Abstract
Sodium/proton (Na+/H+) antiporters, located at the plasma membrane in every cell, are vital for cell homeostasis1. In humans, their dysfunction has been linked to diseases, such as, hypertension, heart failure and epilepsy and they are well-established drug targets2. The best understood model system for Na+/H+ antiport is NhaA from Escherichia coli1,3, where both EM and crystal structures are available4-6. NhaA is made up of two distinct domains, a Core domain and a Dimerisation domain. In the NhaA crystal structure a cavity is located between the two domains providing access to the ion-binding site from the inward-facing surface of the protein1,4. Like many Na+/H+ antiporters, the activity of NhaA is regulated by pH, only becoming active above pH 6.5, where a conformational change is thought to occur7. To date, the only reported NhaA crystal structure is of the low pH inactivated form4. Here, we describe the active-state structure of a Na+/H+ antiporter, NapA from Thermus thermophilus at 3 Å resolution, solved from crystals grown at pH 7.8. In the NapA structure, the Core and Dimerisation domains are in different positions to those seen in NhaA and a negatively charged cavity has now opened to the outside. The extracellular cavity allows access to a strictly conserved aspartate residue thought to directly coordinate ion-binding1,8,9, a role supported here by molecular dynamics simulations. To alternate access to this ion-binding site, however, requires a surprisingly large rotation of the Core domain, some 20° against the Dimerisation interface. We conclude that despite their fast transport rates of up to 1500 ions/sec3, Na+/H+ antiporters operate by a two-domain rocking bundle model, revealing themes relevant to secondary-active transporters in general.
Na+/H+ antiporters are secondary active transporters that are conserved across all biological kingdoms to maintain the internal pH, cell volume and sodium concentration of the cell1, a mechanism first proposed by West and Mitchell10. Na+/H+ antiporters are members of the large monovalent cation proton antiporter (CPA) superfamily that includes, among others, the CPA1 and CPA2 clades2. It is generally thought that Na+/H+ antiporters from the CPA1 clade catalyse electroneutral sodium-proton exchange (SLC9A1-9/Nhe1-9, in mammals), whereas CPA2 members are thought to be electrogenic (SLC9B1-2/Nha1-2, in mammals)2, with stoichiometries of 2H+: 1Na+ and 3H+: 2Na+ ions reported11,12. Like all secondary-active transporters, Na+/H+ antiporters are thought to operate by an alternating access mechanism, however, the different conformational states of the transport cycle have yet to be determined. The inward-facing structure of the well-characterised bacterial CPA2 protein E. coli NhaA is the only representative crystal structure4.
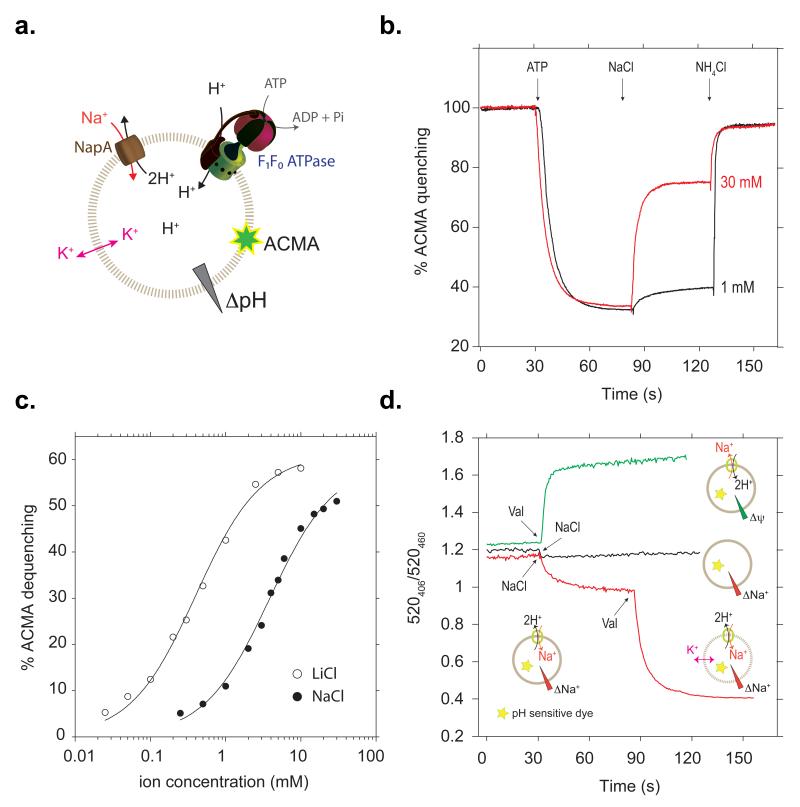
Using fluorescent-based methods13,14 we screened members of the CPA2 clade2 for their suitability for structural studies. NapA from T. thermophilus, which has 21% sequence identity to human Nha2, was thus identified (see Methods, Supplementary Fig. 1a and 2). Although the overall sequence homology to the better characterised NhaA is low, <15% identity, residues identified to be important for transport in NhaA and mammalian homologues are nonetheless well conserved (Supplementary Fig. 2). Using isolated inside-out membrane vesicles it was previously shown that T. thermophilus NapA is active above pH 6, with maximum activity for sodium at pH 815, similar to E. coli NhaA16. To measure activity in an isolated system, we co-reconstituted purified NapA and E. coli F0F1 ATP synthase into liposomes (see Methods). After establishment of a pH gradient by the addition of ATP, proton efflux was monitored in response to Na+ or Li+ addition (Fig. 1a and b). In this experimental setup, the apparent KM for Na+ or Li+ was determined to be 4.0 ± 0.3 and 0.41 ± 0.04 mM respectively, similar to the affinities from inside-out vesicles15 (Fig. 1c). In experiments with NapA proteoliposomes trapped with a water-soluble pH sensitive dye, dissipation of the membrane potential (Δψ) stimulated exchange activity in the presence of a Na+ gradient, confirming the electrogenic nature of NapA (Fig. 1d, lower trace). Furthermore, in the absence of a H+ or Na+ gradient, NapA transport activity was solely driven by Δψ (Fig. 1d, upper trace). Taken together, T. thermophilus NapA has a similar antiport profile to E. coli NhaA and consequently functional and structural studies of the two proteins can complement one another.
Fig. 1. NapA Na+/H+ transport activity is electrogenic.
a, Experimental setup for determination of Na+/Li+ affinity. The ATP synthase and NapA are co-reconstituted in liposomes. Free K+ diffusion by valinomycin suppresses the effect of Δψ. b, Representative ACMA fluorescence traces for Na+/H+ antiporter activity. ATP-driven H+-pumping establishes a ΔpH (acidic inside) as monitored by quenching of fluorescence. H+-efflux is initiated by addition of NaCl/LiCl and further NH4Cl addition collapses the proton gradient. c, Apparent binding affinity for Na+ (closed circles) and Li+ (open circles) in NapA (pH 7.5). d, Na+-gradient (~120 mV) driven Na+/H+-exchange was initiated by addition of NaCl (red). Addition of valinomycin at 90s leads to further H+-efflux; this releases the inhibitory membrane potential established during the first transport phase. No Na+-gradient driven transport was observed in liposomes without NapA (black). Δψ driven Na+/H+-exchange transport was initiated by establishing a K+/valinomycin diffusion potential (green). No activity was observed in liposomes without NapA (data not shown).
