.1111/1462-2920.12213 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Pallecchi L., Bartoloni A., Paradisi F., Rossolini G. M. (2008). Antibiotic resistance in the absence of antimicrobial use: mechanisms and implications. Expert Rev. Anti Infect. Ther. 6 725–732 10.1586/14787210.6.5.725 [DOI] [PubMed] [Google Scholar]
- Patel S. N., Memari N., Shahinas D., Toye B., Jamieson F. B., Farrell D. J. (2013). Linezolid resistance in Enterococcus faecium isolated in Ontario, Canada. Diagn. Microbiol. Infect. Dis. 77 350–353 10.1016/j.diagmicrobio.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Pesapane R., Ponder M., Alexander K. A. (2013). Tracking pathogen transmission at the human-wildlife interface: banded mongoose and Escherichia coli. Ecohealth 10 115–128 10.1007/s10393-013-0838-2 [DOI] [PubMed] [Google Scholar]
- Picard B., Garcia J. S., Gouriou S., Duriez P., Brahimi N., Bingen E., et al. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67 546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B., Goullet P. (1989). Correlation between electrophoretic types B1 and B2 of carboxylesterase B and sex of patients in Escherichia coli urinary tract infections. Epidemiol. Infect. 103 97–103 10.1017/S0950268800030399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L., Radhouani H., Coelho C., Martins da Costa P., Simoes R., Brandao R. M., et al. (2010). Genetic detection of extended-spectrum beta-lactamase-containing Escherichia coli isolates from birds of prey from Serra da Estrela Natural Reserve in Portugal. Appl. Environ. Microbiol. 76 4118–4120 10.1128/AEM.02761-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeta P., Costa D., Igrejas G., Rodrigues J., Torres C. (2007a). Phenotypic and genotypic characterization of antimicrobial resistance in faecal enterococci from wild boars (Sus scrofa). Vet. Microbiol. 125 368–374 10.1016/j.vetmic.2007.06.003 [DOI] [PubMed] [Google Scholar]
- Poeta P., Costa D., Igrejas G., Rojo-Bezares B., Saenz Y., Zarazaga M., et al. (2007b). Characterization of vanA-containing Enterococcus faecium isolates carrying Tn5397-like and Tn916/Tn1545-like transposons in wild boars (Sus Scrofa). Microb. Drug Resist. 13 151–156 10.1089/mdr.2007.759 [DOI] [PubMed] [Google Scholar]
- Poeta P., Radhouani H., Igrejas G., Goncalves A., Carvalho C., Rodrigues J., et al. (2008). Seagulls of the Berlengas natural reserve of Portugal as carriers of fecal Escherichia coli harboring CTX-M and TEM extended-spectrum beta-lactamases. Appl. Environ. Microbiol. 74 7439–7441 10.1128/AEM.00949-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeta P., Radhouani H., Pinto L., Martinho A., Rego V., Rodrigues R., et al. (2009). Wild boars as reservoirs of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli of different phylogenetic groups. J. Basic Microbiol. 49 584–588 10.1002/jobm.200900066 [DOI] [PubMed] [Google Scholar]
- Porrero M. C., Mentaberre G., Sanchez S., Fernandez-Llario P., Gomez-Barrero S., Navarro-Gonzalez N., et al. (2013). Methicillin resistant Staphylococcus aureus (MRSA) carriage in different free-living wild animal species in Spain. Vet. J. 198 127–130 10.1016/j.tvjl.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Prakash V. P., Rao S. R., Parija S. C. (2005). Emergence of unusual species of enterococci causing infections, South India. BMC Infect. Dis. 5:14 10.1186/1471-2334-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo G. M., Karaolis D. K., Lan R., Reeves P. R. (1997). Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65 2685–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhouani H., Igrejas G., Carvalho C., Pinto L., Goncalves A., Lopez M., et al. (2011a). Clonal lineages, antibiotic resistance and virulence factors in vancomycin-resistant enterococci isolated from fecal samples of red foxes (Vulpes vulpes). J. Wildl. Dis. 47 769–773 10.7589/0090-3558-47.3.769 [DOI] [PubMed] [Google Scholar]
