Abstract
In the title compound, C23H16N2·0.5C6H6, the benzimidazole unit [maximum deviation = 0.0258 (6) Å] and the naphthalene ring system [maximum deviation = 0.0254 (6) Å] are both essentially planar and make a dihedral angle of 61.955 (17)°. The imidazole ring makes dihedral angle of 61.73 (4)° with the phenyl ring. An intramolecular C—H⋯N hydrogen bond generates an S(6) ring motif. In the crystal, seven weak C—H⋯π interactions involving the fused ring system, the benzene solvent molecule, the imidazole phenyl rings are observed, leading to a three-dimensional architecture.
Related literature
For linear and non-linear optical properties and the thermal stability of benzimidazole-based chromophores, see: Cross et al. (1995 ▶). For imidazole as a component of vitamin B12, purine and caffeine, see: Brown (2005 ▶). For commercial and therapeutic applications of substituted benzimidazole derivatives, see: Spasov et al. (1999 ▶). For related crystal structures, see: Jayamoorthy et al. (2012 ▶, 2013 ▶); Rosepriya et al. (2011 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶).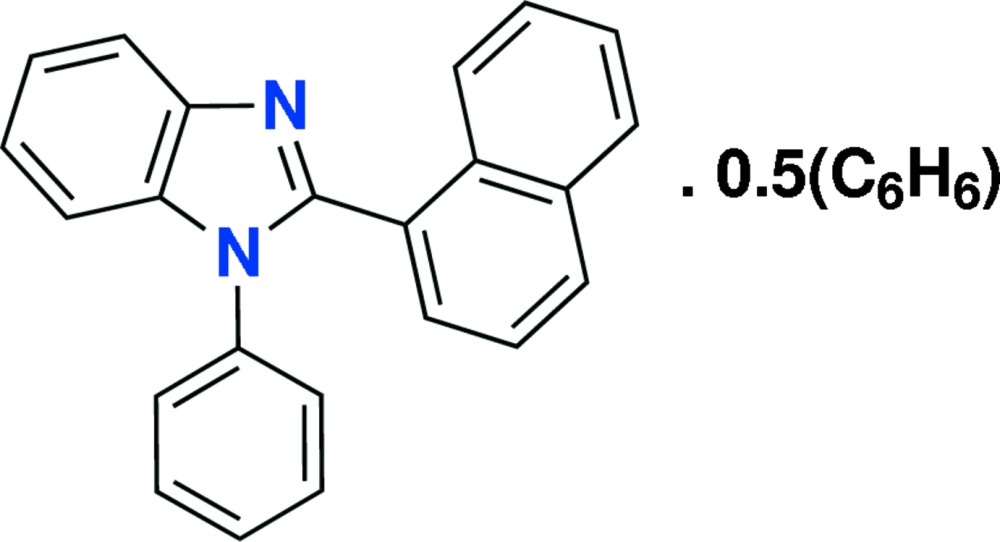
Experimental
Crystal data
C23H16N2·0.5C6H6
M r = 359.43
Triclinic,

a = 8.5529 (3) Å
b = 9.4517 (3) Å
c = 11.8936 (3) Å
α = 86.334 (2)°
β = 89.838 (2)°
γ = 75.051 (3)°
V = 926.94 (5) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 123 K
0.72 × 0.59 × 0.42 mm
Data collection
Agilent Xcalibur Ruby Gemini diffractometer
Absorption correction: analytical [CrysAlis PRO (Agilent, 2012 ▶), using a multifaceted crystal model (Clark & Reid, 1995 ▶)] T min = 0.963, T max = 0.977
57784 measured reflections
12045 independent reflections
9086 reflections with I > 2σ(I)
R int = 0.063
Refinement
R[F 2 > 2σ(F 2)] = 0.057
wR(F 2) = 0.160
S = 1.05
12045 reflections
253 parameters
H-atom parameters constrained
Δρmax = 0.48 e Å−3
Δρmin = −0.42 e Å−3
Data collection: CrysAlis PRO (Agilent, 2012 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS2013 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL2013 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL2013 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681303331X/jj2179sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681303331X/jj2179Isup2.hkl
Supporting information file. DOI: 10.1107/S160053681303331X/jj2179Isup3.cdx
Supporting information file. DOI: 10.1107/S160053681303331X/jj2179Isup4.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2, Cg3, Cg4 and Cg8 are the centroids of the N1/C2/N3/C9/C8 imidazole ring, the C4–C9 fused benzene ring, the C11–C16 phenyl ring, the C21–C24,C30/C29 fused benzene ring and the C1A,C2A,C3A′,C1A′,C2A′,C3A benzene ring, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C28—H28⋯N3 | 0.95 | 2.61 | 3.2113 (10) | 121 |
| C7—H7⋯Cg4i | 0.95 | 2.75 | 3.6019 (8) | 150 |
| C15—H15⋯Cg8ii | 0.95 | 2.99 | 3.6981 (9) | 132 |
| C15—H15⋯Cg8iii | 0.95 | 2.99 | 3.6981 (9) | 132 |
| C22—H22⋯Cg1iv | 0.95 | 2.91 | 3.6478 (8) | 136 |
| C24—H24⋯Cg3v | 0.95 | 2.76 | 3.4888 (9) | 134 |
| C26—H26⋯Cg2iii | 0.95 | 2.87 | 3.5801 (9) | 133 |
| C27—H27⋯Cg1iii | 0.95 | 2.97 | 3.7258 (8) | 137 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Acknowledgments