The structure of NapA at pH 7.8 was solved by multiple isomorphous replacement with anomalous scattering in combination with multi-crystal averaging. The highest resolution data corresponds to a triple mutant of NapA, where three cysteine residues, which have no apparent effect on functional activity, were introduced to facilitate phasing (see Methods, Supplementary Table 1 and 2, Supplementary Fig. 1b). The structure was refined at a resolution of 3 Å, Rfactor 22.3% and Rfree 24.8% (Supplementary Table 1, Supplementary Fig. 3, see Methods). Electron density maps at 3.7 Å of wild type NapA at pH 9.0 show no clear structural differences between the wild type and the triple cysteine mutant.
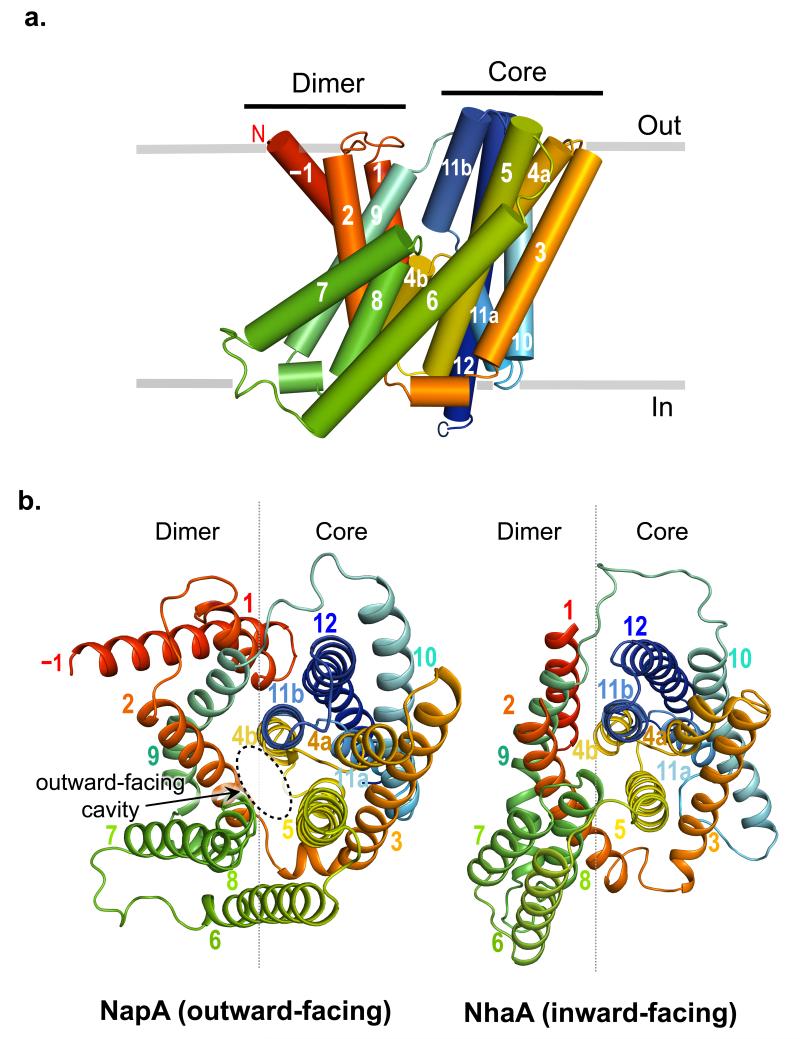
NapA is built from 13 transmembrane helices (TM) with a Nout-Cin topology (Fig. 2 and Supplementary Fig. 4). Relative to NhaA it has an additional helix at the N-terminus. To facilitate comparison to NhaA we refer to the first helix as TM–1 (Figs. 2a and Supplementary Fig. 4). TMs −1 to 5 and TMs 7 to 12 are topologically similar but oppositely orientated in the plane of the membrane (Supplementary Fig. 4). These 6-TM topology inverted repeats intertwine to form a Core (ion-translocation) and Dimerisation (interface) domain and are linked together by TM6 (Fig. 2b and Supplementary Fig 4). The NapA and NhaA4 structures are very similar (Fig. 2b). As there is a change in the position of the Core relative to the Dimerisation domain, however, the similarity is best seen when the two domains are superposed separately; root mean square deviation (r.m.s.d.) of 1.8 Å for 134/148 pairs of Cα atoms of the Core domain TMs and 1.9 Å for 62/88 of the Dimerisation domain TMs (Supplementary Fig. 5a and b). The change in position of the Core relative to the Dimerisation domain, which may reflect the difference in the pH at which the transporters were crystallized, gives rise to a large negatively charged cavity that is now open to the outside in contrast to the inward-facing funnel seen in NhaA (Fig. 2b and Fig. 3b). The interaction between TM2 and TMs 4 and 5 tightly close the cytoplasmic side of the cavity.
Fig. 2. Outward-facing NapA structure.
a, Cartoon representation of NapA as viewed in the plane of the membrane. TMs −1 to 12 have been colored from red at the N-terminus to blue at the C-terminus with the position of the membrane depicted in grey. b, Outward-facing NapA structure (left) and inward-facing NhaA structure (right) as viewed from the extracellular side with the removal of the β-hairpin domain located between TMs 1 and 2 from NhaA to facilitate visual comparison. The Dimerisation and Core domain boundaries are represented by a grey line.
