Abstract
Objective
Preimplantation genetic diagnosis (PGD) is an assisted reproductive technique for couples carrying genetic risks. Charcot-Marie-Tooth (CMT) disease is the most common hereditary neuropathy, with a prevalence rate of 1/2,500. In this study, we report on our experience with PGD cycles performed for CMT types 1A and 2F.
Methods
Before clinical PGD, we assessed the amplification rate and allele drop-out (ADO) rate of multiplex fluorescent polymerase chain reaction (PCR) followed by fragment analysis or sequencing using single lymphocytes. We performed six cycles of PGD for CMT1A and one cycle for CMT2F.
Results
Two duplex and two triplex protocols were developed according to the available markers for each CMT1A couple. Depending on the PCR protocols, the amplification rates and ADO rates ranged from 90.0% to 98.3% and 0.0% to 11.1%, respectively. For CMT2F, the amplification rates and ADO rates were 93.3% and 4.8%, respectively. In case of CMT1A, 60 out of 63 embryos (95.2%) were diagnosed and 13 out of 21 unaffected embryos were transferred in five cycles. Two pregnancies were achieved and three babies were delivered without any complications. In the case of CMT2F, a total of eight embryos were analyzed and diagnosed. Seven embryos were diagnosed as unaffected and four embryos were transferred, resulting in a twin pregnancy. Two healthy babies were delivered.
Conclusion
This is the first report of successful pregnancy and delivery after specific PGD for CMT disease in Korea. Our PGD procedure could provide healthy babies to couples with a high risk of transmitting genetic diseases.
Keywords: Charcot-Marie-Tooth disease, Preimplantation genetic diagnosis, Allele drop-out, Polymorphic marker
Introduction
Preimplantation genetic diagnosis (PGD) for numerical and structural chromosomal abnormalities and single gene disorders has been successfully used as an alternative to prenatal diagnosis of inherited diseases. PGD can be used to differentiate between unaffected and affected embryos before embryo transfer in human IVF-ET programs. Therefore, PGPGD can enable avoiding the initiation of affected pregnancies [1,2].
Charcot-Marie-Tooth (CMT) disease is a sensorineural peripheral polyneuropathy. Affecting approximately 1 in 2,500 individuals, CMT disease is the most common inherited disorder of the peripheral nervous system. Autosomal dominant, autosomal recessive, and X-linked inherited modes have been recognized [3-5]. Autosomal dominant demyelinating CMT1 is a genetically heterogeneous disorder and can be caused by mutations in different genes. Cases of CMT1 are characterized clinically by severely reduced motor nerve conduction velocity (NCV), distal muscle atrophy, marked hand and foot deformity, and weakness. The majority of patients, 70% to 85% of cases, with CMT1A have a 1.5-Mb duplication in the p11-p12 region of chromosome 17, including the peripheral myelin protein 22 (PMP22) gene [5-8]. On the other hand, CMT2 is an axonal (non-demyelinating) peripheral neuropathy characterized by distal muscle weakness and atrophy, mild sensory loss, and normal or near-normal NCV. CMT2 is clinically similar to CMT1, although typically less severe. The subtypes of CMT2 are similar clinically and can be distinguished only by molecular genetic findings. Mutations in the small heat shock protein HSPB1 (HSP27) are a cause of CMT2F and distal hereditary motor neuropathy [6,9,10].
In this study, we describe the first successful pregnancies and births after PGD for CMT disease using multiplex fluorescent polymerase chain reaction (PCR) with linked polymorphic markers for CMT1A, or nested PCR followed by direct sequencing for CMT2F. Clinical application involved six cycles for four couples in the case of CMT1A and one cycle for a couple with CMT2F. Using these protocols, only unaffected normal embryos were transferred, resulting in three pregnancies and the delivery of five healthy babies without any complications.
Methods
1. Patient description
PGD was requested by four couples for CMT1A and one couple for CMT2F (Table 1). In couple 1, the CMT1A duplication was detected in the male partner, who had inherited the mutation from his affected father. In couple 2, the CMT1A duplication was detected in the female partner using PCR analysis with linked polymorphic markers located within the duplication. The patient had inherited the mutation from her affected mother. In couple 3, the CMT1A duplication was detected in the male partner. In couple 4, the CMT1 duplication was present in the female partner, who had inherited the mutation from her affected mother. In the couple with CMT2F, couple 5, the mutation in the heat shock 27 kD protein 1 (HSPB1, c.404C>T) was detected in the male partner, who had inherited the mutation from his affected mother. All of these couples opted for PGD to avoid an affected pregnancy. Due to the retrospective nature of this study, Institutional Review Board approval was not required.