SMP thanks Annamaliar College of Engineering, Mudaiyur, for providing constant support for this research. RJB acknowledges the NSF–MRI program (grant No. CHE-0619278) for funds to purchase an X-ray diffractometer.
supplementary crystallographic information
1. Comment
Benzimidazole based chromophores have received increasing attention due to their distinctive linear, non-linear optical properties and also due to their excellent thermal stability in guest-host systems (Cross et al., 1995). They are a component of vitamin B12 (Brown, 2005) and are related to the DNA base purine and the stimulant caffeine. Substituted benzimidazole derivatives have found commercial applications in veterinarian medicine as anthelmintic agents and in diverse human therapeutic areas as an antiulcer and antihistaminic (Spasov et al., 1999). Therefore, the preparation of benzimidazoles has gained considerable attention in recent years. Jayamoorthy et al., (2012, 2013) and Rosepriya et al., (2011) have reported closely related structures of benzimidazole derivatives. We are interested to use 2-(naphthalen-1-yl)-1-phenyl-1H-benzimidazole as ligand to study its photophysical properties.
The assymmetric unit of the title compound, C23H16N2·0.5C6H6, (Fig. 1), contains a 2-(naphthalen-1-yl)-1-phenyl-1H-benzimidazole molecule and a hemibenzene solvent molecule. The benzimidazole unit is essentially planar [maximum deviation of -0.0258 (6) Å for C8]. The naphthalene unit is also essentially planar [maximum deviation of -0.0254 (6) Å for C23]. The benzimidazole unit makes dihedral angle of 61.955 (17)° with the naphthalene unit. The imidazole ring makes dihedral angle of 61.73 (4)° with the phenyl group attached to N1.
An intramolecular C28—H28···N3 hydrogen bond (Fig. 1) generates an S(6) ring motif (Bernstein et al., 1995). The packing of the title compound, viewed along the a axis is shown in Fig. 2. In the crystal, seven weak C7—H7···π interactions involving the fused benzene ring, C15—H15···π interactions involving the solvent benzene ring, C22—H22···π interaction involving the imidazole ring, C24—H24···π interaction involving the phenyl ring, C26—H26···π interaction involving the fused benzene ring, C27—H27···π interaction involving the imidazole ring, are observed, leading to a three dimensional architecture (Fig. 3, Table 1).
2. Experimental
To the pure N-phenyl-o-phenylenediamine (17 mmol, 3.128 g) in ethanol (10 ml), napthaldehyde (17 mmol, 1.9 ml) and ammonium acetate (3 g) was added about 1 h by maintaining the temperature at 353 K. The reaction mixture was refluxed for 48 hrs and the completion of reaction was monitored by TLC, finally the reaction extracted by dichloromethane. The solid separated was purified by column chromatography using benzene as the eluent. Yield: 2.65 g (50%). The compound was dissolved in benzene and ethyl acetate (9:1) mixture and allowed to slow evaporation for two days, to obtain crystals suitable for X-ray diffraction studies.
3. Refinement
The H atoms were positioned geometrically and allowed to ride on their parent atoms, with C—H = 0.95 Å. Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound, with displacement ellipsoids drawn at the 50% probability level. H atoms are shown as small spheres of arbitrary radius. The dashed line indicates the intramolecular C—H···N hydrogen bond. Symmetry code: (i) -x + 1, -y, -z + 1.