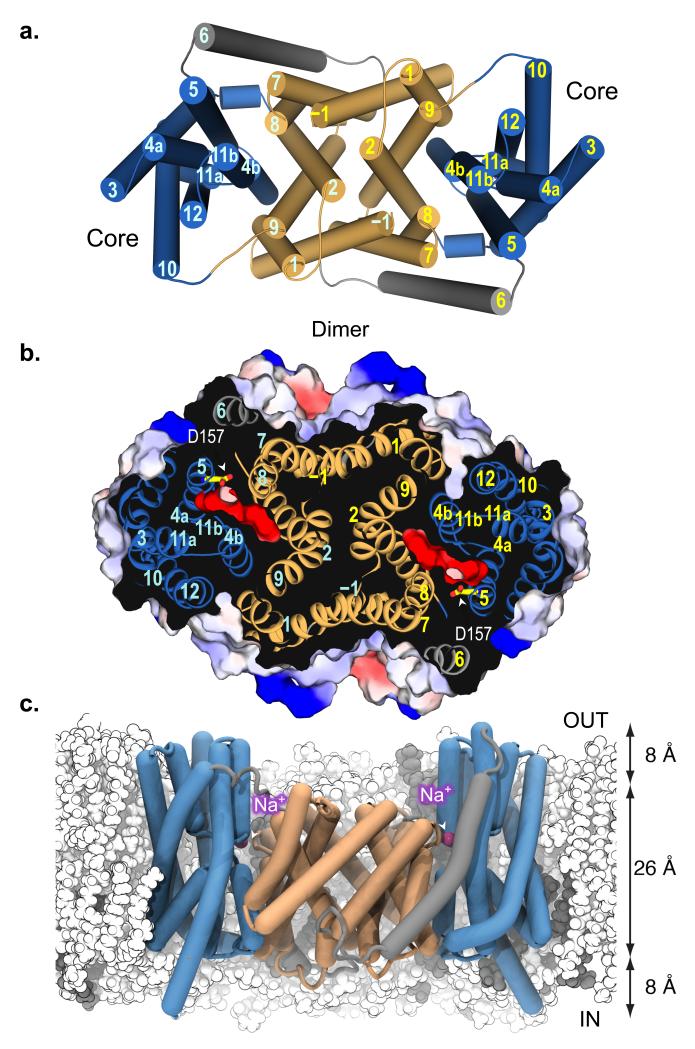
Fig. 3. Structure of NapA dimer and proposed Na+/(Li+) binding site.
a, Cartoon representation of the NapA dimer structure with TMs for one monomer labeled in cyan and the other yellow. The TMs of the Dimerisation domains are colored in pale orange and the Core ion-translocation domains in sky blue, connected together by TM6 shown in grey. b, Electrostatic surface representation showing the location of the negatively charged extracellular cavity as a section through the protein from above (same orientation as in Fig. 3a). The proposed ion-binding aspartate, Asp157, is illustrated as a stick model. c, In MD simulations, the NapA dimer is stable in a 4 POPE (white) : 1 POPG (gray) lipid membrane. Sodium ions (magenta spheres) spontaneously bind to Asp157 at the bottom of the outward facing cavity. The approximate thickness of the hydrophobic core and headgroup regions of the membrane are indicated.
NapA, like other bacterial and mammalian Na+/H+ antiporters6,17,18 purifies as a dimer and is clearly dimeric in the structure with an extensive interface burying a surface area of 1800 Å2 (Fig. 3 and Supplementary Fig. 1c). In molecular dynamics (MD) simulations the dimer sits entirely within a model membrane bilayer (Fig. 3c). The backbone r.m.s.d. from the starting conformation of each monomer increased to about 2.0 Å over the time course of the simulation (Supplementary Fig. 6), indicating a slow relaxation of the crystal structure in the native-like membrane environment. The dimer has a crystallographic 2-fold operator approximately parallel to the transmembrane helices, and it is largely made up of tight hydrophobic helix-helix packing between TM–1 on one monomer and TM7 on the other. There are also contacts between the ends of TM2 and TM9 (Fig. 3a and b). NapA lacks the β-hairpin domain that makes most of the contact between protomers at the extracellular membrane surface in NhaA4,19. The dimer interface in NapA more closely resembles the dimer interface modeled in the 7 Å electron crystallography structure of NhaP120, a CPA1 member from Methanococcus jannaschii. NhaP1 also has 13 TMs built up from two 6-TM topology repeat units.
The substrate-binding cavity is open to the extracellular side. The cavity begins above the Dimerisation interface and funnels between the Dimerisation and Core domains, ending in the middle of the membrane (Fig. 3). It is considerably more open than the inward-facing cavity of NhaA4 and is negatively charged, being lined with glutamate residues. Although there is a similar distribution of charged residues in NhaA and NapA, few, including those that have been predicted to be involved in pH sensing1, are conserved between the two proteins (Supplementary Fig. 7). Near the base of the cavity are two highly conserved aspartates, Asp156 and Asp157, located on TM5 (Supplementary Fig. 2 and 7). The residues equivalent to these aspartates in NhaA, Asp163 and Asp164 (Supplementary Fig. 2 and 7) likely coordinate ion binding based on their position, conservation with mammalian Na+/H+ antiporters2, phenotypes of mutants1,21, isothermal titration calorimetry (ITC) experiments9 and MD simulations8. In NapA, mutation of either residue to alanine15 or asparagine abolishes transport activity completely (Supplementary Fig. 1d and Supplementary Table 2). With NapA crystals grown at pH 7.8, the aspartates are likely to be deprotonated. Consistent with this, Asp157 is orientated towards the centre of the cavity rather than hydrogen bonding with the backbone of TM4 as seen for Asp164 in NhaA, an interaction that requires Asp164 to be protonated (Supplementary Fig. 5c). Using solid-state membrane electrophysiology it was previously shown that NhaA transports cations equally well in either direction and the transport activity profile fits a simple H+ vs Na+ kinetic binding model to a single common site22. To investigate if Na+ ions would bind as expected we carried out equilibrium MD simulations of outward-facing NapA in a model membrane bilayer. Simulations were carried out with both Asp157 and Asp156 deprotonated, as they are likely to be in the crystal structure, and also as combinations of their neutral and charged forms. With both aspartates charged, Na+ ions spontaneously entered the negatively charged extracellular cavity to bind to Asp157 (Fig. 3c and Supplementary Fig. 8). Na+ ions were concentrated at Asp157 (Supplementary Fig. 8) and multiple distinct binding and unbinding events could be observed, which is qualitatively consistent with weak binding. In contrast, Na+ ion binding was not observed when Asp157 was protonated and was drastically reduced when Asp156 was neutral (Supplementary Fig. 8). In MD simulations of inward-facing NhaA, Na+ binds to the equivalent aspartate (Asp164)8, which is positioned at the base of the cytoplasmic cavity.
Asp156 and Asp157 on TM5 are located next to the point at which the two antiparallel discontinuous helices, TMs 4a-b and 11a-b, cross over in the Core domain (Supplementary Fig. 5c). Although discontinuous helices are a common feature for ion-binding in transporters23 the antiparallel crossing over of TMs is a unique feature of the NhaA fold4,24. It results in the positive dipole ends of TMs 4a and 11a facing one another and likewise the negative dipole ends of TMs 4b and 11b. In NhaA, the dipoles are proposed to be neutralised by the side-chains of an aspartate (Asp133; TM4a-4b) and a lysine (Lys300; TM10) respectively (Supplementary Fig. 5c). The lysine (Lys305) is retained in NapA but the aspartate is replaced by a serine, as it is in human Nha2 (Supplementary Fig. 5c and 7). From the structure, however, it appears the side-chain of Glu333 from TM11b can take a similar position to the carboxylate of Asp133 (Supplementary Fig. 5c and Supplementary Fig. 7). The positions of these two residues are pseudo-symmetrically related; the functional significance of this swap is unclear. While the mutation of Glu333 to alanine affected the apparent affinity for Li+ only slightly (< 3-fold), the affinity for Na+ was severely decreased (> 15-fold), which is in agreement with the results obtained from an Asp133 to alanine mutation in the NhaA protein (Supplementary Table 2)25. In contrast, the mutation of the highly conserved Lys305 to alanine severely decreased both Na+ and Li+ affinity (>20-fold, Supplementary Table 2). It was recently speculated that Lys300 in NhaA could have more than a stabilising role and that it could also form part of the pH activation mechanism, as mutation to arginine changed the pH at which NhaA becomes active9. In the NapA structure, Lys305 forms a salt-bridge with Asp156 (Supplementary Fig. 5c and 8a). Interestingly we have recently observed a similar interaction in a different crystal form of NhaA at low pH (O.B et al, manuscript in preparation). We would expect, however, that upon cation binding to Asp156 and Asp157 the salt-bridge is disrupted. As such, the interaction observed here supports the role of lysine in pH activation.
