Abstract
In the title compound, C26H37N5OS, the piperazine ring adopts a chair conformation. The triazole ring forms dihedral angles of 67.85 (9) and 59.41 (9)° with the piperazine and benzene rings, respectively, resulting in an approximate V-shaped conformation for the molecule. An intramolecular C—H⋯O hydrogen bond generates an S(6) ring motif. The crystal structure features C—H⋯π interactions, producing a two-dimensional supramolecular architecture.
Related literature
For the pharmacological activity of adamantane derivatives and adamantyl-1,2,4-triazoles, see: Togo et al. (1968 ▶); El-Emam et al. (2004 ▶, 2013 ▶); Al-Deeb et al. (2006 ▶); Kadi et al. (2007 ▶, 2010 ▶). For related adamantyl-1,2,4-triazole structures, see: Al-Abdullah et al. (2013 ▶); Al-Tamimi, Alafeefy et al. (2013 ▶); Al-Tamimi, Al-Abdullah et al. (2013 ▶); El-Emam et al. (2012 ▶). For the synthesis of the starting material, see: El-Emam & Ibrahim (1991 ▶). For ring conformations and ring puckering analysis, see: Cremer & Pople (1975 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶).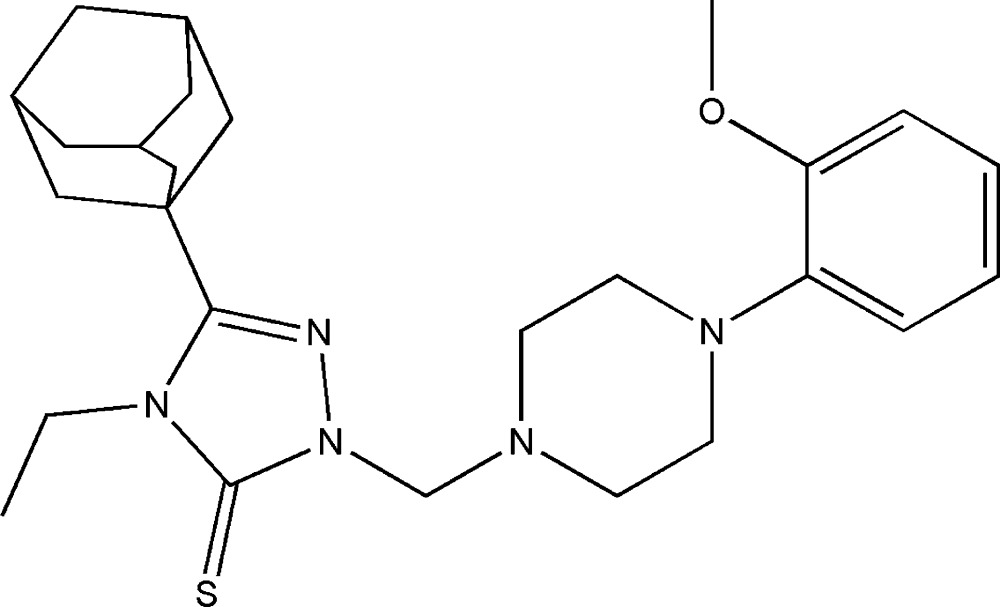
Experimental
Crystal data
C26H37N5OS
M r = 467.67
Monoclinic,
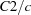
a = 19.8170 (3) Å
b = 11.9384 (3) Å
c = 21.7807 (4) Å
β = 107.886 (2)°
V = 4903.90 (17) Å3
Z = 8
Cu Kα radiation
μ = 1.39 mm−1
T = 296 K
0.98 × 0.62 × 0.41 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.344, T max = 0.599
15455 measured reflections
4029 independent reflections
3606 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.043
wR(F 2) = 0.115
S = 1.05
4029 reflections
308 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.19 e Å−3
Δρmin = −0.27 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813032789/rz5099sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813032789/rz5099Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536813032789/rz5099Isup3.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg is the centroid of the C1–C6 benzene ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C11—H11A⋯O1 | 0.97 | 2.26 | 2.903 (2) | 123 |
| C18—H18A⋯Cg i | 0.97 | 2.81 | 3.748 (2) | 162 |
Symmetry code: (i) 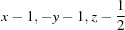 .
.
Acknowledgments
The financial support of the Deanship of Scientific Research and the Research Center for Female Scientific and Medical Colleges, King Saud University, is greatly appreciated. CSCK thanks Universiti Sains Malaysia for a postdoctoral research fellowship.
supplementary crystallographic information
1. Comment
Derivatives of adamantane have long been known for their diverse biological activities including antiviral activity against influenza (Togo et al., 1968) and HIV viruses (El-Emam et al., 2004). Moreover, adamantane derivative were reported to exhibit marked antibacterial and anti-inflammatory activities (Kadi et al., 2007, 2010; El-Emam et al., 2013). In continuation of our interest in the chemical and pharmacological properties of adamantane derivatives, and as part of our on-going structural studies of adamantane derivatives (Al-Abdullah et al., 2013); Al-Tamimi, Alafeefy et al., 2013; Al-Tamimi, Al-Abdullah et al., 2013; El-Emam et al., 2012), we have synthesized the title compound (I) as a potential chemotherapeutic agent.