Table 1.
Clinical characteristics of the five couples who underwent PGD for CMT1A and CMT2F
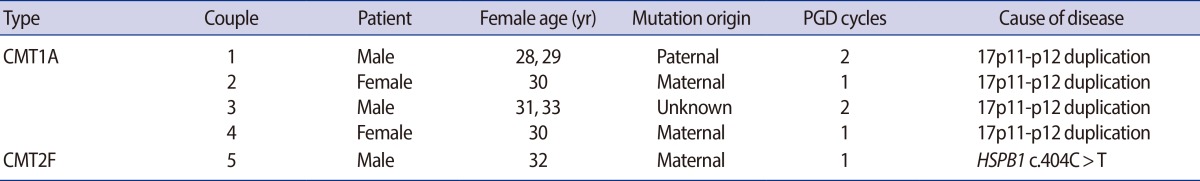
PGD, preimplantation genetic diagnosis; CMT1A, Charcot-Marie-Tooth disease type 1A; CMT2F, Charcot-Marie-Tooth disease type 2F.
2. Isolation and preparation of single lymphocytes
To determine the efficiency of single-cell PCR and the allele drop-out (ADO) rate, primers used for the linked polymorphic markers and detection of the causative mutation were tested using single lymphocytes (Table 2). Peripheral blood was collected in heparinized tubes from the male partner, female partner, and/or relatives, and lymphocytes were isolated from 10 mL of blood using Ficoll-Paque density gradient separation (Ficoll-Paque PLUS, Amersham Biosciences AB, Uppsala, Sweden) according to the manufacturer's protocol. The cell layer containing lymphocytes was removed and diluted with sterile phosphate-buffered saline (PBS) to adjust cell density for single cell isolation. Lymphocytes were handled with a mouth-controlled fine heat-polished glass micropipette. The lymphocytes were selected and retrieved individually under visual control through an inverted microscope. Single lymphocytes were loaded into 0.2 mL thin-wall PCR tubes containing 3.0 μL lysis buffer (200 mmol/L KOH and 50 mmol/L dithiothreitol). The samples were stored at-70℃ before analysis.
Table 2.
Sequences of oligonucleotide primers used for PCR
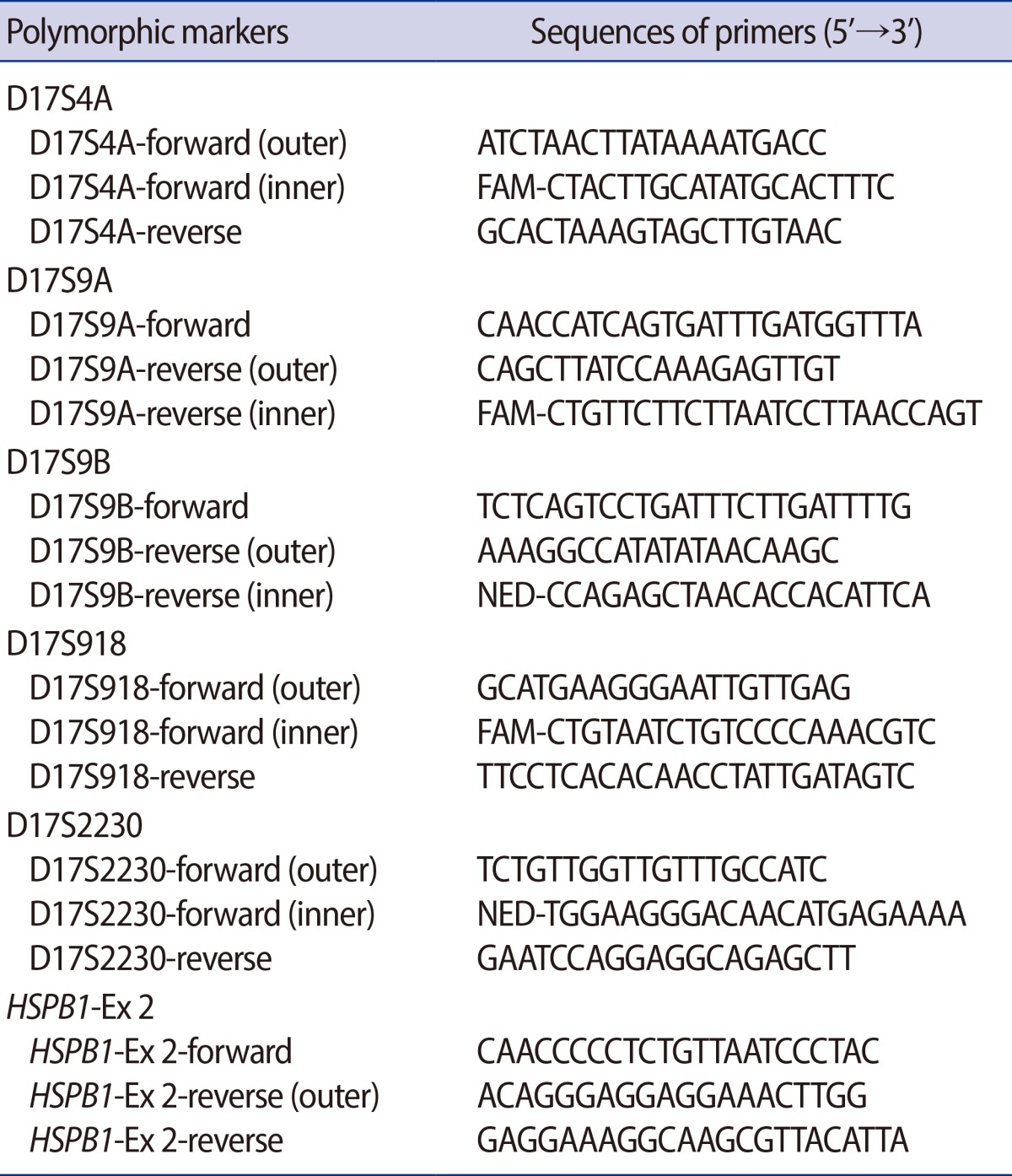
PCR, polymerase chain reaction; outer, primers used for the first 'outer' PCR reactions; inner, primers used for nested PCR reactions.