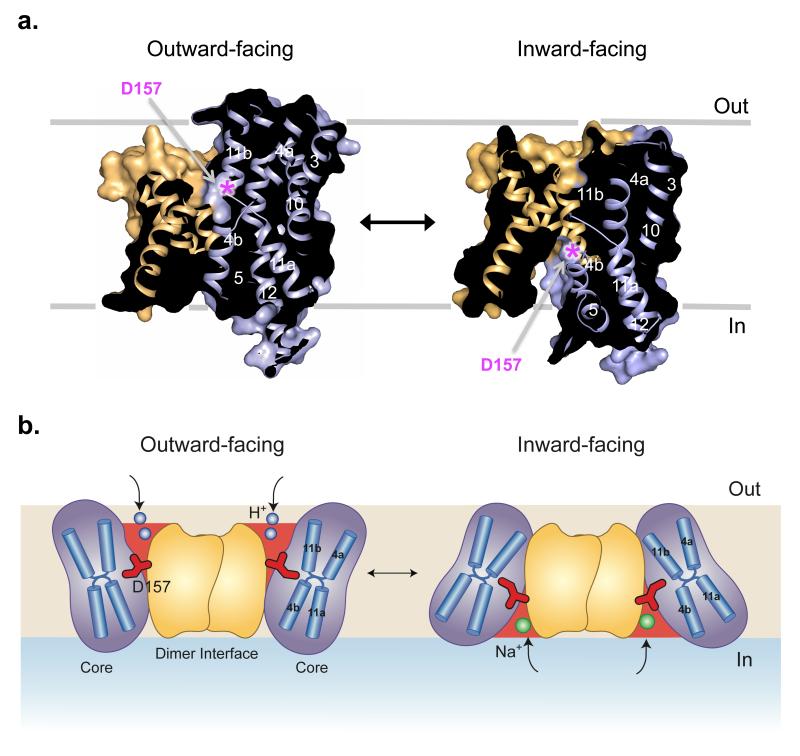
Previously, it was proposed that the exceptionally fast transport of Na+/H+ antiporters would primarily involve local rearrangements of the finely electrostatically balanced discontinuous helices in the Core domain1,4. However, the major structural difference between NapA and NhaA is in the position of the Core domain relative to the Dimerisation domain (Supplementary Fig. 5a and b). With reference to the Dimerisation interface, which seems likely to remain stable during transport across the membrane19,26, the Core domain in the NhaA structure is rotated by 21° relative to the Core of NapA (Fig. 4a and b). This large rotation of the Core domain closes the cavity seen on the outside of NapA and opens the cytoplasmic funnel on the inside, as observed in NhaA (Supplementary Video 1 and 2). During this process the two cation binding aspartates, which are in line with the tip of TM 8 in the Dimerisation domain of the outward-facing NapA structure, are shifted 10 Å towards the cytoplasmic surface of the transporter (Fig. 4a). This elevator movement of a substrate-binding Core domain, in this case to carry Na+ (Li+) ions from one side of the membrane to the other in exchange for protons, resembles that of the transport mechanism seen in the glutamate transporter GltPh27. A two domain transport mechanism was also recently predicted in NhaA, based on a consideration of the two symmetry related inverted repeats as well as elastic network models and biochemical cross-linking28. Such a mechanism was also proposed for the bile acid sodium symporter ASBTNM24, which is a structural homologue of NhaA and NapA.
Fig. 4. Alternating access model of sodium-proton antiport.
a, Surface representation showing a section through the outward-facing NapA structure (left) and inward-facing NapA model (right), (see Methods and Supplementary Video 1 and 2). The position of Asp157 is denoted with a pink asterisk and the helices have been coloured as in Figure 3. For the sake of clarity only one molecule is shown. b, Schematic of the proposed transport mechanism that illustrates the conformational changes with the Core moving against the Dimerisation domain. Protons (shown in blue) bind to the Core domain in the outward-facing state (left) causing it to switch to the inward-facing state (right). On the inside, protons are exchanged for sodium (green) and the Core domain moves back to the outside. Asp157 (shown in red), which is critical for binding both one of the protons and the sodium ion, moves ~ 10 Å during this process. Other residues involved in ion binding are not shown.
In summary, the structure of NapA is consistent with a single ion-translocation site mechanism, of which the strictly conserved Asp157 (Asp164 in NhaA) is ideally positioned for binding ions in outward and inward-facing states (Fig. 4b). To provide alternating access to the aspartate, however, requires a surprisingly large movement of the Core domain that twists around the Dimerisation interface, in essence creating a two-domain rocking bundle model. Although further structures are needed to clarify how ion binding and release is coupled to these global changes, the NapA structure provides a clear example that the extent of the conformational change may not necessarily correlate with the rate of transport or the size of the substrate transported, as previously assumed. Thus, the NapA structure also reveals fresh mechanistic insights relevant to all types of ion-coupled transporters.
METHODS
Thermus thermophilus NapA sequence (uniprot accession number: Q72IM4); residues progressively substituted to cysteine are underlined and in bold and additional C-terminal residues retained after TEV cleavage are shown in italics (see next section for cloning details).
