Abstract
The spontaneous incidence of chloramphenicol (Cam) resistant mutant bacteria is at least ten-fold higher in cultures of enterohemorrhagic E. coli O157:H7 strain EDL933 than in E. coli K-12. It is at least 100-fold higher in the dam (DNA adenine methyltransferase) derivative of EDL933, compared to the dam strain of E. coli K-12, thereby preventing the use of Cam resistance as a marker in gene replacement technology. Genome sequencing of Cam-resistant isolates of EDL933 and its dam derivatives showed that the marR (multiple antibiotic resistance) gene was mutated in every case but not in the Cam-sensitive parental strains. As expected from mutation in the marR gene, the Cam-resistant bacteria were also found to be resistant to tetracycline and nalidixic acid. The marR gene in strain EDL933 is annotated as a shorter open reading frame than that in E. coli K-12 but the longer marR+ open reading frame was more efficient at complementing the marR antibiotic-resistance phenotype of strain EDL933. Beta-lactamase-tolerant derivatives were present at frequencies 10–100 times greater in cultures of marR derivatives of strain EDL933 than the parent strain. Spontaneous mutation frequency to rifampicin, spectinomycin and streptomycin resistance was the same in E. coli O157:H7 and E. coli K-12 strains.
Keywords: Escherichia coli O157, multiple antibiotic resistance, mutation, genes, repressor protein, DNA
1. Introduction
Enterohemorrhagic Escherichia coli (EHEC) serotype O157:H7 are frequently the cause of human food-borne illness in the U.S., Europe, and Japan (for review, see [1–3]). Ingestion of only 100–200 organisms can produce debilitating diarrheal disease. Potentially fatal hemolytic uremic syndrome occurs in approximately 2–7% of cases and is due to the production of Shiga toxin. In addition to their effects on human health, these infections also have an economic consequence in lost sick days and medical costs. Furthermore, when found in human food such as tainted meat or bagged vegetables, EHEC contamination leads to large-scale recalls with the potential of bankrupting established commercial enterprises.
In addition to the four normal bases, the DNA of E. coli O157:H7 and E. coli K-12 also contains N6-methyladenine. This modified base is produced by the postreplication action of a DNA methyltransferase, encoded by the dam (DNA adenine methyltransferase) gene, at GATC sequences in double-stranded DNA. Studies in E. coli K-12 have shown that N6-methyladenine is involved in (a) controlling the frequency of initiation of chromosome replication at oriC (b) regulation of transcription at promoters containing GATC sequences and (c) strand discrimination during post-replicative mismatch repair [4–6]. The misdirection of mismatch repair in dam mutant bacteria leads to a mutator phenotype; that is, mutations are introduced into the parental DNA strand due to lack of direction normally imparted by methylation. Dam methylation has also been shown to affect the levels of virulence gene products in several pathogenic bacterial species [7].
Resistance to antibiotics can occur by mutation using a variety of mechanisms [8–11]. Among these is the acquisition of multiple antibiotic resistance conferred by mutation of the marRAB (multiple antibiotic resistance) genes [10, 12, 13]. The marR gene encodes a repressor while marB produces a probable periplasmic protein of unknown function [13–15]. The marA product is a transcription factor that activates or represses at least 60 chromosomal genes [16–19]. The inactivation of MarR results in an increased level of MarA, which upregulates the expression of acrAB and tolC resulting in increased number of AcrAB-TolC efflux pumps thereby leading to drug-resistance [20, 21]. The increased level of MarA also upregulates the micF gene, which produces a small regulatory antisense RNA that ultimately reduces expression of the ompF gene, thereby increasing permeability to antibiotics [22]. As a result, loss of a functional marR gene leads to broad spectrum antibiotic resistance in a number of bacteria including Salmonella, Shigella, Klebsiella and Yersinia species [23]. Here, we report that the higher than expected presence of chloramphenicol-resistant (CamR) variants in cultures of E. coli O157:H7 is due to either an increased mutation rate in the marR gene or to a normal mutation rate plus selection due to a high level of MarA. Antibiotic resistance not associated with marR, does not occur at high frequency.
2. Materials and methods
2.1 Bacterial strains and plasmids
E. coli O157:H7 strain EDL933 and KM69 have been described [24, 25]. It was previously demonstrated that deletion of the dam gene in EDL933 (KM69) resulted in loss of the 933W prophage [25]. The dam gene was restored to the chromosome of this construct, generating strain KM80, as follows: The dam gene of EHEC was cloned by gap repair following transformation of pKM210 into cells expressing the phage lambda Red recombineering functions. Plasmid pKM210 has been described before [24] and contains a NotI site flanked by upstream (1.3 kb) and downstream (1.2 kb) regions of the EHEC dam gene; the cassette is flanked by SacI and SphI sites. A NotI digest of pKM210 was electroporated into EDL933 containing pKM200, a λ Red-producing CamR plasmid [26]. After electroporation, the cells were grown for 1.5 hours in Luria broth and dilutions of the transformants were plated in Luria broth plates containing ampicillin (100 µg/ml). Gap repair of pKM210 resulted in levitation of the dam gene from the EDL933 chromosome at high efficiency, as most of the colonies tested positive for the EHEC dam gene. The resulting construct (pKM263) was digested with SacI and SphI, to liberate the recombineering fragment, and electroporated into KM68 (Δdam::cat-sacB) containing pKM208, a λ Red-producing AmpR plasmid [26]. Following electroporation, the cells were diluted into 5 ml L and allowed to grow overnight at 37°C. Dilutions of the culture were plated on L plates containing 10% sucrose and colonies were allowed to grow overnight. The next day, colonies were re-streaked on Cam plates. A total of nine out of 63 SucR colonies were found to be CamS, suggesting loss of the cat-sacB cassette ; 4/4 of these candidates tested positive by PCR for the restoration of the dam gene to the EHEC chromosome. All four strains were also AmpS, suggesting they did not contain an intact pKM263, and had spontaneously lost pKM208. These last points were verified by the absence of both pKM263 and pKM208 in a minilysate preparation of KM80. The dam+ phenotype of KM80 was verified by the restoration of wild type levels of spontaneous mutation resistance to rifampicin (data not shown).