3. In vitro fertilization and embryo biopsy procedures
Ovarian stimulation was performed as previously described [11]. After ovarian stimulation, the follicles were aspirated and fertilized by ICSI. The fertilized zygotes were cultured separately in G-1 medium ver. 5 (Vitrolife Sweden AB, Kungsbacka, Sweden) for 3 days at 37℃ in an atmosphere of 6% CO2. On the third day of culture, the embryos were biopsied in Ca2+/Mg2+-free medium (Biopsy medium, Medicult, Jyllinge, Denmark). Acid Tyrode's solution (Medicult) was applied to create a small hole in the zona pellucida. Biopsy was performed by gentle aspiration using a polished micropipette. One or two blastomeres among six to nine cells were biopsied using a single pipette with an inner diameter of 30 μm. After the blastomere biopsy procedure, the embryos were repeatedly washed, transferred to G-2 medium ver. 5 (Vitrolife Sweden AB), and cultured at 37℃ with 6% CO2 in air. Each blastomere was washed twice with two drops of G-2 medium ver. 5 and transferred into sterile 0.2-mL PCR tubes containing 3.0 μL of alkaline lysis buffer. For each embryo biopsied, blank negative controls were prepared using the wash drops. The embryos were cultured under standard culture conditions until the diagnosis was accomplished. Embryos with normal genotypes were selected and transferred on the fourth or fifth day of culture. After a positive β-human chorionic gonadotropin test, clinical pregnancy was determined based on the presence of a fetal heartbeat and gestational sac.