MHGAEHLLEIFYLLLAAQVMAFIFKRLNQPVVIGEVLAGVLVGPALLGLVHEGEILEFLAELGAVFLLFMVGLETRLKDILAVGKEAFLVAVLGVALPFLGGYLYGLEIGFETLPALFLGTALVATSVGITARVLQELGVLSRPYSRIILGAAVIDDVLGLIVLAVVNGVAETGQVEVGAITRLIVLSVVFVGLAVFLSTLIARLPLERLPVGSPLGFALALGVGMAALAASIGLAPIVGAFLGGMLLSEVREKYRLEEPIFAIESFLAPIFFAMVGVRLELSALASPVVLVAGTVVTVIAILGKVLGGFLGALTQGVRSALTVGVGMAPRGEVGLIVAALGLKAGAVNEEEYAIVLFMVVFTTLFAPFALKPLIAWTERERAAKEGSENLYFQ
Expression screening, mutagenesis, protein purification and characterisation
NhaA homologues were cloned as GFP–His8 fusions into the vector pWaldoGFPe13, as fluorescence from the C-terminal GFP fusion is a reliable reporter of membrane-integrated expression29. The monodispersity of expressed fusions were screened in a number of different detergents by fluorescence-detection size-exclusion chromatography (FSEC)30. NapA from T. thermophilus was selected as a suitable candidate showing stability in a wide range of detergents including the harsh detergent n-dodecyl-N,N-dimethylamine-N-oxide (LDAO)14. Expression levels of the protein were initially low in standard culture conditions, but improved significantly using MemStar, which is a new strategy for boosting expression of membrane proteins in E. coli (C.L. et al, manuscript in preparation). In brief, Lemo21(DE3) cells31 were grown at 37 °C in PASM-5052 media32, with and without selenomethionine incorporation, and induced with 0.4 mM IPTG at an OD600 of 0.5 for overnight incubation at 25 °C.
Wild type NapA and mutants generated by Quickchange™ protocol (Agilent technologies) were purified essentially as previously described33. Membranes were isolated from 5-L E. coli cultures and solubilised in 1% DDM (Generon) for 2 h in buffer containing 1xPBS, 150 mM NaCl and 10 mM imidazole. The suspension was cleared by ultracentrifugation at 120,000g for 1 h. The sample was mixed with 1 mL of Ni-NTA Superflow resin (Qiagen) per 1 mg of GFP–His8 and incubated for 2 h at 4 °C. Slurry was loaded onto a glass Econo-Column (Bio-Rad) and washed in 1xPBS buffer containing 0.1% DDM, 150 mM NaCl and 20 mM imidazole for 20 column volumes. Bound material was washed for a further 20 column volumes in the same buffer containing 50 mM imidazole. The NapA–GFP–His8 fusion was eluted in two column volumes of 1xPBS buffer containing 0.6% NM (Generon), 150 mM NaCl and 250 mM imidazole. The eluted protein was dialysed overnight in the presence of stoichiometric amounts of His6-tagged tobacco etch virus protease in 1.5-L of buffer containing 20 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.5% NM. Dialysed sample was passed through a 5-mL Ni-NTA His-Trap column (GE Healthcare), and the flow-through containing NapA was collected. Protein was concentrated to 10 mg. mL−1 using concentrators with a relative molecular mass cut-off of 100kDa, and was loaded onto a Superdex 200 10/300 gel filtration column (GE Healthcare) equilibrated in 20 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.45% NM (Anatrace). The protein peak was collected and concentrated to 10 mg. mL−1 for crystallisation.
Purified NapA was loaded onto a Superdex 200 10/300 size exclusion column (GE Healthcare) coupled to a Viscotek TDAmax tetra detector array (Malvern) with GPCmax solvent pump and integrated auto-sampler, using the OmniSEC software for data analysis. The SEC-UV/LS/refractive index (RI) system was equilibrated in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl and 0.03% DDM at a flow rate of 0.3 mL/min. Standard gel filtration molecular weight markers were used for calibration and all proteins were analysed under the same experimental conditions. Data were collected from the RI, Right Angle LS (RALS) and UV280 detectors. The oligomeric state of NapA from the NapA-detergent micelle was calculated using methods described previously34.
NapA transport activity
In this setup (Fig. 1a), the described Na+/H+ antiport with inverted membrane vesicles was mimicked3,15. For this, we co-reconstituted E. coli FoF1 ATP synthase and T. thermophilus NapA in the same vesicles. While ATP synthase under these conditions has been shown to orient unidirectionally35, NapA most probably has a heterogeneous orientation, which might affect apparent KM values. Furthermore, a mixture of liposomes containing either none, one or two enzymes is expected. Only the liposomes containing ATP synthase will lead to initial ACMA quenching, but only those containing both ATP synthase and NapA will lead to dequenching after addition of the coupling ion. Additionally, it is expected that a new equilibrium is reached after every addition, which might influence the extent of dequenching. This is demonstrated by addition of NH4Cl at the end of every measurement, where any remaining ΔpH is dissipated. In brief, purified NapA wild type and mutants were co-reconstituted with purified ATP synthase from E. coli with an ~2:1 molar ratio (NapA : ATP synthase) in MME buffer (10 mM MOPS-NaOH, pH 7.5, 2.5 mM MgCl2, 100 mM KCl) as described for ATP synthase35,36. Typically, 50 μL proteoliposomes were diluted into 1.5 mL MME buffer containing 3 nM 9-aino-6-chloro-2-methoxyacridine (ACMA) and 140 nM valinomycin. Fluorescence was monitored at 480 nm using an excitation wavelength of 410 nm in a fluorescence spectrophotometer (Cary Eclipse, Agilent Technologies). An outward-directed pH gradient (acidic inside) was established by the addition of 2 mM ATP, as followed by a change in ACMA fluorescence. After ~2 min equilibration, the activity of NapA wildtype and mutants thereof was assessed by the dequenching of ACMA fluorescence after addition of the indicated concentrations of NaCl or LiCl. Addition of 20 mM NH4Cl leads to near complete dequenching. Each experiment was performed in triplicate.
Na+/H+ antiport activity by NapA is electrogenic
In this setup, we followed ΔpNa or Δψ driven H+ transport as a consequence of electrogenic Na+/H+ exchange activity. In this sensitive assay, liposomes containing the highly soluble and membrane impermeable pH sensitive dye pyranine were used to follow H+ influx or efflux. A Na+ gradient was established by the addition of NaCl, while an electrical membrane potential was established with a K+/valinomycin diffusion potential.
Reconstitution of NapA into liposomes containing pyranine was essentially performed as described37. Briefly, to a 500 μL liposome (40-80 nm) suspension (soy bean lipids, type II, SIGMA, 20 mg/ml) in buffer A (10 mM MOPS-PO4, pH 7.5), 45 μL cholate (20 % stock solution) and 17 μL NapA (50 μM, purification buffer) was added and incubated for 30 min at RT with occasional mild mixing. The cholate was removed via a PD-10 gel filtration column (GE Healthcare) equilibrated with buffer A and the proteoliposomes in the void volume were collected (~1.2 mL). They were diluted to 8 mL with buffer A, collected via ultracentrifugation (200,000g, 4 °C, 30 min) and resuspended in 250 μL buffer A. 125 μL of proteoliposomes was then mixed with 1 mM pyranine (0.1 M stock solution) and the desired Na2SO4 and K2SO4 concentrations, frozen in liquid nitrogen, thawed in water and briefly sonicated in a bath type sonicator (2 × 5 s). The freeze/sonication procedure was repeated once. The external pyranine was subsequently removed via a prepacked G25 gel filtration column (GE Healthcare) and the proteoliposomes, equilibrated in buffer A, were collected from the void volume of the column.