Fig. 2.
The packing of the title compound, viewed along the a axis.
Fig. 3.
Part of the crystal structure of compound, showing the formation of C—H···π interactions. Interactions involving benzene solvent Cg8 are not shown.
Crystal data
| C23H16N2·0.5C6H6 | Z = 2 |
| Mr = 359.43 | F(000) = 378 |
| Triclinic, P1 | Dx = 1.288 Mg m−3 |
| Hall symbol: -P 1 | Melting point: 392 K |
| a = 8.5529 (3) Å | Mo Kα radiation, λ = 0.71069 Å |
| b = 9.4517 (3) Å | Cell parameters from 15125 reflections |
| c = 11.8936 (3) Å | θ = 3.0–41.0° |
| α = 86.334 (2)° | µ = 0.08 mm−1 |
| β = 89.838 (2)° | T = 123 K |
| γ = 75.051 (3)° | Prism, colourless |
| V = 926.94 (5) Å3 | 0.72 × 0.59 × 0.42 mm |
Data collection
| Agilent Xcalibur Ruby Gemini diffractometer | 12045 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 9086 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.063 |
| Detector resolution: 10.5081 pixels mm-1 | θmax = 41.1°, θmin = 3.0° |
| ω scans | h = −15→15 |
| Absorption correction: analytical [CrysAlis PRO (Agilent, 2012), using a multifaceted crystal model (Clark & Reid, 1995)] | k = −17→17 |
| Tmin = 0.963, Tmax = 0.977 | l = −21→21 |
| 57784 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.057 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.160 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.076P)2 + 0.0921P] where P = (Fo2 + 2Fc2)/3 |
| 12045 reflections | (Δ/σ)max < 0.001 |
| 253 parameters | Δρmax = 0.48 e Å−3 |
| 0 restraints | Δρmin = −0.42 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.67234 (7) | 0.54147 (6) | 0.17737 (5) | 0.0196 (1) | |
| N3 | 0.58580 (7) | 0.34162 (7) | 0.23145 (5) | 0.0218 (1) | |
| C2 | 0.54214 (8) | 0.48307 (7) | 0.20135 (6) | 0.0197 (1) | |
| C4 | 0.86569 (9) | 0.16857 (8) | 0.25577 (6) | 0.0230 (2) | |
| C5 | 1.02936 (9) | 0.16196 (8) | 0.24850 (6) | 0.0241 (2) | |
| C6 | 1.08319 (9) | 0.28681 (9) | 0.21552 (6) | 0.0237 (2) | |
| C7 | 0.97488 (9) | 0.42165 (8) | 0.18733 (6) | 0.0222 (2) | |
| C8 | 0.81081 (8) | 0.42675 (7) | 0.19437 (6) | 0.0194 (1) | |
| C9 | 0.75441 (8) | 0.30379 (7) | 0.22843 (6) | 0.0200 (2) | |
| C11 | 0.66581 (8) | 0.69261 (7) | 0.15057 (5) | 0.0190 (1) | |
| C12 | 0.72504 (9) | 0.73392 (8) | 0.04813 (6) | 0.0217 (2) | |
| C13 | 0.71614 (9) | 0.88153 (8) | 0.02178 (6) | 0.0247 (2) | |
| C14 | 0.64697 (9) | 0.98608 (8) | 0.09676 (7) | 0.0253 (2) | |
| C15 | 0.58828 (9) | 0.94403 (8) | 0.19913 (7) | 0.0259 (2) | |
| C16 | 0.59863 (9) | 0.79665 (8) | 0.22698 (6) | 0.0233 (2) | |