4. Cell lysis and multiplex nested PCR procedure
The cells were lysed by incubation with alkaline lysis buffer at 65℃ for 10 minutes. The buffer was neutralized by the addition of 5 μL of neutralization buffer (900 mmol/L Tris-HCl, 300 mmol/L KCl, 200 mmol/L HCl) before proceeding to PCR. The PCR strategy consisted of initial multiplex PCR followed by multiplex nested fluorescent PCR specific for linked markers, or nested PCR followed by direct sequencing for mutated loci of the causative gene. After cell lysis and neutralization, 1.5 mmol/L MgCl2, 200 μmol/L of each dNTP (Roche Diagnostics GmbH, Mannheim, Germany), 1 IU Neotherm DNA Polymerase (GeneCraft Co., Munster, Germany) and 2 pmol of each outer primer were added to each tube in a total volume of 30 μL. The first round of PCR involved denaturation at 96℃ for 10 minutes to reduce ADO [12], followed by 25 cycles consisting of 96℃ for 30 seconds, 58℃ to 62℃ for 30 seconds, and 72℃ for 40 seconds with a final extension step of 10 minutes at 72℃ using a GeneAmp PCR System 2700 (Applied Biosystems, Foster City, CA, USA). For the second round of DNA amplification, 0.5 μL of the primary PCR product was added to another tube containing 2 μL of PCR buffer (50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 8.3), 1.5 mmol/L MgCl2, 200 μmol/L of each dNTP, 2 IU Neotherm DNA polymerase (GeneCraft Co.), and 1-5 pmol of each specific inner primer pair in a total volume of 20 μL, and the amplification was performed for 40 cycles as described above. To detect PCR products from conventional PCR, 3 μL of each PCR product was subjected to electrophoresis on a 2% agarose gel. The sequences of the oligonucleotide primers used for PCR are shown in Table 2.
5. Fragment analysis of the amplified products
One microliter of the PCR product was added to 9 μL of genetic analysis grade Hi-Di Formamide (Applied Biosystems) and 0.2 μL of GeneScan-500 LIZ Size Standard (Applied Biosystems). After boiling for 3 minutes at 96℃, the mixtures were capillary-electrophoresed in an ABI 3130 automatic genetic analyzer (Applied Biosystems). The results were analyzed by GeneScan Analysis software ver. 3.7 (Applied Biosystems).
6. Direct DNA sequencing analysis
For DNA sequence analyses, the PCR products were purified using a purification kit (Bioneer, Daejeon, Korea). The purified PCR products (40 ng) were sequenced by direct cycle sequencing using fluorescent-labeled dideoxy terminators (Big Dye Terminator Cycle Sequencing Ready Reaction Kit, Applied Biosystems) according to the manufacturer's protocol. The reaction conditions were 25 PCR cycles consisting of denaturation for 10 seconds at 96℃, annealing for 5 seconds at 50℃, and extension for 4 minutes at 60℃ using a GeneAmp PCR System 2700. The samples were cleared by ethanol precipitation to remove unincorporated dye terminator. The precipitated pellets were resuspended in 15 μL of Hi-Di Formamide (Applied Biosystems), denatured at 96℃ for 3 minutes, and run on an ABI Prism 3130 Avant automated genetic analyzer (Applied Biosystems). The sequences were compared with wild type controls using Seqscape Software (Applied Biosystems) for mutation analysis.
Results
1. Pre-clinical test using single lymphocytes
As a result of linkage analysis for the couples with CMT1A, the polymorphic markers D17S4A, D17S9A, D17S9B, D17S918, or D17S2230 were identified as fully-or semi-informative markers according to the genotypes of the couple (Table 3). Couples 2 and 3 were informative for two markers, whereas couples 1 and 4 had three informative markers. The efficiency and accuracy of the different PCR protocols was evaluated in pre-clinical tests on single lymphocytes collected from each couple. In couple 1, the amplification rates of the D17S4A, D17S918, and D17S2230 markers were 96.7%, 98.3%, and 98.3%, and the ADO rates were 2.0%, 1.7%, and 0.0%, respectively. In couple 2, the amplification rates of D17S4A and D17S2230 were 90.0% and 93.3%, and the ADO rates were 11.1% and 0.0%, respectively. In couples 3 and 4, PCR with polymorphic markers D17S9A, D17S9B, and D17S2230 (couple 4 only) resulted in amplification rates ranging from 97.6% to 100.0%, and the ADO rates ranged from 0.0% to 5.1% (Table 3). The lowest amplification rate and the highest ADO rate were obtained for D17S4A in couple 2.
Table 3.