Pyranine fluorescence measurements monitoring pH changes on the inside of the proteoliposomes were performed as described37. Typically, an amount of 20 μL liposomes containing the desired Na+ and K+ concentrations was mixed with 2.5 mL buffer containing the same buffer with the appropriate salt concentrations. After 30 s, exchange activity was either initiated by the addition of 50 mM Na+ to the outside (0.5 mM Na+ on the inside), establishing an inwardly-directed Na+ gradient (~120 mV), and leading to H+ efflux. Accordingly, in a system in the absence of Na+ gradient (50 mM Na+ on both sides), but in the presence of a K+ gradient (100 mM K+ inside, 1 mM K+ outside liposomes), addition of valinomycin (10 nM) established a membrane potential of ~120 mV (inside negative) driving H+ influx (and Na+ efflux).
Crystallisation and preliminary screening
Crystals were grown at 20 °C using the hanging drop vapor diffusion method. A 1 μ L aliquot of pure protein was mixed 1:1 with reservoir solution containing 0.001 M zinc sulphate, 0.05 M HEPES pH 7.8 and 22–36% PEG 600 (mutants) or 0.05 M glycine pH 9.0, 0.05 M magnesium acetate, 26% PEG 400 (wild type). Crystals appeared overnight and reached maximum size after 3–4 days. For specified crystals, coverslides were transferred for overnight incubation with reservoir solution containing 2% increments of PEG 400. Dehydrated crystals in 30-36% PEG 400 were finally soaked with 1 μ L reservoir solution containing 1% NM and 40% PEG 400 followed by flash freezing in liquid nitrogen prior to data collection.
Data were collected at the European Synchrotron Radiation Facility and Diamond Light Source. The majority of the crystals were triclinic, however very occasionally crystals with different space groups were observed. The highest resolution data were collected from an orthorhombic crystal of the triple mutant that had been reannealed on the beamline. This crystal was grown with 0.025 % dichloromethane (Hampton Research) as an additive.
Structure determination
To obtain phases cysteine mutants were introduced into the protein to enable derivatisation with mercury. Three positions were chosen (Met20, Val166 and Val326) and single, double and triple mutants created as described above. Mercury-derivatised protein was prepared by incubation of the protein at 20 °C for 1 h with 2.5 mM mercury acetate. The structure was solved using MIRAS from four triclinic crystals (native, mercury derivatised and selenomethionine) as shown in Supplementary Table 1. Data were processed using the Xia2 pipeline38 to XDS39, with further processing using the CCP4 suite of programs40. Heavy atom sites were located from anomalous difference Patterson maps of the double mutant using the program RSPS41. Phases were calculated and refined in SHARP42. Further mercury and selenomethionine sites were located in residual maps. The crystals contain four molecules in the asymmetric unit. Non-crystallographic symmetry operators were determined from the heavy atom positions and averaging was carried out in DM43 with a mask calculated in O44. This map was then used as a search model for molecular replacement in Phaser45 for the P21 and C2221 data sets. Based on the operators obtained, multi-crystal averaging in DMMULTI43 using the P1 (4 molecules in au), P21 (2 molecules in au) and C2221 (1 molecule in au) data sets was carried out. This gave maps of sufficient quality to see all 13 helices of the NapA subunit. Model building was carried out in O44 and COOT46 and was facilitated by the positions of the mercury and selenomethionine peaks. Refinement of the atomic coordinates and individual B-factors using the C2221 data at 2.9 Å was carried out in PHENIX47. Secondary structure restraints were applied and refinement was interspersed with rebuilding using O and COOT. Peaks in anomalous difference maps indicated the presence of 2 zinc ions bound to the protein on the periplasmic surface to residues at the N-terminus of TM-1 and the loop between TM1 and 2. The final refinement statistics are shown in Supplementary Table 1.
Maps were calculated based on the final refined model for the data from the wild type selenomethionine derivatised crystals (Supplementary Table 1). At 3.7 Å resolution no clear differences were observed and the positions of the peaks in the anomalous difference maps were consistent with the positions of the methionines in the structure (data not shown).
Superpositions were carried out in Lsqman48 such that all matching Cα pairs were less than 3.8 Å apart after superposition. Figures were drawn using Pymol49 except those showing electron density, which were made using the CCP4mg50. The inward-facing model of NapA was created by superposing the Core and Dimerisation domains of NapA onto the corresponding domains of NhaA separately. The only adjustments that were made to the model were to the polypeptide chain between the two domains. No modifications were made to alleviate the clashes between the loop between TM −1 and TM 1 and TM 4a. The video was made in Lsqman48 by morphing between the outward-facing crystal structure and the inward-facing model displayed using Pymol48.
Molecular dynamics simulations
Molecular dynamics (MD) simulations of the NapA dimer in a mixed 1-palmitoyl-2-oleoylphosphatidylethanolamine (POPE)/1-palmitoyl-2-oleoylphosphatidylglycerol (POPG) bilayer were carried out with the Gromacs simulation package51, either version 4.5.5 or a development version of 4.6. All simulations employed the CHARMM force field including CMAP52,53 with the original TIP3P water model54 and updated CHARMM parameters for POPE and POPG lipids (CHARMM)55, as implemented in Gromacs51. The ratio of POPE to POPG molecules was about 4:1 in order to approximate the major components of the E. coli membrane. We employed a multi-scale approach to embed the dimer into the membrane56. The protein was first simulated in a coarse grained representation57 and the membrane was allowed to self-assemble around the protein from a random mixture of lipids and water in the simulation box58. After 200 ns simulation with an integration time step of 20 fs the bilayer had assembled around the NapA dimer. In the second step of the multi-scale approach, the system was converted to the CHARMM atomistic representation with the CG2AT protocol56 and the original crystal structure inserted in place of the back-translated protein. The simulation system consisted of an orthorhombic simulation box of size 114 Å × 114 Å × 91 Å containing 118,832 atoms in 768 protein residues, 215 POPC and 57 POPG lipids, 180 Na+, 109 Cl− ions and 24,575 water molecules. The free NaCl concentration was about 250 mM in all simulations in order to approximate the outward-open transporter facing an environment of elevated salt-stress.
Most titratable residues were predicted by PROPKA59 to be in their default charge states at pH 7.8 and were simulated as such (including deprotonated Asp156 and Asp157, with predicted pKa as 3.2 and 5.7). The pKa of the buried Lys305 was predicted to be 9.5, with its charged form partially stabilised by a salt-bridge with the (charged) Asp156. Therefore, Lys305 was simulated in its protonated (positively charged) form. For a number of residues the predicted value was within 1 unit of the environmental pH value we wished to simulate and for those residues we rationalised our choices as follows: The pKa of Glu333 was predicted as 7.0. Glu333 might help to stabilise the helix dipoles of helices TM 4a and TM 11a (similar to Asp133 in NhaA) but such a charge-dipole interaction is not encoded in the empirical rules of the PROPKA algorithm59. Furthermore, simulations clearly showed that Glu333 protrudes into the outward facing cavity and is solvated. We, therefore, adopted a charged Glu333. His6 and His51, which reside on the surface and do not seem to have any specific mechanistic role, had predicted pKa of 7.8 and 7.1 and were modeled in their dissociated (neutral) form.