Cam-resistant derivatives of KM80 and KM69 were isolated as spontaneous colonies arising on L broth plates with 10 µg/ml Cam after plating aliquots of overnight cultures. Individual colonies were purified by streaking three more times on the same medium.
E. coli K-12 strains AG100 (wildtype) and AG102 (marR1) [27, 28] were kindly provided by Dr. J.L Rosner (NIDDK, NIH, Bethesda, MD USA).
Plasmids pMQ583 and pMQ584, containing the short (378 bp) and long (435 bp) ORFs of marR respectively, were constructed by PCR amplification from EDL933 and cloning into the EcoRI and HindIII sites of pBAD18 [29]. The plasmids were introduced into the marR mutant bacteria by electroporation [26] and transformants were grown in L broth with carbenicillin and spotted onto L broth-carbenicillin plates containing Cam (10 µg/ml) or tetracycline (10 µg/ml) or nalidixic acid (25 µg/ml). The plates were incubated at 37°C overnight.
Ampicillin-resistant plasmids, pMQ315 (mutS), pMQ350 (mutL), pRH71-17 (mutH) have been described previously [30–32] and were introduced into KM69 by electroporation. Ten transformants of each plasmid were grown to saturation in L broth with carbenicillin and 10 ul spotted onto L broth plates containing 25 µg nalidixic acid and 50 µg carbenicillin per ml. The plates were incubated at 37°C overnight.
E. coli K-12 strain MM294 [33] was used as a recipient in an attempt to clone Cam-resistance genes from EDL933.
2.2 Media
Bacterial strains were cultivated in either L broth (10 g/l tryptone, 5 g/l yeast extract, 0.5 g/l NaCl) or Difo Brain Heart Infusion (20 g/l) and solidified, when required, with 16 g Difco Agar per l. Media were supplemented with ampicillin, carbenicillin, tetracycline, Cam, nalidixic acid and rifampicin at 100, 50, 10, 10, 25 and 100 µg/ml respectively.
2.3 Deep sequencing of bacterial strains
DNA was isolated from seven strains of E. coli by lysis in 1% SDS and 1mg/ml Proteinase K overnight at 55°C followed by 25:24:1 phenol:chloroform:isoamyl alcohol extraction and ethanol precipitation. Ten µg of purified DNA was sheared with a Covaris M220 ultrasonicator to a median size range of 250 bp. Deep sequencing libraries were prepared as per standard Illumina protocols (http://www.illumina.com). Single read 36 bp sequences were generated for strains GM7275, GM9236, and GM9237 on Illumina GAII sequencer and 100 bp sequences were generated for strains GM7274, GM9241, GM9242, and GM9243 on Illumina HiSeq 2000 sequencer with a mean coverage of 42 times. Perfectly matching reads were mapped to to E. coli O157:H7 EDL933 genome (Genebank AE005174.2) using the Bowtie software package [34]. Locations of single nucleotide polymorphisms and indels were determined using Seqmonk (www.bioinformatics.babraham.ac.uk) by identifying regions which contain no mapping sequences.
2.4 DNA fragment synthesis and sequencing
The marR gene was synthesized using the polymerase chain reaction (PCR) with Herculase II DNA polymerase (Agilent Technologies) and primers GCTATGAATGGTAATAGCGTCAG and GACTTATACTTGCCTGGGCA as described by the manufacturer except that 2 µl of cells from an overnight culture were included. The DNA fragment was gel-purified using Qiagen columns and reagents and sequenced by GeneWiz Corporation.
2.5 Minimal Inhibitory Concentrations (MICs) and Fitness
The MIC of a strain was determined by adding about 10,000 cells to 1 ml of L broth containing two-fold dilutions of Cam and incubating the tubes overnight at 37°C. To measure fitness, about 1000 wildtype and marR cells were introduced together into 1 ml of L broth and after overnight incubation dilutions were spread on L broth plates, with and without 10 µg/ml Cam, to assay for viability.
3. Results
3.1 Strain construction
We reported previously that dam mutants from E. coli O157:H7 strain EDL933 were generated at a very low frequency using bacteriophage lambda Red recombination of DNA fragments into the bacterial chromosome (“recombineering”) [25]. The few dam mutant strains that were isolated from EDL933 also were deleted for the 933W prophage. In order to use an isogenic wildtype control strain in the experiments described here, KM69 (Δdam Δ933W) was converted to dam+ as follows to yield strain KM80 (Δ933W). The dam+ gene from EDL933 was levitated into a suitable vector, cut with restriction enzymes and electroporated into Δdam::cat-sacB mutant bacteria expressing bacteriophage lambda Red recombination enzymes. Recombinant cells, in which the dam+ gene has replaced the Δdam::cat-sacB allele, should be resistant to sucrose since the cat-sacB fusion results in sucrose-sensitivity. Strain KM80 (Δ933W) was isolated as such a sucrose-resistant clone and confirmed to have an intact dam gene.