In the crystal structure of the title compound (Fig. 1), the piperazine (N1–N2/C8–C11) ring adopts a chair conformation with puckering parameters: Q = 0.5783 (18) Å, θ = 178.03 (17)°, and φ = 25 (5)° (Cremer & Pople, 1975). The dihedral angle between the piperazine ring and the triazole ring (N3–N5/C13/C14) is 67.85 (9)°. The triazole ring forms a dihedral angle of 59.41 (9)° with the benzene ring (C1—C6), resulting in an approximate V-shape conformation of the molecule. An intramolecular C–H···O hydrogen bond generates an S(6) ring motif (Bernstein et al., 1995). The crystal structure features an intermolecular C–H···π interaction with a H18A···Cg distance of 2.81 Å, where Cg is the centroid of the benzene ring (C1—C6).
2. Experimental
A mixture of 527 mg (2 mmol) of 3-(1-adamantyl)-4-ethyl-4H-1,2,4- triazole-5-thiol (El-Emam & Ibrahim, 1991), 1-(2-methoxyphenyl)piperazine (383 mg, 2 mmol) and 37% formaldehyde solution (1 ml) in ethanol (8 ml) was heated under reflux for 15 min until a clear solution was obtained. Stirring was continued for 12 h at room temperature and the mixture was allowed to stand overnight. Cold water (5 ml) was added slowly and the mixture was stirred for 20 min. The precipitated crude product were filtered, washed with water, dried, and crystallized from ethanol to yield 860 mg (92%) of the title compound (C26H37N5OS) as colourless needle crystals. M.p.: 477–479 K. Single plate-shaped crystals suitable for X-ray analysis were obtained by slow evaporation of a CHCl3:EtOH solution (1:1 v/v; 5 ml) at room temperature.
1H NMR (CDCl3, 500.13 MHz): δ 1.32 (t, 3H, CH2CH3, J = 7.0 Hz), 1.71–1.76 (m, 6H, Adamantane-H), 1.98–2.12 (m, 9H, Adamantane-H), 3.08 (s, 8H, Piperazine-H), 3.81 (s, 3H, OCH3), 4.15 (q, 2H, CH2CH3, J = 7.0 Hz), 5.15 (s, 2H, CH2), 6.79–7.01 (m, 4H, Ar—H). 13C NMR (CDCl3, 125.76 MHz): δ 13.76 (CH2CH3), 27.92, 35.32, 36.48, 39.83 (Adamantane-C), 43.83 (CH2CH3), 47.40, 50.18 (Piperazine-C), 55.48 (OCH3), 72.58 (CH2), 111.43, 118.38, 121.12, 123.55, 152.13, 152.26 (Ar—C), 156.57 (Triazole C-5), 167.34 (C=S).
3. Refinement
The H atoms bound to atom C12 were located in a difference Fourier map and refined freely. All other H atoms were positioned geometrically [C—H = 0.93–1.01 Å] and refined using a riding model with Uiso(H) = 1.2 Ueq(C) or 1.5 Ueq(C) for methyl H atoms. A rotating group model was used for the methyl groups.
Figures
Fig. 1.
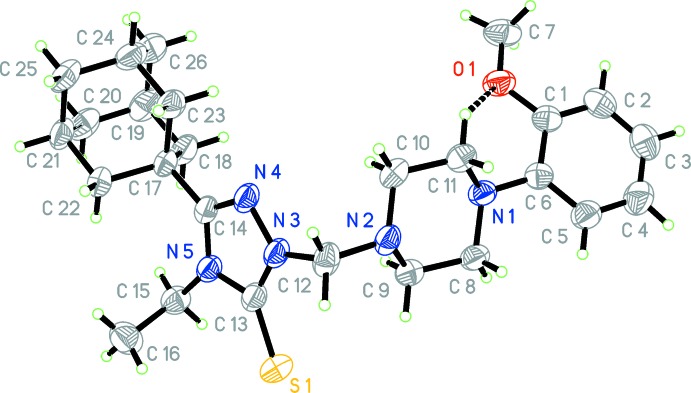
The molecular structure of the title compound with 50% probability displacement ellipsoids. The intramolecular hydrogen bond is shown as a dashed line.