- Radhouani H., Igrejas G., Pinto L., Goncalves A., Coelho C., Rodrigues J., et al. (2011b). Molecular characterization of antibiotic resistance in enterococci recovered from seagulls (Larus cachinnans) representing an environmental health problem. J. Environ. Monit. 13 2227–2233 10.1039/c0em00682c [DOI] [PubMed] [Google Scholar]
- Radhouani H., Poeta P., Pinto L., Miranda J., Coelho C., Carvalho C., et al. (2010c). Proteomic characterization of vanA-containing Enterococcus recovered from Seagulls at the Berlengas Natural Reserve, W Portugal. Proteome Sci. 8 48 10.1186/1477-5956-8-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhouani H., Igrejas G., Gonçalves A., Estepa V., Sargo R., Torres C., et al. (2012a). Molecular characterization of extended-spectrum-beta-lactamase-producing Escherichia coli isolates from red foxes in Portugal. Arch. Microbiol. 195 141–144 10.1007/s00203-012-0853-7 [DOI] [PubMed] [Google Scholar]
- Radhouani H., Poeta P., Goncalves A., Pacheco R., Sargo R., Igrejas G. (2012b). Wild birds as biological indicators of environmental pollution: antimicrobial resistance patterns of Escherichia coli and enterococci isolated from common buzzards (Buteo buteo). J. Med. Microbiol. 61 837–843 10.1099/jmm.0.038364-0 [DOI] [PubMed] [Google Scholar]
- Radhouani H., Igrejas G., Goncalves A., Pacheco R., Monteiro R., Sargo R., et al. (2013). Antimicrobial resistance and virulence genes in Escherichia coli and enterococci from red foxes (Vulpes vulpes). Anaerobe 23 82–86 10.1016/j.anaerobe.2013.06.013 [DOI] [PubMed] [Google Scholar]
- Radhouani H., Pinto L., Coelho C., Goncalves A., Sargo R., Torres C., et al. (2010a). Detection of Escherichia coli harbouring extended-spectrum β-lactamases of the CTX-M classes in faecal samples of common buzzards (Buteo buteo). J. Antimicrob. Chemother. 65 171–173 10.1093/jac/dkp403 [DOI] [PubMed] [Google Scholar]
- Radhouani H., Pinto L., Coelho C., Sargo R., Araujo C., Lopez M., et al. (2010b). MLST and a genetic study of antibiotic resistance and virulence factors in vanA-containing Enterococcus from buzzards (Buteo buteo). Lett. Appl. Microbiol. 50 537–541 10.1111/j.1472-765X.2010.02807.x [DOI] [PubMed] [Google Scholar]
- Radhouani H., Poeta P., Igrejas G., Goncalves A., Vinue L., Torres C. (2009). Antimicrobial resistance and phylogenetic groups in isolates of Escherichia coli from seagulls at the Berlengas nature reserve. Vet. Rec. 165 138–142 10.1136/vr.165.5.138 [DOI] [PubMed] [Google Scholar]
- Rankin D. J., Rocha E. P., Brown S. P. (2011). What traits are carried on mobile genetic elements, and why? Heredity (Edinb.) 106 1–10 10.1038/hdy.2010.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A., Buckley M. (2011). Microbial Evolution. Washington, DC: American Academy of Microbiology [Google Scholar]
- Ruiz-Garbajosa P., Bonten M. J., Robinson D. A., Top J., Nallapareddy S. R., Torres C., et al. (2006). Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44 2220–2228 10.1128/JCM.02596-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati M., Campanile F., Stefani S. (2012). Genomic diversification of enterococci in hosts: the role of the mobilome. Front. Microbiol. 3:95 10.3389/fmicb.2012.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T., Silva N., Igrejas G., Rodrigues P., Micael J., Rodrigues T., et al. (2013). Dissemination of antibiotic resistant Enterococcus spp. and Escherichia coli from wild birds of Azores Archipelago. Anaerobe 24 25–31 10.1016/j.anaerobe.2013.09.004 [DOI] [PubMed] [Google Scholar]