3.2 Mutation frequency to chloramphenicol resistance
The spontaneous mutation frequencies of KM69 (dam) to various drug resistances are shown in Table 1. The result for Cam was surprising because the high level of spontaneous resistance was not encountered with E. coli K-12 dam strains. Furthermore, the frequency of spontaneous CamR mutant bacteria from the isogenic wildtype strain, KM80 (Δ933W), was also higher than expected compared to E. coli K-12 (Table 1). This spontaneous frequency to CamR during recombineering of a Δ933W derivative of EDL933 was observed previously (Murphy and Campellone, unpublished observations). Re-streaking on fresh Cam plates identified recombinants in which the CamR gene was recombined into the chromosome, as the spontaneous CamR mutants (not recombined) grew poorly upon re-streaking. In this study, authentic recombinant colonies of KM80 could also be found by re-streaking (data not shown). However, with KM69 (dam), the number of spontaneous CamR colonies far outnumbered true recombinants, so that additional streaking of the colonies (or changing the Cam concentration) could not reliably distinguish between spontaneous versus recombinant CamR colonies following recombineering. For further study, and characterization of the source of this increased frequency to CamR, five independently-isolated CamR strains were obtained from each of KM69 (designated GM9236-9240) and KM80 (GM9241-9245).
Table 1.
Frequencies of E. coli K-12 and EHEC to antibiotic resistance.
| Strain | Background | Genotype | CamR Frequency | NalR Mutant Frequency |
TetR Mutant Frequency |
CarbR Frequency |
|---|---|---|---|---|---|---|
| AB1157 | K-12 | wild | <10−8 | 2 × 10−8 | <10−8 | <10−8 |
| GM3819 | K-12 | Δdam-16 | <10−8 | 51 × 10−8 | <10−8 | <10−8 |
| KM80 | EDL933 | wild | 1 – 10 × 10−8 | 3 × 10−8 | 2 × 10−8 | 1 – 4 × 10−8 |
| KM69 | EDL933 | Δdam | 208 – 1500 × 10−8 | 152 × 10−8 | 105 × 10−8 | 2 – 12 × 10−8 |
| GM9241 | EDL933 | marR | R | R | R | 24 – 558 × 10−8 |
Mutant frequency is expressed as the number of antibiotic resistant colonies per 108 cells. The numbers for Nal and Tet are the average of ten independent cultures. For Cam and Carb, the range of values from ten independent cultures are shown. R = resistant; Carb= carbenicillin, Nal = nalidixic acid, Tet = tetracycline
3.3 Attempted cloning of chloramphenicol resistance
In order to identify the mechanism of Cam-resistance in EDL933, DNA was isolated from GM9236, digested with either EcoRI or HindIII and ligated to pBAD18 vector DNA. The ligation mixtures yielded many ampicillin-resistant transformants of strain MM294, but no colonies that were both CamR and ampicillin-resistant were obtained. The failure to clone the drug-resistance was due to the responsible gene being recessive to its wildtype allele in the cloning host (see Discussion).
3.4 Whole genome sequencing
The failure to clone CamR from GM9236 provoked us to use whole genome sequencing of representative Cam-sensitive (KM69, KM80) and resistant (GM9236, GM9237, GM9241, GM9242 and GM9243) bacteria. Chromosomal DNA libraries were constructed from each strain and subjected to Illumina sequencing technology. The DNA sequences were aligned to the EDL933 genome. Every CamR mutant bacterial clone had a mutation in the marR gene while the CamS parental strains did not.
3.5 Sequencing of the marR gene
Identification of a mutated marR gene as the cause for CamR in the whole genome sequenced strains, prompted us to sequence this gene in all CamR mutant and parental bacteria using PCR cloning and sequencing. The results are shown in Table 2. As expected, the marR mutation identification by sequencing of PCR products from these strains confirmed those obtained by whole genome sequencing.
Table 2.
Type and location of mutations in the marR gene.
| Strain | Parental genotype |
Mutation in marR | Location in chromosome |
Amino acid change |
|---|---|---|---|---|
| GM9236 | dam | GGG to GAG | 1,944,735 | G69R |
| GM9237 | dam | AT to GC | 1,944,803 | L46P |
| GM9238 | dam | TTT to TT | 1,944,811–813 | F43 fs |
| GM9239 | dam | GGG to GGA | 1,944,734 | G69E |
| GM9240 | dam | CGCC to CACC | 1,944,710 | R77Q |
| GM9241 | wt | IS629 | 1,944,792–3 | |
| GM9242 | wt | Deletion | 1,944,603–618 | C108 |
| GM9243 | wt | AT to CT | 1,944,707 | L78R |
| GM9244 | wt | AT to GC | 1,944,842 | L33P |
| GM9245 | wt | Deletion | 1,944,698–716 | R73P |
The amino acid sequence numbering is that of the annotated E. coli K-12 gene. fs = frameshift, wt = wildtype.
The marR mutations identified from CamR bacteria derived from the wildtype strain (KM80) comprised an IS (insertion sequence) element insertion, two deletions, a transition and a transversion (Table 2). In contrast, with one exception, the mutations in marR from the dam mutant derivative (KM69) occur in runs of iterated bases and are due to transition mutations (AT to GC and GC to AT). These mutation spectra in wildtype and dam mutant strains are precisely those previously described using other mutational targets [35, 36].
The N-terminal 41 amino acids of the MarR protein have been proposed to be responsible for dimerization and amino acids 31–132 as a DNA binding winged helix-turn-helix (HTH) domain [37]. Mutant L33P should affect the oligomeric state while all other mutations listed in Table 2 should prevent DNA binding.