Crystal data
| C26H37N5OS | F(000) = 2016 |
| Mr = 467.67 | Dx = 1.267 Mg m−3 |
| Monoclinic, C2/c | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: -C 2yc | Cell parameters from 4154 reflections |
| a = 19.8170 (3) Å | θ = 4.3–69.2° |
| b = 11.9384 (3) Å | µ = 1.39 mm−1 |
| c = 21.7807 (4) Å | T = 296 K |
| β = 107.886 (2)° | Plate, colourless |
| V = 4903.90 (17) Å3 | 0.98 × 0.62 × 0.41 mm |
| Z = 8 |
Data collection
| Bruker APEXII CCD diffractometer | 4029 independent reflections |
| Radiation source: fine-focus sealed tube | 3606 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.033 |
| φ and ω scans | θmax = 65.0°, θmin = 4.3° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −22→23 |
| Tmin = 0.344, Tmax = 0.599 | k = −9→14 |
| 15455 measured reflections | l = −25→21 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.115 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0627P)2 + 2.6908P] where P = (Fo2 + 2Fc2)/3 |
| 4029 reflections | (Δ/σ)max < 0.001 |
| 308 parameters | Δρmax = 0.19 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.04659 (3) | 0.66541 (4) | 0.36270 (3) | 0.06079 (18) | |
| O1 | −0.25953 (6) | 1.15940 (11) | 0.28999 (6) | 0.0511 (3) | |
| N1 | −0.12211 (7) | 1.09436 (12) | 0.31028 (6) | 0.0392 (3) | |
| N2 | −0.03369 (7) | 0.95212 (11) | 0.40765 (7) | 0.0401 (3) | |
| N3 | −0.04059 (7) | 0.74873 (11) | 0.42680 (7) | 0.0410 (3) | |
| N4 | −0.09869 (7) | 0.71981 (11) | 0.44558 (7) | 0.0402 (3) | |
| N5 | −0.06803 (7) | 0.58126 (11) | 0.39235 (6) | 0.0380 (3) | |
| C1 | −0.21966 (9) | 1.22602 (15) | 0.26341 (8) | 0.0414 (4) | |
| C2 | −0.24588 (10) | 1.32048 (17) | 0.22721 (9) | 0.0531 (5) | |
| H2A | −0.2916 | 1.3448 | 0.2233 | 0.064* | |
| C3 | −0.20535 (12) | 1.37922 (18) | 0.19688 (10) | 0.0612 (5) | |
| H3A | −0.2239 | 1.4422 | 0.1723 | 0.073* | |
| C4 | −0.13806 (12) | 1.34470 (18) | 0.20304 (10) | 0.0609 (5) | |
| H4A | −0.1110 | 1.3826 | 0.1816 | 0.073* | |
| C5 | −0.11006 (10) | 1.25299 (16) | 0.24133 (9) | 0.0502 (4) | |
| H5A | −0.0636 | 1.2317 | 0.2460 | 0.060* | |
| C6 | −0.14894 (9) | 1.19165 (14) | 0.27302 (8) | 0.0392 (4) | |
| C7 | −0.33410 (10) | 1.17746 (19) | 0.27037 (11) | 0.0604 (5) | |
| H7A | −0.3564 | 1.1195 | 0.2879 | 0.091* | |
| H7B | −0.3523 | 1.1760 | 0.2241 | 0.091* | |
| H7C | −0.3439 | 1.2490 | 0.2859 | 0.091* | |
| C8 | −0.05394 (9) | 1.05442 (15) | 0.30672 (9) | 0.0442 (4) | |
| H8A | −0.0175 | 1.1089 | 0.3266 | 0.053* | |
| H8B | −0.0560 | 1.0466 | 0.2619 | 0.053* | |
| C9 | −0.03495 (9) | 0.94278 (15) | 0.34052 (9) | 0.0447 (4) | |
| H9A | −0.0695 | 0.8868 | 0.3187 | 0.054* | |
| H9B | 0.0112 | 0.9190 | 0.3388 | 0.054* | |
| C10 | −0.10279 (9) | 0.98856 (14) | 0.41065 (8) | 0.0407 (4) | |
| H10A | −0.1018 | 0.9943 | 0.4553 | 0.049* | |
| H10B | −0.1385 | 0.9339 | 0.3894 | 0.049* | |
| C11 | −0.12140 (9) | 1.10112 (15) | 0.37793 (8) | 0.0420 (4) | |
| H11A | −0.1677 | 1.1245 | 0.3796 | 0.050* | |
| H11B | −0.0869 | 1.1565 | 0.4006 | 0.050* | |
| C12 | −0.00463 (9) | 0.85666 (15) | 0.44685 (9) | 0.0449 (4) | |