Amp and ADO rate for the different PCR protocols according to the genotypes of the four couples who underwent PGD for CMT1A
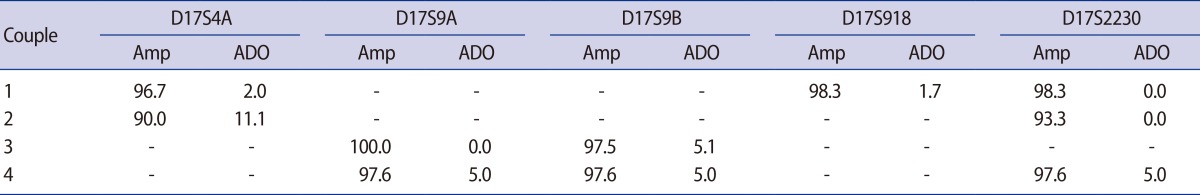
Values are presented as %.
Amp, amplification rate; ADO, allele drop-out rate; PCR, polymerase chain reaction; PGD, preimplantation genetic diagnosis; CMT1A, Charcot-Marie-Tooth disease type 1A.
In the case of CMT2F, PCR analysis of single lymphocytes resulted in an amplification rate of 93.3% (42/45), and none of the negative controls showed a positive band. ADO was detected in 2 out of 42 lymphocytes (4.8%).
2. Clinical PGD program for CMT disease
After pre-clinical testing, the clinical PGD program was applied for five couples (Table 4). In the clinical PGD program, a single blastomere was biopsied for embryos with fewer than six cells and two blastomeres were biopsied for embryos with six or more cells.
Table 4.
Clinical outcomes of seven PGD cycles for CMT1A and CMT2F
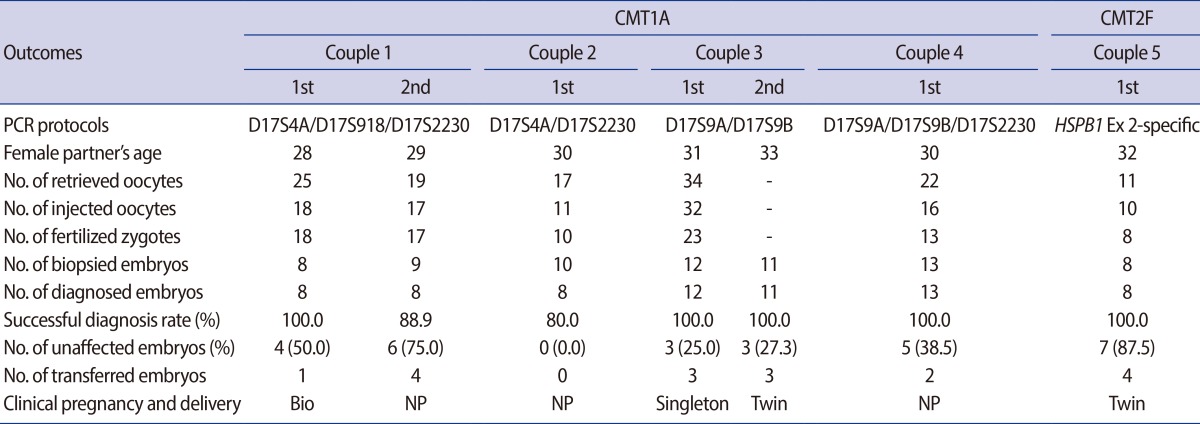
PGD, preimplantation genetic diagnosis; CMT1A, Charcot-Marie-Tooth disease type 1A; CMT2F, Charcot-Marie-Tooth disease type 2F; Bio, biochemical pregnancy; NP, not pregnant.
For patients with CMT1A, a total of 63 embryos were analyzed and 60 embryos (95.2%) were successfully diagnosed. Twenty-one (35.0%) out of 60 embryos were diagnosed as unaffected. Six cycles with biopsy resulted in five transfer cycles, in which 13 embryos were transferred (a mean of 2.6 per transferred cycle). In one cycle for couple 2, embryo transfer was not possible because no unaffected embryos were available. In couple 1, one unaffected healthy embryo was transferred into the mother's uterus and resulted in biochemical pregnancy in the first PGD cycle, and four unaffected embryos were transferred in the second PGD cycle. However, no pregnancy was achieved. In couple 3, two consecutive PGD cycles resulted in two pregnancies. In both pregnancies, PGD was confirmed by amniocentesis. This patient delivered a singleton baby and twin babies from the two PGD cycles. For couple 4, 13 embryos were biopsied and diagnosed. Two out of five unaffected embryos were transferred into the mother's uterus. However, pregnancy was not achieved.