3.6 Multiple antibiotic resistance
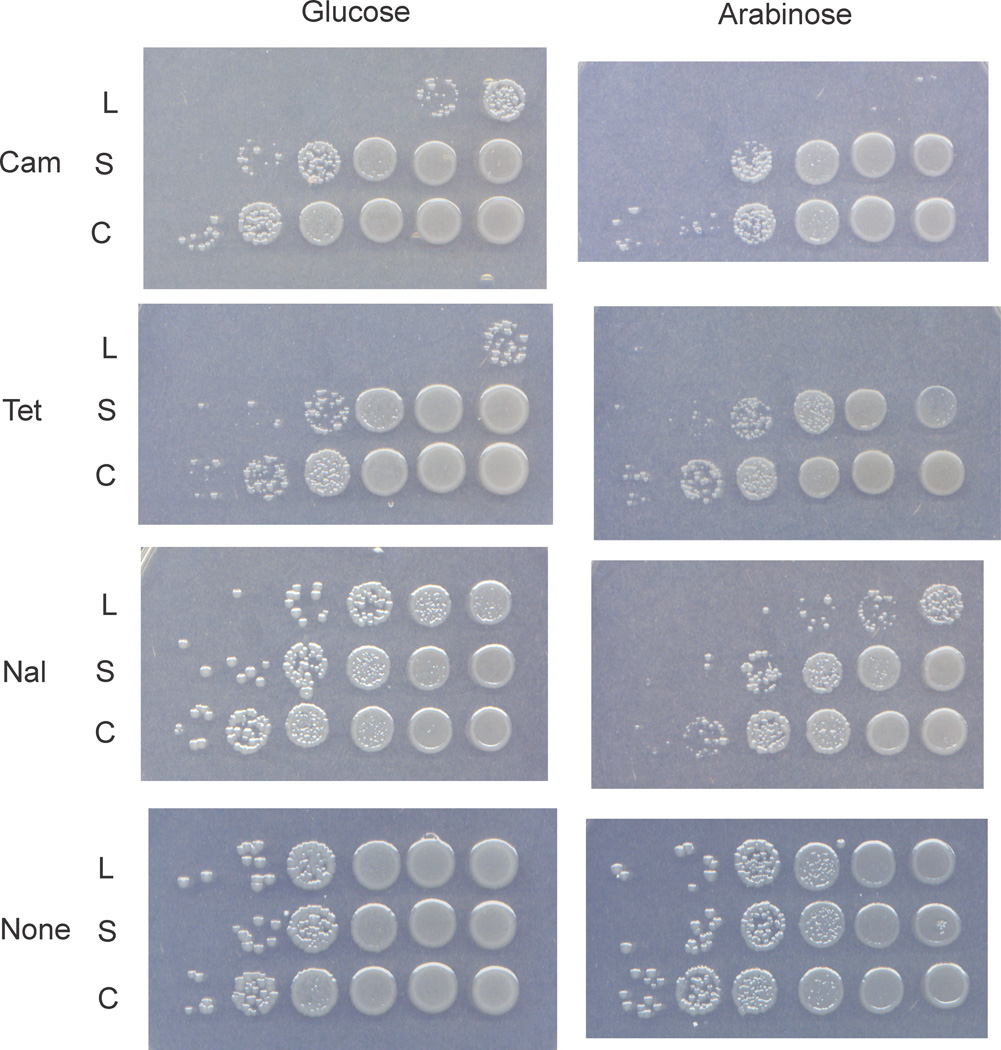
The identification of marR as the basis for Cam-resistance in EDL933 wildtype and dam mutant strains indicated that there should also be resistance to other antibiotics. This indeed is the case and all CamR bacteria were also resistant to tetracycline (10 µg/ml) and the quinolone antibiotic, nalidixic acid (25 µg/ml) (Fig. 1).
Fig 1.
marR+ complementation of GM9242 (marR). The marR gene was cloned into pBAD18 in which expression of the gene is reduced when the cell containing it is grown with glucose and expression is induced when the cell is grown in the presence of arabinose. Two constructs were used: the long marR ORF (L) as annotated in E. coli K-12 and the short ORF (S) as annotated in E. coli O157::H7 EDL933. The control strain (C) contained the vector plasmid. Cells grown overnight were diluted in ten-fold increments and 10 µl drops placed on appropriate medium. The plates were incubated overnight at 37°C. Cam = chloramphenicol, Tet = Tetracycline, Nal = Nalidixic acid.
3.7 Complementation of marR
The marR gene in E. coli K-12 is annotated to encode an additional 19 N-terminal amino acids compared with marR in EDL933. We constructed plasmids pMQ583 and pMQ584 containing the shorter (378 bp) and longer (435 bp) predicted open reading frames respectively of the EDL933 marR sequence under the control of the arabinose BAD promoter. Strain GM9242 (Mar−) was transformed with each marR plasmid and the vector control and tested for resistance/sensitivity to Cam, tetracycline and nalidixic acid. Expression of marR+ in GM9242, in the presence of arabinose, reversed the antibiotic resistance to sensitivity for all three antibiotics (Fig. 1). For Cam and tetracycline, the reduction of viability was greater than 105-fold but less for nalidixic acid. Even in the absence of arabinose (and the presence of glucose), there was antibiotic sensitivity indicating that the level of basal transcription in the absence of inducer was sufficient to allow some MarR production and reversal of phenotype. All the other EDL933 CamR strains were transformed with the longer marR open reading frame plasmid and shown to become antibiotic sensitive (data not shown).
The plasmid containing the longer marR open reading frame was more efficient at reversing the antibiotic resistance phenotype than the shorter version. We suggest that the annotation of marR in EDL933 be changed in view of these results.