| C13 | −0.02030 (9) | 0.66608 (14) | 0.39407 (8) | 0.0419 (4) | |
| C14 | −0.11453 (8) | 0.61806 (13) | 0.42412 (8) | 0.0361 (4) | |
| C15 | −0.06398 (9) | 0.47423 (15) | 0.36034 (9) | 0.0460 (4) | |
| H15A | −0.0532 | 0.4888 | 0.3205 | 0.055* | |
| H15B | −0.1098 | 0.4374 | 0.3494 | 0.055* | |
| C16 | −0.00816 (11) | 0.39690 (17) | 0.40219 (12) | 0.0619 (5) | |
| H16A | −0.0079 | 0.3279 | 0.3796 | 0.093* | |
| H16B | −0.0187 | 0.3818 | 0.4416 | 0.093* | |
| H16C | 0.0375 | 0.4319 | 0.4120 | 0.093* | |
| C17 | −0.17657 (8) | 0.55496 (13) | 0.43342 (8) | 0.0364 (4) | |
| C18 | −0.23590 (9) | 0.54186 (17) | 0.36848 (9) | 0.0499 (5) | |
| H18A | −0.2510 | 0.6151 | 0.3501 | 0.060* | |
| H18B | −0.2181 | 0.5003 | 0.3384 | 0.060* | |
| C19 | −0.29894 (10) | 0.4796 (2) | 0.37923 (10) | 0.0603 (5) | |
| H19A | −0.3360 | 0.4702 | 0.3378 | 0.072* | |
| C20 | −0.27523 (11) | 0.36486 (17) | 0.40858 (10) | 0.0565 (5) | |
| H20A | −0.3155 | 0.3246 | 0.4142 | 0.068* | |
| H20B | −0.2563 | 0.3216 | 0.3799 | 0.068* | |
| C21 | −0.21867 (10) | 0.37929 (14) | 0.47347 (9) | 0.0475 (4) | |
| H21A | −0.2040 | 0.3053 | 0.4924 | 0.057* | |
| C22 | −0.15471 (9) | 0.43877 (14) | 0.46354 (8) | 0.0410 (4) | |
| H22A | −0.1357 | 0.3944 | 0.4354 | 0.049* | |
| H22B | −0.1180 | 0.4469 | 0.5047 | 0.049* | |
| C23 | −0.20662 (11) | 0.62237 (15) | 0.47925 (10) | 0.0507 (5) | |
| H23A | −0.2210 | 0.6960 | 0.4612 | 0.061* | |
| H23B | −0.1702 | 0.6319 | 0.5204 | 0.061* | |
| C24 | −0.27071 (11) | 0.56134 (16) | 0.48944 (11) | 0.0560 (5) | |
| H24A | −0.2893 | 0.6053 | 0.5186 | 0.067* | |
| C25 | −0.24790 (12) | 0.44675 (16) | 0.51877 (10) | 0.0559 (5) | |
| H25A | −0.2118 | 0.4550 | 0.5603 | 0.067* | |
| H25B | −0.2881 | 0.4081 | 0.5254 | 0.067* | |
| C26 | −0.32823 (11) | 0.54824 (19) | 0.42461 (13) | 0.0687 (6) | |
| H26A | −0.3431 | 0.6214 | 0.4060 | 0.082* | |
| H26B | −0.3691 | 0.5107 | 0.4307 | 0.082* | |
| H12B | 0.0444 (11) | 0.8413 (14) | 0.4491 (9) | 0.041 (5)* | |
| H12A | −0.0064 (10) | 0.8708 (16) | 0.4920 (10) | 0.047 (5)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0474 (3) | 0.0637 (3) | 0.0818 (4) | −0.0027 (2) | 0.0354 (3) | 0.0117 (3) |
| O1 | 0.0367 (6) | 0.0607 (8) | 0.0589 (7) | 0.0015 (5) | 0.0190 (6) | 0.0018 (6) |
| N1 | 0.0369 (7) | 0.0470 (8) | 0.0371 (7) | 0.0033 (6) | 0.0162 (6) | 0.0050 (6) |
| N2 | 0.0353 (7) | 0.0400 (7) | 0.0438 (7) | −0.0084 (6) | 0.0102 (6) | 0.0071 (6) |
| N3 | 0.0369 (7) | 0.0382 (7) | 0.0488 (8) | −0.0090 (6) | 0.0146 (6) | 0.0065 (6) |
| N4 | 0.0393 (7) | 0.0368 (7) | 0.0465 (8) | −0.0088 (6) | 0.0163 (6) | 0.0046 (6) |
| N5 | 0.0357 (7) | 0.0383 (7) | 0.0413 (7) | −0.0050 (5) | 0.0139 (6) | 0.0039 (6) |
| C1 | 0.0408 (8) | 0.0461 (9) | 0.0374 (8) | 0.0006 (7) | 0.0119 (7) | −0.0064 (7) |
| C2 | 0.0489 (10) | 0.0551 (11) | 0.0510 (10) | 0.0109 (8) | 0.0091 (8) | −0.0003 (9) |
| C3 | 0.0687 (13) | 0.0524 (11) | 0.0572 (11) | 0.0101 (10) | 0.0115 (10) | 0.0121 (10) |