| C21 | 0.37491 (8) | 0.57825 (7) | 0.19356 (6) | 0.0194 (2) | |
| C22 | 0.31678 (9) | 0.65122 (8) | 0.09166 (6) | 0.0230 (2) | |
| C23 | 0.16256 (9) | 0.75196 (9) | 0.08347 (6) | 0.0252 (2) | |
| C24 | 0.07012 (9) | 0.78075 (8) | 0.17773 (7) | 0.0243 (2) | |
| C25 | 0.02654 (9) | 0.73198 (9) | 0.38062 (7) | 0.0270 (2) | |
| C26 | 0.07884 (10) | 0.65771 (10) | 0.48214 (7) | 0.0291 (2) | |
| C27 | 0.23194 (10) | 0.55553 (10) | 0.49096 (6) | 0.0271 (2) | |
| C28 | 0.32981 (9) | 0.52890 (8) | 0.39875 (6) | 0.0225 (2) | |
| C29 | 0.27828 (8) | 0.60281 (7) | 0.29210 (5) | 0.0187 (2) | |
| C30 | 0.12426 (8) | 0.70669 (8) | 0.28362 (6) | 0.0210 (2) | |
| C1A | 0.55334 (12) | 0.12597 (10) | 0.47928 (7) | 0.0319 (2) | |
| C2A | 0.39180 (12) | 0.13022 (10) | 0.46027 (8) | 0.0329 (2) | |
| C3A | 0.66115 (11) | −0.00416 (11) | 0.51904 (7) | 0.0315 (2) | |
| H4 | 0.83010 | 0.08400 | 0.27860 | 0.0276* | |
| H5 | 1.10668 | 0.07119 | 0.26616 | 0.0289* | |
| H6 | 1.19612 | 0.27882 | 0.21245 | 0.0285* | |
| H7 | 1.01084 | 0.50599 | 0.16443 | 0.0266* | |
| H12 | 0.77109 | 0.66228 | −0.00334 | 0.0261* | |
| H13 | 0.75746 | 0.91078 | −0.04763 | 0.0297* | |
| H14 | 0.63978 | 1.08678 | 0.07795 | 0.0303* | |
| H15 | 0.54107 | 1.01593 | 0.25013 | 0.0311* | |
| H16 | 0.56028 | 0.76719 | 0.29741 | 0.0279* | |
| H22 | 0.38129 | 0.63341 | 0.02635 | 0.0276* | |
| H23 | 0.12295 | 0.79973 | 0.01263 | 0.0302* | |
| H24 | −0.03168 | 0.85133 | 0.17208 | 0.0292* | |
| H25 | −0.07625 | 0.80110 | 0.37523 | 0.0323* | |
| H26 | 0.01210 | 0.67503 | 0.54642 | 0.0350* | |
| H27 | 0.26770 | 0.50460 | 0.56147 | 0.0325* | |
| H28 | 0.43288 | 0.46046 | 0.40632 | 0.0270* | |
| H1A | 0.58997 | 0.21206 | 0.46507 | 0.0383* | |
| H2A | 0.31790 | 0.21918 | 0.43320 | 0.0394* | |
| H3A | 0.77144 | −0.00685 | 0.53211 | 0.0378* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0184 (2) | 0.0182 (2) | 0.0204 (2) | −0.0019 (2) | 0.0024 (2) | 0.0005 (2) |
| N3 | 0.0201 (2) | 0.0205 (2) | 0.0231 (3) | −0.0031 (2) | 0.0015 (2) | 0.0016 (2) |
| C2 | 0.0188 (2) | 0.0205 (3) | 0.0184 (2) | −0.0031 (2) | 0.0016 (2) | 0.0005 (2) |
| C4 | 0.0238 (3) | 0.0194 (3) | 0.0233 (3) | −0.0013 (2) | 0.0001 (2) | 0.0004 (2) |
| C5 | 0.0226 (3) | 0.0222 (3) | 0.0236 (3) | 0.0011 (2) | −0.0003 (2) | −0.0009 (2) |
| C6 | 0.0192 (3) | 0.0260 (3) | 0.0234 (3) | −0.0009 (2) | 0.0011 (2) | −0.0025 (2) |
| C7 | 0.0197 (3) | 0.0226 (3) | 0.0230 (3) | −0.0034 (2) | 0.0023 (2) | −0.0016 (2) |
| C8 | 0.0183 (2) | 0.0191 (3) | 0.0186 (2) | −0.0013 (2) | 0.0016 (2) | −0.0009 (2) |