3.8 High frequency carbenicillin resistance in marR mutant bacteria
During the marR complementation experiments described above, it was noticed that there were a substantial number of carbenicillin-(or ampicillin) tolerant colonies on the control plates during transformation of the marR mutant strains. This feature was not observed with the wildtype strains (KM69, KM80) transformed with the same plasmid (data not shown). Further experiments showed that, indeed, the marR cultures had a high frequency of carbenicillin-tolerant variants relative to the wildtype parent (Table 1).
The carbenicillin-tolerant colonies were extremely heterogeneous in colony size and properties. At one end of the spectrum, colonies from the primary plate grew well when re-streaked on agar with carbenicillin and retained carbenicillin-resistance even after growth without the drug. At the other end of the spectrum were colonies that failed to grow when re-streaked on carbenicillin medium. Between these two extremes were colonies that retained carbenicillin-tolerance for a random number of passages on carbenicillin medium. Other colonies lost resistance to 50 µg carbenicillin per ml but were able to grow in lower concentrations of the drug (data not shown). It was also found that carbenicillin-tolerant colonies can be isolated from an E. coli K-12 marR strain but the frequency is at least 100-fold less than for E. coli O157:H7 marR bacteria (data not shown). The properties described above are remarkably similar to those described by George and Levy [27, 28] for marR-mediated drug-resistance in E. coli K-12 suggesting a similar mechanistic basis (i.e., increased MarA level).
The carbenicillin-tolerant phenotype of E. coli O157:H7 marR bacteria was completely suppressed by expression of MarR from the pMQ584 plasmid (data not shown). Beta-lactamase resistance in E. coli is often caused by increased ampC promoter activity [38] but no such alterations were found in several of the carbenicillin-tolerant marR mutant strains (data not shown).
An increased MarA level is a consequence of loss of MarR repressor activity. We conclude that in a sub-set of the cells in the population, MarA promotes carbenicillin-tolerance.
3.9 Expression of multicopy mismatch repair genes in KM69 (Δdam)
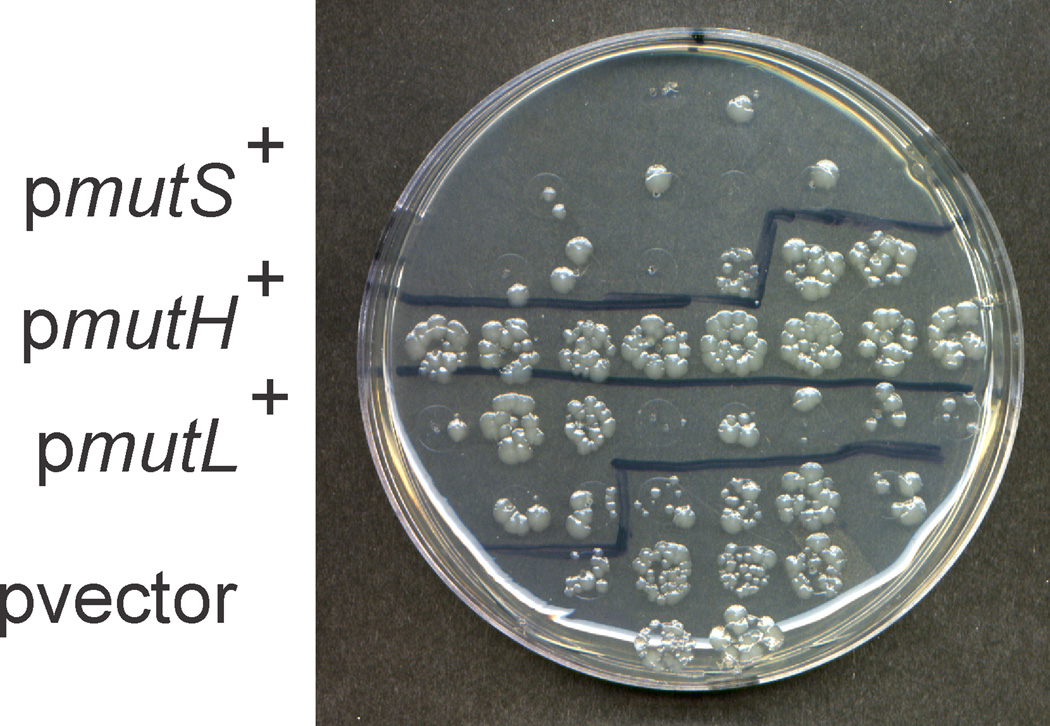
E. coli K-12 dam mutants contain single- and double-strand breaks in their genomes that are produced by misdirected mismatch repair in which mutations are introduced into the parental strand [4, 7]. Jacquelin et al [39] have shown that multicopy mismatch repair genes (mutH, mutL, or mutS) in E. coli K-12 dam bacteria reduce the mutation frequency relative to the control. We repeated this experiment in KM69 and the results shown in Fig. 2 indicate that the presence of multicopy mutS and mutL, but not mutH, dramatically reduced the formation of nalidixic acidresistant colonies We conclude that mismatch repair is responsible for the increased multiple antibiotic resistance frequency in KM69 (Δdam) by introducing the “correction” into the parental DNA strand of a dam mutant strain.
Fig 2.
Effect of overproduction of mismatch repair proteins on mutant frequency to nalidixic acid in KM69 (Δdam). Ten independent cultures of strain KM69 with multicopy plasmids encoding mutS, mutL, mutH and the vector were grown to saturation in L broth and 10 ul from each culture was spotted on an L broth plate containing 25 µl nalidixic acid per ml. The plate was incubated overnight at 37°C.