| C4 | 0.0680 (13) | 0.0610 (12) | 0.0575 (12) | −0.0006 (10) | 0.0249 (10) | 0.0158 (10) |
| C5 | 0.0469 (10) | 0.0566 (11) | 0.0510 (10) | 0.0035 (8) | 0.0206 (8) | 0.0090 (9) |
| C6 | 0.0403 (8) | 0.0435 (9) | 0.0340 (8) | 0.0006 (7) | 0.0116 (6) | −0.0011 (7) |
| C7 | 0.0373 (10) | 0.0740 (13) | 0.0698 (13) | −0.0014 (9) | 0.0162 (9) | −0.0158 (11) |
| C8 | 0.0399 (9) | 0.0523 (10) | 0.0457 (9) | 0.0031 (7) | 0.0208 (7) | 0.0087 (8) |
| C9 | 0.0384 (9) | 0.0479 (9) | 0.0513 (10) | 0.0024 (7) | 0.0188 (7) | 0.0068 (8) |
| C10 | 0.0394 (8) | 0.0461 (9) | 0.0380 (8) | −0.0102 (7) | 0.0140 (7) | 0.0000 (7) |
| C11 | 0.0431 (9) | 0.0474 (9) | 0.0376 (8) | −0.0020 (7) | 0.0157 (7) | 0.0006 (7) |
| C12 | 0.0367 (9) | 0.0431 (9) | 0.0485 (10) | −0.0124 (7) | 0.0038 (7) | 0.0081 (8) |
| C13 | 0.0344 (8) | 0.0443 (9) | 0.0453 (9) | −0.0033 (7) | 0.0101 (7) | 0.0122 (8) |
| C14 | 0.0375 (8) | 0.0347 (8) | 0.0366 (8) | −0.0046 (6) | 0.0120 (6) | 0.0058 (7) |
| C15 | 0.0457 (9) | 0.0468 (9) | 0.0474 (9) | −0.0047 (8) | 0.0171 (8) | −0.0041 (8) |
| C16 | 0.0531 (11) | 0.0473 (10) | 0.0829 (15) | 0.0033 (8) | 0.0174 (10) | −0.0013 (10) |
| C17 | 0.0390 (8) | 0.0335 (8) | 0.0391 (8) | −0.0079 (6) | 0.0154 (7) | 0.0025 (7) |
| C18 | 0.0422 (9) | 0.0618 (11) | 0.0441 (9) | −0.0080 (8) | 0.0109 (7) | 0.0140 (9) |
| C19 | 0.0403 (10) | 0.0835 (14) | 0.0527 (11) | −0.0191 (10) | 0.0079 (8) | 0.0068 (11) |
| C20 | 0.0600 (12) | 0.0545 (11) | 0.0612 (12) | −0.0278 (9) | 0.0280 (9) | −0.0116 (10) |
| C21 | 0.0615 (11) | 0.0347 (8) | 0.0530 (10) | −0.0109 (8) | 0.0277 (9) | 0.0040 (8) |
| C22 | 0.0487 (9) | 0.0362 (8) | 0.0393 (8) | −0.0065 (7) | 0.0151 (7) | 0.0033 (7) |
| C23 | 0.0579 (11) | 0.0366 (9) | 0.0684 (12) | −0.0104 (8) | 0.0351 (9) | −0.0055 (9) |
| C24 | 0.0626 (12) | 0.0443 (10) | 0.0778 (14) | −0.0117 (9) | 0.0464 (11) | −0.0081 (9) |
| C25 | 0.0698 (12) | 0.0532 (11) | 0.0568 (11) | −0.0191 (9) | 0.0372 (10) | −0.0017 (9) |
| C26 | 0.0457 (11) | 0.0653 (13) | 0.1038 (18) | −0.0044 (9) | 0.0356 (11) | 0.0186 (13) |
Geometric parameters (Å, º)
| S1—C13 | 1.6674 (18) | C11—H11B | 0.9700 |
| O1—C1 | 1.369 (2) | C12—H12B | 0.98 (2) |
| O1—C7 | 1.423 (2) | C12—H12A | 1.01 (2) |
| N1—C6 | 1.423 (2) | C14—C17 | 1.508 (2) |
| N1—C8 | 1.457 (2) | C15—C16 | 1.512 (3) |
| N1—C11 | 1.472 (2) | C15—H15A | 0.9700 |
| N2—C12 | 1.434 (2) | C15—H15B | 0.9700 |
| N2—C10 | 1.457 (2) | C16—H16A | 0.9600 |
| N2—C9 | 1.459 (2) | C16—H16B | 0.9600 |
| N3—C13 | 1.349 (2) | C16—H16C | 0.9600 |
| N3—N4 | 1.3790 (19) | C17—C23 | 1.538 (2) |
| N3—C12 | 1.472 (2) | C17—C22 | 1.539 (2) |
| N4—C14 | 1.305 (2) | C17—C18 | 1.545 (2) |
| N5—C13 | 1.378 (2) | C18—C19 | 1.532 (2) |
| N5—C14 | 1.383 (2) | C18—H18A | 0.9700 |
| N5—C15 | 1.470 (2) | C18—H18B | 0.9700 |
| C1—C2 | 1.383 (3) | C19—C20 | 1.524 (3) |
| C1—C6 | 1.413 (2) | C19—C26 | 1.529 (3) |
| C2—C3 | 1.378 (3) | C19—H19A | 0.9800 |
| C2—H2A | 0.9300 | C20—C21 | 1.520 (3) |
| C3—C4 | 1.362 (3) | C20—H20A | 0.9700 |
| C3—H3A | 0.9300 | C20—H20B | 0.9700 |