- Sato G., Oka C., Asagi M., Ishiguro N. (1978). Detection of conjugative R plasmids conferring chloramphenicol resistance in Escherichia coli isolated from domestic and feral pigeons and crows. Zentralbl. Bakteriol. Orig. A 241 407–417 [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L. (2009). Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458 342–345 10.1038/nature07670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierack P., Romer A., Jores J., Kaspar H., Guenther S., Filter M., et al. (2009). Isolation and characterization of intestinal Escherichia coli clones from wild boars in Germany. Appl. Environ. Microbiol. 75 695–702 10.1128/AEM.01650-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S., Darlu P., Clermont O., Wieser A., Magistro G., Hoffmann C., et al. (2009). Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog. 5:e1000257 10.1371/journal.ppat.1000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura P. A., Francois M., Gagnon C., Sauve S. (2009). Review of the occurrence of anti-infectives in contaminated wastewaters and natural and drinking waters. Environ. Health Perspect. 117 675–684 10.1289/ehp.11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. (1986). Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Whittam T. S. (1987). “Genetic structure and variation in natural populations of Escherichia coli,” in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology eds Neidhardt F. C., Ingraham J. L., Low K. B., Magasanik B., Schaechter M., Umbarger H. E. (Washington: American Society for Microbiology) 1625–1648 [Google Scholar]
- Sherley M., Gordon D. M., Collignon P. J. (2000). Variations in antibiotic resistance profile in Enterobacteriaceae isolated from wild Australian mammals. Environ. Microbiol. 2 620–631 10.1046/j.1462-2920.2000.00145.x [DOI] [PubMed] [Google Scholar]
- Silva N., Igrejas G., Figueiredo N., Goncalves A., Radhouani H., Rodrigues J., et al. (2010). Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci. Total Environ. 408 4871–4876 10.1016/j.scitotenv.2010.06.046 [DOI] [PubMed] [Google Scholar]
- Silva N., Igrejas G., Rodrigues P., Rodrigues T., Goncalves A., Felgar A. C., et al. (2011a). Molecular characterization of vancomycin-resistant enterococci and extended-spectrum beta-lactamase-containing Escherichia coli isolates in wild birds from the Azores Archipelago. Avian Pathol. 40 473–479 10.1080/03079457.2011.599061 [DOI] [PubMed] [Google Scholar]
- Silva N., Igrejas G., Vaz J., Araujo C., Cardoso L., Rodrigues J., et al. (2011b). Virulence factors in enterococci from partridges (Alectoris rufa) representing a food safety problem. Foodborne Pathog. Dis. 8 831–833 10.1089/fpd.2010.0781 [DOI] [PubMed] [Google Scholar]
- Silva N., Igrejas G., Felgar A., Goncalves A., Pacheco R., Poeta P. (2012). Molecular characterization of vanA-containing Enterococcus from migratory birds: song thrush (Turdus philomelos). Braz. J. Microbiol. 43 1026–1029 10.1590/S1517-83822012000300026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões R. R., Poirel L., Da Costa P. M., Nordmann P. (2010). Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli. Emerg. Infect. Dis. 16 110–112 10.3201/eid1601.090896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer R. S., Ward M. P., Maldonado G. (2006). Can landscape ecology untangle the complexity of antibiotic resistance? Nat. Rev. Microbiol. 4 943–952 10.1038/nrmicro1553 [DOI] [PubMed] [Google Scholar]
- Skurnik D., Bonnet D., Bernede-Bauduin C., Michel R., Guette C., Becker J. M., et al. (2008). Characteristics of human intestinal Escherichia coli with changing environments. Environ. Microbiol. 10 2132–2137 10.1111/j.1462-2920.2008.01636.x [DOI] [PubMed] [Google Scholar]
- Skurnik D., Le Menac’h A., Zurakowski D., Mazel D., Courvalin P., Denamur E., et al. (2005). Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob. Agents Chemother. 49 3062–3065 10.1128/AAC.49.7.3062-3065.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik D., Ruimy R., Andremont A., Amorin C., Rouquet P., Picard B., et al. (2006). Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57 1215–1219 10.1093/jac/dkl122 [DOI] [PubMed] [Google Scholar]