3.10 Mutant frequency to streptomycin, spectinomycin and rifampicin resistance
In order to test if the increase in spontaneous marR mutability is a general feature of E. coli O157:H7, we compared the frequencies of streptomycin, spectinomycin and rifampicin resistance in E. coli K-12 and KM80 strains. The streptomycin and spectinomycin mutant frequencies were the same in both strains – 1.2 × 10−9 and for rifampicin resistance, 1.4 × 10−8 for both.
3.11 Minimal Inhibitory Concentration (MIC) and fitness
The MIC for CamR of KM80 and AG100, the wildtype EHEC and K-12 strains respectively, in our L broth is 0.6 – 1.2 µg/ml and the MIC of the marR mutant strains GM4292 and AG102 is 20 – 25 µg/ml. The doubling time for both EHEC and K-12 wildtype and marR strains was the same (about 27 min).
To study fitness, we inoculated about 1000 wildtype and marR cells together into L broth and grew the cells until the culture was saturated. Dilutions were spread on L broth plates with and without 10 µg/ml Cam. For both EHEC and K-12, the number of colonies on the Cam plates was about one-third that of the control plates. We conclude that the fitness for both EHEC and K-12 marR strains is the same.
4. Discussion
There is clearly something different about MarR in EHEC versus E. coli K-12. This difference could be at the gene or protein level. The EHEC MarR protein has two amino acid changes compared with that in K-12 which might alter the affinity of the protein for its recognition sequence. Alternatively, the absolute level of the MarR protein may be different in the two strains leading to the changes we have found. In addition, it may be that when MarR is inactive, the MarA levels are different in the two strains leading to an alternate activation/repression program of chromosomal genes. This alternate program might permit cells to survive antibiotic challenge more forcefully in EHEC than K-12. At present, we do not know which of these alternatives, if any, is the correct one.
At the gene level, the increased numbers of marR mutant bacteria in cultures of E. coli O157:H7 versus E. coli K-12 could be due to an increase in mutation frequency or to a normal mutation frequency plus a selective advantage during growth of the culture or to both of these possibilities. The mutant frequencies to streptomycin, spectinomycin and rifampicin resistance are similar in both EHEC and K-12 strains suggesting that the increase in the number of marR variants in EHEC may be due to a selective advantage during growth. However, we found that differences in doubling time or fitness could not explain the increase. That the MICs of the wildtype and marR strains was the same for both EHEC and K-12 indicates that there is no difference in intrinsic resistance when selection was at 10 µg/ml Cam. In wildtype KM80, the increased marR mutant frequency is unlikely to reflect an increase in a specific mutational pathway or a specific DNA repair system (including the SOS response) because the increased spontaneous mutant frequency is non-specific in the types of mutations recovered. The marR genes are located in the same chromosomal surroundings (synteny) in the two strains thereby excluding the possibility of alternate chromosomal location as an explanation.
An alternative explanation that the EHEC marR locus is more mutable than rpsL, rpsE and rpoB, is more problematical because it is expected that genes should be equally mutable irrespective of their location. It has been reported that mutability is correlated with the level of gene expression [40, 41]. This explanation is rendered unlikely for our results because the rpsL and rpsE genes are more highly expressed than marR in E. coli O157:H7 [24] but the mutant frequency for the ribosomal genes is at the same level in both EHEC and K-12. A recent study found that the frequency of spontaneous mutation in E. coli K-12 was more closely correlated with chromosome supercoiling than gene expression [42]. The K-12 chromosome is organized into about 400 domains with dynamic boundaries and this number correlates with the average length of supercoiled loops (see [43] for review). In both K-12 and EHEC, the marR, rpoB and the two ribosomal genes would be in separate supercoiled loops. If, however, the extent of supercoiling in the marR loop is different in EHEC and K-12, then a difference in spontaneous mutagenesis might result. It should be noted that the MarR mutability phenotype is too variable to allow Luria-Delbruck fluctuation analysis.
The marR mutants arise during growth in L broth before exposure to Cam. It is possible that stress responses, such as those mediated by LexA (SOS) [44] or RpoE [45] or RpoS [46], in EHEC are of a greater magnitude in either duration and/or strength than those in K-12. If so, there would be a greater survival of potential CamR cells and an apparent increase in mutation frequency.
At present, we cannot distinguish between the possibilities of an increase in mutation frequency or to a normal mutation frequency plus a selective advantage or to both. Whatever the mechanism, the high background of marR cells is a hindrance for recombineering experiments in EHEC strains. Our current working hypothesis is that mutation in the marR gene activates MarA but that the cascade of genes activated or repressed is different in EHEC and K-12, which may lead to a selective advantage to the former upon exposure to antibiotics. If the mutation rate of marR is indeed higher due to supercoiling effects it would amplify this process.
Although the sample size of the marR sequenced strains is small, it is interesting that no mutation was recovered twice. This result suggests that there is no mutational “hotspot” in the EDL933 marR gene and that the gene can be inactivated at a large number of sites, at least between amino acid residues 33–108.
The initial attempt to identify the cause of increased mutagenesis in strain EDL933 by random cloning of DNA fragments encoding Cam resistance was not successful. In retrospect, this is consistent with the mutagenic target being the MarR repressor. Since the plasmid-encoded marR− allele is recessive to host-encoded marR+, drug-resistant colonies cannot be formed.
The crystal structure of E. coli K-12 MarR protein (144 amino acids) has been solved at 2.3 A resolution using the 435 bp long open reading frame [37]. The EDL933 marR sequence is 98% identical but is annotated as a shorter 378 bp reading frame missing the N-terminal 19 amino acids. This truncation removes part of the first alpha helix of the protein and, as we show in Fig. 1, is not as effective in complementing the marR defect in GM9242 as is the longer reading frame. Clearly, the longer open reading frame is the correct sequence for E. coli O157:H7 marR gene based on our results and those for E. coli K-12 and the structural data of the protein.