| C4—C5 | 1.385 (3) | C21—C25 | 1.519 (3) |
| C4—H4A | 0.9300 | C21—C22 | 1.525 (2) |
| C5—C6 | 1.390 (2) | C21—H21A | 0.9800 |
| C5—H5A | 0.9300 | C22—H22A | 0.9700 |
| C7—H7A | 0.9600 | C22—H22B | 0.9700 |
| C7—H7B | 0.9600 | C23—C24 | 1.538 (2) |
| C7—H7C | 0.9600 | C23—H23A | 0.9700 |
| C8—C9 | 1.513 (2) | C23—H23B | 0.9700 |
| C8—H8A | 0.9700 | C24—C25 | 1.519 (3) |
| C8—H8B | 0.9700 | C24—C26 | 1.526 (3) |
| C9—H9A | 0.9700 | C24—H24A | 0.9800 |
| C9—H9B | 0.9700 | C25—H25A | 0.9700 |
| C10—C11 | 1.513 (2) | C25—H25B | 0.9700 |
| C10—H10A | 0.9700 | C26—H26A | 0.9700 |
| C10—H10B | 0.9700 | C26—H26B | 0.9700 |
| C11—H11A | 0.9700 | ||
| C1—O1—C7 | 117.79 (15) | N5—C14—C17 | 127.20 (14) |
| C6—N1—C8 | 115.28 (13) | N5—C15—C16 | 112.40 (15) |
| C6—N1—C11 | 114.41 (13) | N5—C15—H15A | 109.1 |
| C8—N1—C11 | 110.33 (13) | C16—C15—H15A | 109.1 |
| C12—N2—C10 | 114.93 (14) | N5—C15—H15B | 109.1 |
| C12—N2—C9 | 114.58 (15) | C16—C15—H15B | 109.1 |
| C10—N2—C9 | 109.92 (13) | H15A—C15—H15B | 107.9 |
| C13—N3—N4 | 112.65 (13) | C15—C16—H16A | 109.5 |
| C13—N3—C12 | 126.97 (15) | C15—C16—H16B | 109.5 |
| N4—N3—C12 | 120.21 (15) | H16A—C16—H16B | 109.5 |
| C14—N4—N3 | 104.94 (13) | C15—C16—H16C | 109.5 |
| C13—N5—C14 | 108.06 (14) | H16A—C16—H16C | 109.5 |
| C13—N5—C15 | 120.98 (14) | H16B—C16—H16C | 109.5 |
| C14—N5—C15 | 130.96 (13) | C14—C17—C23 | 108.67 (13) |
| O1—C1—C2 | 123.40 (16) | C14—C17—C22 | 111.92 (13) |
| O1—C1—C6 | 116.34 (15) | C23—C17—C22 | 108.02 (14) |
| C2—C1—C6 | 120.23 (17) | C14—C17—C18 | 110.51 (13) |
| C3—C2—C1 | 121.06 (18) | C23—C17—C18 | 108.05 (15) |
| C3—C2—H2A | 119.5 | C22—C17—C18 | 109.55 (13) |
| C1—C2—H2A | 119.5 | C19—C18—C17 | 109.64 (14) |
| C4—C3—C2 | 119.73 (19) | C19—C18—H18A | 109.7 |
| C4—C3—H3A | 120.1 | C17—C18—H18A | 109.7 |
| C2—C3—H3A | 120.1 | C19—C18—H18B | 109.7 |
| C3—C4—C5 | 119.8 (2) | C17—C18—H18B | 109.7 |
| C3—C4—H4A | 120.1 | H18A—C18—H18B | 108.2 |
| C5—C4—H4A | 120.1 | C20—C19—C26 | 109.87 (17) |
| C4—C5—C6 | 122.32 (18) | C20—C19—C18 | 109.84 (17) |
| C4—C5—H5A | 118.8 | C26—C19—C18 | 109.18 (18) |
| C6—C5—H5A | 118.8 | C20—C19—H19A | 109.3 |
| C5—C6—C1 | 116.69 (16) | C26—C19—H19A | 109.3 |
| C5—C6—N1 | 123.09 (15) | C18—C19—H19A | 109.3 |
| C1—C6—N1 | 120.10 (15) | C21—C20—C19 | 109.41 (15) |
| O1—C7—H7A | 109.5 | C21—C20—H20A | 109.8 |
| O1—C7—H7B | 109.5 | C19—C20—H20A | 109.8 |
| H7A—C7—H7B | 109.5 | C21—C20—H20B | 109.8 |
| O1—C7—H7C | 109.5 | C19—C20—H20B | 109.8 |
| H7A—C7—H7C | 109.5 | H20A—C20—H20B | 108.2 |
| H7B—C7—H7C | 109.5 | C25—C21—C20 | 110.15 (17) |
| N1—C8—C9 | 111.00 (14) | C25—C21—C22 | 110.15 (15) |
| N1—C8—H8A | 109.4 | C20—C21—C22 | 109.18 (15) |
| C9—C8—H8A | 109.4 | C25—C21—H21A | 109.1 |
| N1—C8—H8B | 109.4 | C20—C21—H21A | 109.1 |
| C9—C8—H8B | 109.4 | C22—C21—H21A | 109.1 |
| H8A—C8—H8B | 108.0 | C21—C22—C17 | 110.05 (14) |
| N2—C9—C8 | 110.21 (15) | C21—C22—H22A | 109.7 |
| N2—C9—H9A | 109.6 | C17—C22—H22A | 109.7 |
| C8—C9—H9A | 109.6 | C21—C22—H22B | 109.7 |
| N2—C9—H9B | 109.6 | C17—C22—H22B | 109.7 |