In the case of couple 5 with CMT2F, eight embryos were biopsied and successfully diagnosed. Seven embryos were diagnosed as unaffected and four well-developed embryos were transferred into the mother's uterus. Pregnancy was achieved and twin babies were delivered without any complications.
Overall, 17 embryos were transferred in 6 cycles (a mean of 2.8 per transferred cycle). Three pregnancies were achieved and five healthy babies were delivered without any complications. The PGD results for CMT disease in our center represent a delivery rate of 50.0% per transferred cycle and 42.9% per started cycle.
Discussion
Multiplex (fluorescent) nested PCR methods followed by direct sequencing and/or fragment analysis with linked polymorphic markers provided reasonable reliability in single-cell analysis in our PGD center. These protocols have been successfully applied to many clinical PGD cases for couples at high risk of having children with single gene disorders [2,13-18].
In our pre-clinical tests for the polymorphic markers D17S4A, D17S9A, D17S9B, D17S918, and D17S2230, amplification rates ranged from 90.0% to 100.0% and ADO rates ranged from 0.0% to 11.1% (Table 3). The lowest amplification rate (90.0%) and the highest ADO rate (11.1%) were obtained for the D17S4A marker in the duplex nested PCR protocol with D17S4A-D17S2230 markers for couple 2. However, in couple 1, the amplification rate and ADO rate for the same D17S4A marker in the triplex nested PCR protocol using D17S4A-D17S918-D17S2230 markers were 96.7% and 2.0%, respectively. The amplification and ADO rates obtained in this triplex protocol are comparable to those obtained with duplex or other triplex PCR protocols. This result suggests differences according to the complexity of the primer sets and the quality of lymphocytes prepared from each patient. Although our data showed a lower PCR amplification rate and higher ADO rate for D17S4A than for the other markers used, the reliability and accuracy of the diagnosis were improved by combination with additional polymorphic markers and by biopsy of two blastomeres from the same embryo. Amplification failure or misdiagnosis due to ADO at one blastomere or one locus was avoided by extrapolating the results obtained from other blastomeres or other loci [13,18].
In this study, four different multiplex nested fluorescent PCR-based PGD protocols for CMT1A and a protocol of simplex nested PCR followed by direct sequencing for CMT2F are reported. All of these multiplex or simplex primer sets are suitable for clinical application and have been applied in seven clinical PGD cycles for five couples. In the case of CMT1A, 60 out of 63 embryos were successfully diagnosed. Three embryos (4.8%) resulted in no diagnosis for the following reasons: no amplification in two blastomeres (second PGD cycle of couple 1) and unclear and conflicting results (PGD cycle of couple 2). In total, 21 (35.0%) out of 60 embryos were diagnosed as unaffected. On the other hand, in the case of CMT2F, all eight embryos were biopsied and successfully diagnosed. Using these protocols, only healthy embryos were transferred, resulting in three pregnancies and the delivery of five healthy babies without any complications. To our knowledge, this is the first report of successful pregnancies after specific PGD for CMT diseases in Korea.