The E. coli K-12 and annotated EHEC marR gene sequences are shown in Fig. 3. The EHEC sequence has the 57 bp 5’ truncation and seven other differences. Five of these are conserved changes that do not alter the encoded amino acid. Two sequence differences result in changes Gly103Ser and Tyr137His but these still allow the MarR protein to be fully functional (Fig. 1). We have not yet tested if these two amino acid changes in the marR gene affect spontaneous mutation frequency to Cam-resistance. However, none of the mutations in the marR gene identified in Table 2 map to the codons for these two amino acids.
Fig. 3.
Comparison of the marR gene sequence of E. coli K-12 (K) and the annotated E. coli O157:H7 marR sequence (E). The top line is the sequence present in the K-12 marR gene but not in the annotated EHEC sequence.
We assume that a computer program searching for an open reading frame beginning with an ATG and ending with a nonsense codon generated the annotation for the EHEC marR gene. We have found no experimental evidence in the literature to support this annotation. The computer-derived annotation would explain why the structural and genetic data in K-12 were not taken into account. The actual start codon for marR gene is a GTG.
Over-expression of MutS and MutL in KM69 reduced the frequency of nalidixic acid-resistant mutant colonies. A similar observation (but also including MutH) has been made in an E. coli K-12 dam mutant strain. Jacquelin et al [39] argued on the basis of their experimental results, that the reduction in mutant frequency in E. coli K-12 was due to mismatch repair-dependent cell killing. That is, increasing the level of mismatch repair proteins would, in turn, increase the number of DNA double-strand breaks thereby resulting in an increased number of fatalities in the bacterial population and eliminating those cells from the population. This interpretation is consistent with the results obtained in Fig. 2 for E. coli O157:H7.
An unexpected result was the high frequency of carbenicillin-tolerant bacteria in cultures of marR strains. Although promoter-up mutations in the ampC promoter are frequently the cause of such resistance, this is not the case in beta-lactam-tolerant marR mutants (data not shown). The suppression of beta-lactam resistance by episomal marR+ argues that the simplest explanation for their existence is that the high level of MarA promotes changes in the cell envelope that, in a presumably stochastic manner, can render tolerance to beta-lactam antibiotics. In Enterobacter aerogenes, for example, a high level of MarR promotes resistance to beta-lactams by overexpression of OmpX [47, 48]. A similar rationale is probably the basis for the observations of beta-lactam resistance in E. coli that we have made in this report.
E. coli O157:H7 marR mutant strains were first described by Yaron et al [49] who isolated a spontaneous E. coli O157:H7 CamR strain from cattle feces and showed that it had a G to T transversion in marR leading to Glu10Ochre change in the protein. In general, the mutant strain had the same growth characteristics in a variety of media and cultural conditions as the wildtype but was resistant to nalidixic acid, tetracycline and ciprofloxacin in addition to Cam. DNA sequencing of the mar operon indicated that it was 99% identical to that in E. coli K-12. The resistance to antibiotics could be complemented in the mutant strain [50] (although the data were not shown) by an E. coli K-12 marR plasmid constructed by Alekshun and Levy [51].
The relative increase in marR variants in cultures of E. coli O157:H7 and the propensity of such mutant strains to also become carbenicillin-tolerant, indicates how easily such pathogenic bacteria can become resistant to antibiotics. This is in contrast to the laboratory adapted E. coli K-12 strain which has a lower incidence of marR-mediated antibiotic resistance. In the external environment, it may be desirable for bacteria to rapidly select for antibiotic resistant variants especially in niches where antibiotic-producing organisms are present. In addition, the widespread use of antibiotics in veterinary and medical use, would allow rapid selection of antibiotic-resistant variants.
marR gene mutations occur at a higher frequency in E. coli O157:H7 than E. coli K-12
rpsL, rpsE and rpoB mutations occur at the same frequency in both E. coli strains
E. coli O157:H7 marR mutant strains can become tolerant to beta-lactam antibiotics
Acknowledgements
This work was supported by grant R01 GM063790 from the National Institutes of Health. We thank Dr. J.L. Rosner for the gift of E. coli strains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors have no conflicts of interests to declare.