| C8—C9—H9B | 109.6 | H22A—C22—H22B | 108.2 |
| H9A—C9—H9B | 108.1 | C17—C23—C24 | 110.33 (14) |
| N2—C10—C11 | 109.91 (13) | C17—C23—H23A | 109.6 |
| N2—C10—H10A | 109.7 | C24—C23—H23A | 109.6 |
| C11—C10—H10A | 109.7 | C17—C23—H23B | 109.6 |
| N2—C10—H10B | 109.7 | C24—C23—H23B | 109.6 |
| C11—C10—H10B | 109.7 | H23A—C23—H23B | 108.1 |
| H10A—C10—H10B | 108.2 | C25—C24—C26 | 109.76 (16) |
| N1—C11—C10 | 110.43 (14) | C25—C24—C23 | 109.57 (17) |
| N1—C11—H11A | 109.6 | C26—C24—C23 | 109.29 (17) |
| C10—C11—H11A | 109.6 | C25—C24—H24A | 109.4 |
| N1—C11—H11B | 109.6 | C26—C24—H24A | 109.4 |
| C10—C11—H11B | 109.6 | C23—C24—H24A | 109.4 |
| H11A—C11—H11B | 108.1 | C21—C25—C24 | 109.14 (15) |
| N2—C12—N3 | 116.71 (13) | C21—C25—H25A | 109.9 |
| N2—C12—H12B | 113.0 (11) | C24—C25—H25A | 109.9 |
| N3—C12—H12B | 103.6 (10) | C21—C25—H25B | 109.9 |
| N2—C12—H12A | 108.6 (11) | C24—C25—H25B | 109.9 |
| N3—C12—H12A | 106.1 (11) | H25A—C25—H25B | 108.3 |
| H12B—C12—H12A | 108.5 (15) | C24—C26—C19 | 109.21 (16) |
| N3—C13—N5 | 103.80 (14) | C24—C26—H26A | 109.8 |
| N3—C13—S1 | 128.57 (13) | C19—C26—H26A | 109.8 |
| N5—C13—S1 | 127.63 (14) | C24—C26—H26B | 109.8 |
| N4—C14—N5 | 110.55 (13) | C19—C26—H26B | 109.8 |
| N4—C14—C17 | 122.24 (15) | H26A—C26—H26B | 108.3 |
| C13—N3—N4—C14 | −0.07 (17) | N3—N4—C14—N5 | 0.16 (17) |
| C12—N3—N4—C14 | −175.71 (14) | N3—N4—C14—C17 | −178.86 (14) |
| C7—O1—C1—C2 | 10.9 (2) | C13—N5—C14—N4 | −0.20 (18) |
| C7—O1—C1—C6 | −167.17 (15) | C15—N5—C14—N4 | 179.95 (15) |
| O1—C1—C2—C3 | −174.35 (18) | C13—N5—C14—C17 | 178.76 (15) |
| C6—C1—C2—C3 | 3.7 (3) | C15—N5—C14—C17 | −1.1 (3) |
| C1—C2—C3—C4 | −0.7 (3) | C13—N5—C15—C16 | 80.4 (2) |
| C2—C3—C4—C5 | −2.0 (3) | C14—N5—C15—C16 | −99.8 (2) |
| C3—C4—C5—C6 | 1.7 (3) | N4—C14—C17—C23 | −8.7 (2) |
| C4—C5—C6—C1 | 1.2 (3) | N5—C14—C17—C23 | 172.44 (16) |
| C4—C5—C6—N1 | 177.19 (17) | N4—C14—C17—C22 | −127.92 (16) |
| O1—C1—C6—C5 | 174.34 (15) | N5—C14—C17—C22 | 53.2 (2) |
| C2—C1—C6—C5 | −3.8 (2) | N4—C14—C17—C18 | 109.70 (18) |
| O1—C1—C6—N1 | −1.8 (2) | N5—C14—C17—C18 | −69.2 (2) |
| C2—C1—C6—N1 | −179.97 (15) | C14—C17—C18—C19 | −178.89 (16) |
| C8—N1—C6—C5 | −7.3 (2) | C23—C17—C18—C19 | −60.1 (2) |
| C11—N1—C6—C5 | 122.18 (18) | C22—C17—C18—C19 | 57.4 (2) |
| C8—N1—C6—C1 | 168.57 (15) | C17—C18—C19—C20 | −58.9 (2) |
| C11—N1—C6—C1 | −61.93 (19) | C17—C18—C19—C26 | 61.6 (2) |
| C6—N1—C8—C9 | −172.41 (14) | C26—C19—C20—C21 | −59.0 (2) |
| C11—N1—C8—C9 | 56.12 (19) | C18—C19—C20—C21 | 61.1 (2) |
| C12—N2—C9—C8 | −169.87 (13) | C19—C20—C21—C25 | 59.6 (2) |
| C10—N2—C9—C8 | 58.92 (17) | C19—C20—C21—C22 | −61.5 (2) |
| N1—C8—C9—N2 | −57.44 (18) | C25—C21—C22—C17 | −60.62 (19) |
| C12—N2—C10—C11 | 169.35 (14) | C20—C21—C22—C17 | 60.46 (19) |
| C9—N2—C10—C11 | −59.63 (17) | C14—C17—C22—C21 | 178.67 (14) |
| C6—N1—C11—C10 | 171.40 (13) | C23—C17—C22—C21 | 59.08 (18) |
| C8—N1—C11—C10 | −56.68 (17) | C18—C17—C22—C21 | −58.40 (18) |
| N2—C10—C11—N1 | 58.53 (17) | C14—C17—C23—C24 | 179.37 (15) |