Equilibrium MD simulations were performed with periodic boundary conditions at constant temperature T=323 K and pressure P=1 bar using the velocity rescaling algorithm for the thermostat (time constant 0.1 ps)60 and semi-isotropic Parrinello-Rahman barostat (time constant 5.0 ps, compressibility 4.6×10−5 bar−1, coupling every 10 steps)61. Long range corrections for energy and pressure were applied62. Lennard-Jones interactions were switched off between 8 Å and 12 Å while electrostatic interactions were handled by the SPME method63 where Coulomb interactions were computed in real space up to a cutoff of 12 Å and long range interactions beyond the cutoff were calculated in reciprocal space with fast fourier transforms on a grid with spacing 1.2 Å and 4th order splines for fitting of the charge density. The grid-based neighbour list was updated every 5 steps to a distance of 14 Å. Bonds to hydrogen atoms were constrained with the P-LINCS algorithm62 or SETTLE (for water molecules)64. The classical equations of motions were integrated with a leap frog integrator and a time step of 2 fs. Conformations were saved every 1 ps for analysis.
The simulation protocol included an initial energy minimisation of the atomistic system and a 1-ns equilibrium simulation during which the protein heavy atoms were restrained with a harmonic force with force constant of 1000 kJ mol−1 nm−2. An initial unrestrained simulation of the dimer with Asp156 and Asp157 in their default protonation state was run for 100 ns. Eight additional 100-ns simulations were performed in three sets (two simulations in set 1, five in set 2, one in set 3). The starting configuration for each set was generated from the last frame of the initial simulation by exchanging any sodium ion within 3 Å of Asp157 or Asp156 with a random bulk water molecule. Repeats in each set always differed by the seed of the random number generator, thus leading to differing initial assignments of velocities and generation of independent trajectories through the stochastic component of the velocity rescaling thermostat60, as seen from the different r.m.s.d. time series in Supplementary Fig. 6.
Additional simulations were performed to assess the influence of the protonation state of the conserved residues Asp156 and Asp157 on sodium binding as performed previously for NhaA8. Two independent 100-ns simulations were performed for each of (1) Asp156 deprotonated (negatively charged) and Asp157 protonated (neutral), (2) Asp156 protonated and Asp157 deprotonated, and (3) both Asp156 and Asp157 protonated. Charge states were modified with the Gromacs tool pdb2gmx50. These six simulations used the 100-ns frame of the initial simulation (default charge states) with any sodium ions near the aspartates exchanged with a bulk water molecule as a well-equilibrated starting conformation. In each case, a 1-ns position restraint simulation (as above) was followed by a 100-ns production equilibrium simulation.
Simulations were analysed with MDAnalysis65 and Gromacs tools51. In order to calculate the sodium density, data from all nine simulations with the deprotonated aspartates were used at 1-ps intervals. Ion binding and unbinding to each protomer appeared to be independent so that data for both protomers were combined by superpositioning trajectories of both protomer A and protomer B on the coordinates of protomer A from the start of the initial simulation. The density was calculated by histogramming sodium coordinates in cubic volume elements at a resolution of 1 Å in a fixed coordinate system defined by the initial coordinates of protomer A. The thicknesses of bilayer regions were calculated from the distributions of the headgroup phosphate and acyl chain atoms along the membrane normal, using trajectories superpositioned on the dimer.
Images showing simulation data were prepared with VMD66 and the Bendix plugin for curved helices67 or UCSF Chimera68,69.
Supplementary Material
Acknowledgements
We are grateful to Dr. Dirk Slotboom for critical reading of the manuscript and Dr. Nien-Jen Hu for assistance in data collection. Data were collected at the European Synchrotron Radiation Facility and Diamond Light Source, with excellent assistance from beamline scientists. This work was funded by grants from the Medical Research Council (MRC grant, G0900990 to A.C. and D.D.), the Swedish Research Council (to C.vB and D.D.) and the BBSRC (BB/G02325/1 to Prof. So Iwata). The authors are grateful for the use of the Membrane Protein Laboratory funded by the Wellcome Trust (grant 062164/Z/00/Z) at the Diamond Light Source Limited and The Centre for Biomembrane Research (CBR), supported by the Swedish Foundation for Strategic Research. Computer simulations were partially run on XSEDE resources (grant TG-MCB120151, to O.B.). C.L. was a recipient of a BBSRC-funded PhD scholarship, H.K. a Human Frontiers Postdoctoral fellowship, and D.D. acknowledges the support from The Royal Society through the University Research Fellow (URF) scheme.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Padan E. The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem Sci. 2008;33:435–443. doi: 10.1016/j.tibs.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:C223–239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 3.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 4.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 5.Williams KA, Geldmacher-Kaufer U, Padan E, Schuldiner S, Kuhlbrandt W. Projection structure of NhaA, a secondary transporter from Escherichia coli, at 4.0 Å resolution. EMBO J. 1999;18:3558–3563. doi: 10.1093/emboj/18.13.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams KA. Three-dimensional structure of the ion-coupled transport protein NhaA. Nature. 2000;403:112–115. doi: 10.1038/47534. [DOI] [PubMed] [Google Scholar]
- 7.Kozachkov L, Padan E. Conformational changes in NhaA Na(+)/H(+) antiporter. Mol Membr Biol. 2012 doi: 10.3109/09687688.2012.693209. [DOI] [PubMed] [Google Scholar]
- 8.Arkin IT, et al. Mechanism of Na+/H+ antiporting. Science. 2007;317:799–803. doi: 10.1126/science.1142824. [DOI] [PubMed] [Google Scholar]
- 9.Maes M, Rimon A, Kozachkov-Magrisso L, Friedler A, Padan E. Revealing the Ligand Binding Site of NhaA Na+/H+ Antiporter and its pH Dependence. J Biol Chem. 2012 doi: 10.1074/jbc.M112.391128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West IC, Mitchell P. Proton/sodium ion antiport in Escherichia coli. Biochem J. 1974;144:87–90. doi: 10.1042/bj1440087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taglicht D, Padan E, Schuldiner S. Proton-sodium stoichiometry of NhaA, an electrogenic antiporter from Escherichia coli. J Biol Chem. 1993;268:5382–5387. [PubMed] [Google Scholar]
- 12.Pinner E, Padan E, Schuldiner S. Kinetic properties of NhaB, a Na+/H+ antiporter from Escherichia coli. J Biol Chem. 1994;269:26274–26279. [PubMed] [Google Scholar]
- 13.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. Optimization of membrane protein overexpression and purification using GFP fusions. Nat Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- 14.Sonoda Y, et al. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure. 2011;19:17–25. doi: 10.1016/j.str.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furrer EM, Ronchetti MF, Verrey F, Pos KM. Functional characterization of a NapA Na(+)/H(+) antiporter from Thermus thermophilus. FEBS Lett. 2007;581:572–578. doi: 10.1016/j.febslet.2006.12.059. [DOI] [PubMed] [Google Scholar]