References
- 1.Donnenberg MS, Whittam TS. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Invest. 2001;107:539–548. doi: 10.1172/JCI12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.Spears KJ, Roe AJ, Gally DL. A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol. Lett. 2006;255:187–202. doi: 10.1111/j.1574-6968.2006.00119.x. [DOI] [PubMed] [Google Scholar]
- 4.Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 2005;8:154–160. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Low DA, Casadesus J. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 2008;11:106–112. doi: 10.1016/j.mib.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Marinus MG, Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 9.Hawkey PM, Jones AM. The changing epidemiology of resistance. J. Antimicrob. Chemother. 2009;64(Suppl 1):i3–i10. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 10.Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soo VW, Hanson-Manful P, Patrick WM. Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1484–1489. doi: 10.1073/pnas.1012108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hachler H, Cohen SP, Levy SB. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 1991;173:5532–5538. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariza RR, Cohen SP, Bachhawat N, Levy SB, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seoane AS, Levy SB. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin RG, Rosner JL. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin RG, Jair KW, Wolf RE, Jr, Rosner JL. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz C, Levy SB. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob. Agents Chemother. 2010;54:2125–2134. doi: 10.1128/AAC.01420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbosa TM, Levy SB. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 2000;182:3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki T, Ueguchi C, Mizuno T. H-NS regulates OmpF expression through micF antisense RNA in Escherichia coli. J. Bacteriol. 1996;178:3650–3653. doi: 10.1128/jb.178.12.3650-3653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Campellone KG, Roe AJ, Lobner-Olesen A, Murphy KC, Magoun L, Brady MJ, Donohue-Rolfe A, Tzipori S, Gally DL, Leong JM, Marinus MG. Increased adherence and actin pedestal formation by dam-deficient enterohemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 2007;63:1468–1481. doi: 10.1111/j.1365-2958.2007.05602.x. [DOI] [PubMed] [Google Scholar]
- 25.Murphy KC, Ritchie JM, Waldor MK, Lobner-Olesen A, Marinus MG. Dam methyltransferase is required for stable lysogeny of the Shiga toxin (Stx2)-encoding bacteriophage 933W of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 2008;190:438–441. doi: 10.1128/JB.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E coli BMC. Mol. Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George AM, Levy SB. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 1983;155:541–548. doi: 10.1128/jb.155.2.541-548.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aronshtam A, Marinus MG. Dominant negative mutator mutations in the mutL gene of Escherichia coli. Nucleic Acids Res. 1996;24:2498–2504. doi: 10.1093/nar/24.13.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grafstrom RH, Hoess RH. Nucleotide sequence of the Escherichia coli mutH gene. Nucleic Acids Res. 1987;15:3073–3084. doi: 10.1093/nar/15.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu TH, Marinus MG. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J. Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meselson M, Yuan R. DNA restriction enzyme from E coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carraway M, Rewinski C, Wu TH, Marinus MG. Specificity of the Dam-directed mismatch repair system of Escherichia coli K-12. Gene. 1988;74:157–158. doi: 10.1016/0378-1119(88)90275-2. [DOI] [PubMed] [Google Scholar]
- 36.Glickman BW. Spontaneous mutagenesis in Escherichia coli strains lacking 6-methyladenine residues in their DNA: an altered mutational spectrum in dam-mutants. Mutat. Res. 1979;61:153–162. doi: 10.1016/0027-5107(79)90122-2. [DOI] [PubMed] [Google Scholar]
- 37.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 38.Peter-Getzlaff S, Polsfuss S, Poledica M, Hombach M, Giger J, Bottger EC, Zbinden R, Bloemberg GV. Detection of AmpC beta-lactamase in Escherichia coli: comparison of three phenotypic confirmation assays and genetic analysis. J. Clin. Microbiol. 2011;49:2924–2932. doi: 10.1128/JCM.00091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacquelin DK, Martina MA, Argarana CE, Barra JL. Plasmid expression of mutS, -L and/or -H gene in Escherichia coli dam cells results in strains that display reduced mutation frequency. Mutat. Res. 2008;637:197–204. doi: 10.1016/j.mrfmmm.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Hudson RE, Bergthorsson U, Ochman H. Transcription increases multiple spontaneous point mutations in Salmonella enterica. Nucleic Acids Res. 2003;31:4517–4522. doi: 10.1093/nar/gkg651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martincorena I, Seshasayee AS, Luscombe NM. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485:95–98. doi: 10.1038/nature10995. [DOI] [PubMed] [Google Scholar]
- 42.Foster PL, Hanson AJ, Lee H, Popodi EM, Tang H. On the mutational topology of the bacterial genome. G3. (Bethesda.) 2013;3:399–407. doi: 10.1534/g3.112.005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Travers A, Muskhelishvili G. Bacterial chromatin. Curr. Opin. Genet. Dev. 2005;15:507–514. doi: 10.1016/j.gde.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Simmons LA, Foti JJ, Cohen SE, Walker GC. In: The SOS Regulatory Network. Bock A, Curtiss R, Karp PD, Neidhardt FC, Nystrom T, Slauch JM, Squires CL, editors. Washington, DC.: EcoSal, ASM Press; 2008. [Google Scholar]
- 45.Gibson JL, Lombardo MJ, Thornton PC, Hu KH, Galhardo RS, Beadle B, Habib A, Magner DB, Frost LS, Herman C, Hastings PJ, Rosenberg SM. The sigma(E) stress response is required for stress-induced mutation and amplification in Escherichia coli. Mol. Microbiol. 2010;77:415–430. doi: 10.1111/j.1365-2958.2010.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chollet R, Bollet C, Chevalier J, Mallea M, Pages JM, Davin-Regli A. mar Operon involved in multidrug resistance of Enterobacter aerogenes. Antimicrob. Agents Chemother. 2002;46:1093–1097. doi: 10.1128/AAC.46.4.1093-1097.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dupont M, De E, Chollet R, Chevalier J, Pages JM. Enterobacter aerogenes OmpX, a cation-selective channel mar- and osmo-regulated. FEBS Lett. 2004;569:27–30. doi: 10.1016/j.febslet.2004.05.047. [DOI] [PubMed] [Google Scholar]
- 49.Yaron S, White DG, Matthews KR. Characterization of an Escherichia coli O157:H7 marR mutant. Int. J. Food Microbiol. 2003;85:281–291. doi: 10.1016/s0168-1605(02)00547-0. [DOI] [PubMed] [Google Scholar]
- 50.Golding SS, Matthews KR. Intrinsic mechanism decreases susceptibility of Escherichia coli O157:H7 to multiple antibiotics. J. Food Prot. 2004;67:34–39. doi: 10.4315/0362-028x-67.1.34. [DOI] [PubMed] [Google Scholar]
- 51.Alekshun MN, Levy SB. Characterization of MarR superrepressor mutants. J. Bacteriol. 1999;181:3303–3306. doi: 10.1128/jb.181.10.3303-3306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]