| C10—N2—C12—N3 | 69.0 (2) | C22—C17—C23—C24 | −59.0 (2) |
| C9—N2—C12—N3 | −59.8 (2) | C18—C17—C23—C24 | 59.4 (2) |
| C13—N3—C12—N2 | 100.2 (2) | C17—C23—C24—C25 | 60.2 (2) |
| N4—N3—C12—N2 | −84.8 (2) | C17—C23—C24—C26 | −60.1 (2) |
| N4—N3—C13—N5 | −0.05 (17) | C20—C21—C25—C24 | −60.16 (19) |
| C12—N3—C13—N5 | 175.23 (14) | C22—C21—C25—C24 | 60.3 (2) |
| N4—N3—C13—S1 | 179.88 (12) | C26—C24—C25—C21 | 60.3 (2) |
| C12—N3—C13—S1 | −4.8 (2) | C23—C24—C25—C21 | −59.7 (2) |
| C14—N5—C13—N3 | 0.14 (17) | C25—C24—C26—C19 | −60.0 (2) |
| C15—N5—C13—N3 | −179.98 (13) | C23—C24—C26—C19 | 60.2 (2) |
| C14—N5—C13—S1 | −179.79 (12) | C20—C19—C26—C24 | 59.2 (2) |
| C15—N5—C13—S1 | 0.1 (2) | C18—C19—C26—C24 | −61.3 (2) |
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C1–C6 benzene ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C11—H11A···O1 | 0.97 | 2.26 | 2.903 (2) | 123 |
| C18—H18A···Cgi | 0.97 | 2.81 | 3.748 (2) | 162 |
Symmetry code: (i) x−1, −y−1, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: RZ5099).
References
- Al-Abdullah, E. S., Al-Tuwaijri, H. M., El-Emam, A. A., Chidan Kumar, C. S. & Fun, H.-K. (2013). Acta Cryst. E69, o1813–o1814. [DOI] [PMC free article] [PubMed]
- Al-Deeb, O. A., Al-Omar, M. A., El-Brollosy, N. R., Habib, E. E., Ibrahim, T. M. & El-Emam, A. A. (2006). Arzneim. Forsch. Drug Res. 56, 40–47. [DOI] [PubMed]
- Al-Tamimi, A.-M. S., Al-Abdullah, E. S., El-Emam, A. A., Ng, S. W. & Tiekink, E. R. T. (2013). Acta Cryst. E69, o685–o686. [DOI] [PMC free article] [PubMed]
- Al-Tamimi, A.-M. S., Alafeefy, A. M., El-Emam, A. A., Ng, S. W. & Tiekink, E. R. T. (2013). Acta Cryst. E69, o683. [DOI] [PMC free article] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2009). SADABS, APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- El-Emam, A. A., Al-Deeb, O. A., Al-Omar, M. A. & Lehmann, J. (2004). Bioorg. Med. Chem. 12, 5107–5113. [DOI] [PubMed]
- El-Emam, A. A., Alrashood, K. A., Al-Tamimi, A.-M. S., Ng, S. W. & Tiekink, E. R. T. (2012). Acta Cryst. E68, o657–o658. [DOI] [PMC free article] [PubMed]
- El-Emam, A. A., Al-Tamimi, A.-S., Al-Omar, A. A., Alrashood, K. A. & Habib, E. E. (2013). Eur. J. Med. Chem. 68, 96–102. [DOI] [PubMed]
- El-Emam, A. A. & Ibrahim, T. M. (1991). Arzneim. Forsch. Drug Res. 41, 1260–1264. [PubMed]
- Kadi, A. A., Al-Abdullah, E. S., Shehata, I. A., Habib, E. E., Ibrahim, T. M. & El-Emam, A. A. (2010). Eur. J. Med. Chem. 45, 5006–5011. [DOI] [PubMed]
- Kadi, A. A., El-Brollosy, N. R., Al-Deeb, O. A., Habib, E. E., Ibrahim, T. M. & El-Emam, A. A. (2007). Eur. J. Med. Chem. 42, 235–242. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Togo, Y., Hornick, R. B. & Dawkins, A. T. (1968). J. Am. Med. Assoc. 203, 1089–1094.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813032789/rz5099sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813032789/rz5099Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536813032789/rz5099Isup3.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report