- Smith P. (2008). Antimicrobial resistance in aquaculture. Rev. Sci. Tech. 27 243–264 [PubMed] [Google Scholar]
- Sorum H., Sunde M. (2001). Resistance to antibiotics in the normal flora of animals. Vet. Res. 32 227–241 10.1051/vetres:2001121 [DOI] [PubMed] [Google Scholar]
- Sousa M., Torres C., Barros J., Somalo S., Igrejas G., Poeta P. (2011). Gilthead seabream (Sparus aurata) as carriers of SHV-12 and TEM-52 extended-spectrum beta-lactamases-containing Escherichia coli isolates. Foodborne Pathog. Dis. 8 1139–1141 10.1089/fpd.2011.0866 [DOI] [PubMed] [Google Scholar]
- Souza V., Rocha M., Valera A., Eguiarte L. E. (1999). Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65 3373–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Skurnik D., Picard B., Denamur E. (2010). The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8 207–217 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- Thaller M. C., Migliore L., Marquez C., Tapia W., Cedeno V., Rossolini G. M., et al. (2010). Tracking acquired antibiotic resistance in commensal bacteria of Galapagos land iguanas: no man, no resistance. PLoS ONE 5:e8989 10.1371/journal.pone.0008989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele-Bruhn S. (2003). Pharmaceutical antibiotic compounds in soils – a review. J. Plant Nutr. Soil Sci. 166 145–167 10.1002/jpln.200390023 [DOI] [Google Scholar]
- Touchon M., Hoede C., Tenaillon O., Barbe V., Baeriswyl S., Bidet P., et al. (2009). Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogaard A. E., Stobberingh E. E. (2000). Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 14 327–335 10.1016/S0924-8579(00)00145-X [DOI] [PubMed] [Google Scholar]
- van Schaik W., Willems R. J. (2010). Genome-based insights into the evolution of enterococci. Clin. Microbiol. Infect. 16 527–532 10.1111/j.1469-0691.2010.03201.x [DOI] [PubMed] [Google Scholar]
- Wagner A. (2008). Neutralism and selectionism: a network-based reconciliation. Nat. Rev. Genet. 9 965–974 10.1038/nrg2473 [DOI] [PubMed] [Google Scholar]
- Walk S. T., Alm E. W., Calhoun L. M., Mladonicky J. M., Whittam T. S. (2007). Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9 2274–2288 10.1111/j.1462-2920.2007.01341.x [DOI] [PubMed] [Google Scholar]
- Walsh C. (2003). Antibiotics: Actions, Origins, Resistance. Washington: ASM Press [Google Scholar]
- Weng P. L., Ramli R., Shamsudin M. N., Cheah Y. K., Hamat R. A. (2013). High genetic diversity of Enterococcus faecium and Enterococcus faecalis clinical isolates by pulsed-field gel electrophoresis and multilocus sequence typing from a hospital in Malaysia. Biomed Res. Int. 2013 938937 10.1155/2013/938937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner G., Fleige C., Geringer U., van Schaik W., Klare I., Witte W. (2011). IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect. Dis. 11:80 10.1186/1471-2334-11-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems R. J., Top J., van Santen M., Robinson D. A., Coque T. M., Baquero F., et al. (2005). Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11 821–828 10.3201/1106.041204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems R. J, van Schaik W. (2009). Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 4 1125–1135 10.2217/fmb.09.82 [DOI] [PubMed] [Google Scholar]
- Wright G. D. (2010). Antibiotic resistance in the environment: a link to the clinic? Curr.Opin. Microbiol. 13 589–594 10.1016/j.mib.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Xavier D. B., Rosa A. H., Teixeira D. S., Tomaz C. A. B., Titze-de-Almeida R. (2010). Absence of intestinal colonization by vancomycin-resistant enterococci in nonhuman primates. Pesq. Vet. Bras. 30 491–496 10.1590/S0100-736X2010000600004 [DOI] [Google Scholar]