The multiplex PCR protocols with polymorphic markers used in this study can be used for PGD in other couples that suffer from the same CMT diseases, especially CMT1A duplication, although the usefulness for many polymorphic markers needs to be tested in each couple. Moreover, the application of new additional polymorphic markers promotes the development of other multiplex protocols, so that many other couples with CMT1A can be treated with these protocols. Our study provides further evidence that PGD is a reliable and effective clinical technique and a useful option for many couples at high risk of transmitting a genetic disease.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Grace J, El-Toukhy T, Scriven P, Ogilvie C, Pickering S, Lashwood A, et al. Three hundred and thirty cycles of preimplantation genetic diagnosis for serious genetic disease: clinical considerations affecting outcome. BJOG. 2006;113:1393–1401. doi: 10.1111/j.1471-0528.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee HS, Jun JH, Choi HW, Lim CK, Yoo HW, Koong MK, et al. Preimplantation genetic diagnosis for ornithine transcarbamylase deficiency by simultaneous analysis of duplex-nested PCR and fluorescence in situ hybridization: a case report. J Korean Med Sci. 2007;22:572–576. doi: 10.3346/jkms.2007.22.3.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth's disease. Clin Genet. 1974;6:98–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 4.Patel PI, Lupski JR. Charcot-Marie-Tooth disease: a new paradigm for the mechanism of inherited disease. Trends Genet. 1994;10:128–133. doi: 10.1016/0168-9525(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 5.Berciano J, Sevilla T, Casasnovas C, Sivera R, Vilchez JJ, Infante J, et al. Guidelines for molecular diagnosis of Charcot-Marie-Tooth disease. Neurologia. 2012;27:169–178. doi: 10.1016/j.nrl.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Braathen GJ. Genetic epidemiology of Charcot-Marie-Tooth disease. Acta Neurol Scand Suppl. 2012:iv–22. doi: 10.1111/ane.12013. [DOI] [PubMed] [Google Scholar]
- 7.Hoogendijk JE, Hensels GW, Zorn I, Valentijn L, Janssen EA, de Visser M, et al. The duplication in Charcot-Marie-Tooth disease type 1a spans at least 1,100 kb on chromosome 17p11.2. Hum Genet. 1991;88:215–218. doi: 10.1007/BF00206075. [DOI] [PubMed] [Google Scholar]
- 8.Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 9.Tang B, Liu X, Zhao G, Luo W, Xia K, Pan Q, et al. Mutation analysis of the small heat shock protein 27 gene in chinese patients with Charcot-Marie-Tooth disease. Arch Neurol. 2005;62:1201–1207. doi: 10.1001/archneur.62.8.1201. [DOI] [PubMed] [Google Scholar]
- 10.Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 11.Lim CK, Jun JH, Min DM, Lee HS, Kim JY, Koong MK, et al. Efficacy and clinical outcome of preimplantation genetic diagnosis using FISH for couples of reciprocal and Robertsonian translocations: the Korean experience. Prenat Diagn. 2004;24:556–561. doi: 10.1002/pd.923. [DOI] [PubMed] [Google Scholar]
- 12.Ray PF, Handyside AH. Increasing the denaturation temperature during the first cycles of amplification reduces allele dropout from single cells for preimplantation genetic diagnosis. Mol Hum Reprod. 1996;2:213–218. doi: 10.1093/molehr/2.3.213. [DOI] [PubMed] [Google Scholar]
- 13.Lee HS, Choi HW, Lim CK, Park SY, Kim JY, Koong MK, et al. Efficacy of duplex-nested PCR and fluorescent pcr in the preimplantation genetic diagnosis for duchenne muscular dystrophy. Korean J Fertil Steril. 2005;32:17–26. [Google Scholar]
- 14.Lee HS, Choi HW, Lim CK, Koong MK, Kang IS, Yoo HW, et al. Identification of a novel single nucleotide polymorphism of HADHA gene at a referred primer-binding site during pre-diagnostic tests for preimplantation genetic diagnosis. J Korean Med Sci. 2006;21:794–799. doi: 10.3346/jkms.2006.21.5.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sermon K. Current concepts in preimplantation genetic diagnosis (PGD): a molecular biologist's view. Hum Reprod Update. 2002;8:11–20. doi: 10.1093/humupd/8.1.11. [DOI] [PubMed] [Google Scholar]
- 16.De Vos A, Sermon K, Van de Velde H, Joris H, Vandervorst M, Lissens W, et al. Pregnancy after preimplantation genetic diagnosis for Charcot-Marie-Tooth disease type 1A. Mol Hum Reprod. 1998;4:978–984. doi: 10.1093/molehr/4.10.978. [DOI] [PubMed] [Google Scholar]
- 17.De Vos A, Sermon K, De Rijcke M, Goossens V, Henderix P, Van Ranst N, et al. Preimplantation genetic diagnosis for Charcot-Marie-Tooth disease type 1A. Mol Hum Reprod. 2003;9:429–435. doi: 10.1093/molehr/gag054. [DOI] [PubMed] [Google Scholar]
- 18.Lee HS, Kim MJ, Lim CK, Cho JW, Song IO, Kang IS. Multiple displacement amplification for preimplantation genetic diagnosis of fragile X syndrome. Genet Mol Res. 2011;10:2851–2859. doi: 10.4238/2011.November.17.3. [DOI] [PubMed] [Google Scholar]