- 16.Padan E, et al. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+ antiporter. Biochim Biophys Acta. 2004;1658:2–13. doi: 10.1016/j.bbabio.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Rimon A, Tzubery T, Padan E. Monomers of the NhaA Na+/H+ antiporter of Escherichia coli are fully functional yet dimers are beneficial under extreme stress conditions at alkaline pH in the presence of Na+ or Li+ J Biol Chem. 2007;282:26810–26821. doi: 10.1074/jbc.M704469200. [DOI] [PubMed] [Google Scholar]
- 18.Hisamitsu T, Ben Ammar Y, Nakamura TY, Wakabayashi S. Dimerization is crucial for the function of the Na+/H+ exchanger NHE1. Biochemistry. 2006;45:13346–13355. doi: 10.1021/bi0608616. [DOI] [PubMed] [Google Scholar]
- 19.Appel M, Hizlan D, Vinothkumar KR, Ziegler C, Kuhlbrandt W. Conformations of NhaA, the Na+/H+ exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J Mol Biol. 2009;388:659–672. doi: 10.1016/j.jmb.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Goswami P, et al. Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1. EMBO J. 2011;30:439–449. doi: 10.1038/emboj.2010.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwabara N, Inoue H, Tsuboi Y, Nakamura N, Kanazawa H. The fourth transmembrane domain of the Helicobacter pylori Na+/H+ antiporter NhaA faces a water-filled channel required for ion transport. J Biol Chem. 2004;279:40567–40575. doi: 10.1074/jbc.M401132200. [DOI] [PubMed] [Google Scholar]
- 22.Mager T, Rimon A, Padan E, Fendler K. Transport Mechanism and pH Regulation of the Na+/H+ Antiporter NhaA from Escherichia coli: AN ELECTROPHYSIOLOGICAL STUDY. J Biol Chem. 2011;286:23570–23581. doi: 10.1074/jbc.M111.230235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Screpanti E, Hunte C. Discontinuous membrane helices in transport proteins and their correlation with function. J Struct Biol. 2007;159:261–267. doi: 10.1016/j.jsb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Hu NJ, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011;478:408–411. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noumi T, Inoue H, Sakurai T, Tsuchiya T, Kanazawa H. Identification and characterization of functional residues in a Na+/H+ antiporter (NhaA) from Escherichia coli by random mutagenesis. J Biochem. 1997;121:661–670. doi: 10.1093/oxfordjournals.jbchem.a021637. [DOI] [PubMed] [Google Scholar]
- 26.Vinothkumar KR, Smits SH, Kuhlbrandt W. pH-induced structural change in a sodium/proton antiporter from Methanococcus jannaschii. EMBO J. 2005;24:2720–2729. doi: 10.1038/sj.emboj.7600727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462:880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schushan M, et al. A model-structure of a periplasm-facing state of the NhaA antiporter suggests the molecular underpinnings of pH-induced conformational changes. J Biol Chem. 2012;287:18249–18261. doi: 10.1074/jbc.M111.336446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 29.Drew DE, von Heijne G, Nordlund P, de Gier JW. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 2001;507:220–224. doi: 10.1016/s0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- 30.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Wagner S, et al. Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci U S A. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Drew D, et al. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat Protoc. 2008;3:784–798. doi: 10.1038/nprot.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slotboom DJ, Duurkens RH, Olieman K, Erkens GB. Static light scattering to characterize membrane proteins in detergent solution. Methods. 2008;46:73–82. doi: 10.1016/j.ymeth.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Wiedenmann A, Dimroth P, von Ballmoos C. Functional asymmetry of the F(0) motor in bacterial ATP synthases. Mol Microbiol. 2009;72:479–490. doi: 10.1111/j.1365-2958.2009.06658.x. [DOI] [PubMed] [Google Scholar]
- 36.Ishmukhametov RR, Galkin MA, Vik SB. Ultrafast purification and reconstitution of His-tagged cysteine-less Escherichia coli F1Fo ATP synthase. Biochim Biophys Acta. 2005;1706:110–116. doi: 10.1016/j.bbabio.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Wiedenmann A, Dimroth P, von Ballmoos C. Deltapsi and DeltapH are equivalent driving forces for proton transport through isolated F(0) complexes of ATP synthases. Biochim Biophys Acta. 2008;1777:1301–1310. doi: 10.1016/j.bbabio.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Winter G. xia2: an expert system for macromolecular crystallography data reduction. Journal of Appl Crystallogr. 2010;43:186–190. [Google Scholar]
- 39.Kabsch W. XDS. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Knight SD. RSPS version 4.0: a semi-interactive vector-search program for solving heavy-atom derivatives. Acta Crystallogr D Biol Crystallogr. 2000;56:42–47. doi: 10.1107/s0907444999012913. [DOI] [PubMed] [Google Scholar]
- 42.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 43.Cowtan K. ‘dm’: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 44.Jones TA, Kjeldgaard M, Charles WC, Jr., Robert MS. Electron-density map interpretation. Methods in Enzymology. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleywegt GJ, Jones TA. A super position. ESF/CCP4 Newsletter. 1994;31:9. [Google Scholar]
- 49.Delano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 50.Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 51.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theo Comp. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 52.MacKerell A, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 53.MacKerell J, A. D, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comp Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 54.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 55.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stansfeld PJ, Sansom MSP. From Coarse Grained to Atomistic: A Serial Multiscale Approach to Membrane Protein Simulations. J Chem Theo Comp. 2011;7:1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- 57.Bond PJ, Wee CL, Sansom MSP. Coarse-grained molecular dynamics simulations of the energetics of helix insertion into a lipid bilayer. Biochemistry. 2008;47:11321–11331. doi: 10.1021/bi800642m. [DOI] [PubMed] [Google Scholar]
- 58.Scott KA, et al. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–630. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 59.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins: Struct Funct Genet. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 60.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:01410. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 61.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 62.Hess B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J Chem Theo Comp. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 63.Essman U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8592. [Google Scholar]
- 64.Miyamoto S, Kollman PA. SETTLE: An Analytical Version of the SHAKE and RATTLE Algorithms for Rigid Water Models. J Comp Chem. 1992;13:952–962. [Google Scholar]
- 65.Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O. MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. J Comp Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Humphrey W, Dalke A, Schulten K. VMD — Visual Molecular Dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 67.Dahl ACE, Chavent M, Sansom MSP. Bendix: Intuitive helix geometry analysis and abstraction for VMD. Bioinformatics. 2012;28:2193–2194. doi: 10.1093/bioinformatics/bts357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 69.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.