| C9 | 0.0199 (3) | 0.0192 (3) | 0.0190 (3) | −0.0021 (2) | 0.0011 (2) | −0.0003 (2) |
| C11 | 0.0184 (2) | 0.0184 (2) | 0.0186 (2) | −0.0021 (2) | 0.0010 (2) | 0.0001 (2) |
| C12 | 0.0222 (3) | 0.0213 (3) | 0.0198 (3) | −0.0026 (2) | 0.0024 (2) | 0.0003 (2) |
| C13 | 0.0248 (3) | 0.0240 (3) | 0.0238 (3) | −0.0050 (2) | −0.0014 (2) | 0.0050 (2) |
| C14 | 0.0230 (3) | 0.0195 (3) | 0.0315 (3) | −0.0032 (2) | −0.0053 (3) | 0.0023 (2) |
| C15 | 0.0245 (3) | 0.0214 (3) | 0.0303 (3) | −0.0021 (2) | 0.0003 (3) | −0.0062 (2) |
| C16 | 0.0242 (3) | 0.0235 (3) | 0.0215 (3) | −0.0045 (2) | 0.0034 (2) | −0.0040 (2) |
| C21 | 0.0175 (2) | 0.0196 (3) | 0.0198 (3) | −0.0028 (2) | 0.0003 (2) | 0.0004 (2) |
| C22 | 0.0221 (3) | 0.0252 (3) | 0.0199 (3) | −0.0036 (2) | −0.0003 (2) | 0.0019 (2) |
| C23 | 0.0241 (3) | 0.0251 (3) | 0.0238 (3) | −0.0032 (2) | −0.0043 (2) | 0.0041 (2) |
| C24 | 0.0201 (3) | 0.0219 (3) | 0.0282 (3) | −0.0008 (2) | −0.0039 (2) | 0.0000 (2) |
| C25 | 0.0202 (3) | 0.0296 (3) | 0.0287 (3) | −0.0011 (3) | 0.0032 (2) | −0.0065 (3) |
| C26 | 0.0264 (3) | 0.0355 (4) | 0.0246 (3) | −0.0055 (3) | 0.0067 (3) | −0.0059 (3) |
| C27 | 0.0273 (3) | 0.0326 (4) | 0.0199 (3) | −0.0054 (3) | 0.0027 (2) | 0.0001 (3) |
| C28 | 0.0211 (3) | 0.0245 (3) | 0.0201 (3) | −0.0030 (2) | 0.0004 (2) | 0.0004 (2) |
| C29 | 0.0167 (2) | 0.0191 (3) | 0.0194 (3) | −0.0033 (2) | −0.0001 (2) | −0.0008 (2) |
| C30 | 0.0179 (2) | 0.0206 (3) | 0.0233 (3) | −0.0023 (2) | −0.0004 (2) | −0.0027 (2) |
| C1A | 0.0385 (4) | 0.0284 (4) | 0.0274 (3) | −0.0071 (3) | 0.0019 (3) | 0.0013 (3) |
| C2A | 0.0360 (4) | 0.0282 (4) | 0.0290 (4) | 0.0004 (3) | 0.0000 (3) | 0.0030 (3) |
| C3A | 0.0301 (4) | 0.0350 (4) | 0.0271 (3) | −0.0047 (3) | 0.0005 (3) | −0.0001 (3) |
Geometric parameters (Å, º)
| N1—C2 | 1.3861 (9) | C27—C28 | 1.3741 (11) |
| N1—C8 | 1.3892 (9) | C28—C29 | 1.4221 (9) |
| N1—C11 | 1.4304 (8) | C29—C30 | 1.4249 (10) |
| N3—C2 | 1.3180 (9) | C4—H4 | 0.9500 |
| N3—C9 | 1.3943 (9) | C5—H5 | 0.9500 |
| C2—C21 | 1.4798 (10) | C6—H6 | 0.9500 |
| C4—C5 | 1.3878 (11) | C7—H7 | 0.9500 |
| C4—C9 | 1.4022 (10) | C12—H12 | 0.9500 |
| C5—C6 | 1.4066 (11) | C13—H13 | 0.9500 |
| C6—C7 | 1.3903 (11) | C14—H14 | 0.9500 |
| C7—C8 | 1.3941 (11) | C15—H15 | 0.9500 |
| C8—C9 | 1.4055 (9) | C16—H16 | 0.9500 |
| C11—C12 | 1.3897 (10) | C22—H22 | 0.9500 |
| C11—C16 | 1.3939 (10) | C23—H23 | 0.9500 |
| C12—C13 | 1.3928 (10) | C24—H24 | 0.9500 |
| C13—C14 | 1.3892 (11) | C25—H25 | 0.9500 |
| C14—C15 | 1.3892 (12) | C26—H26 | 0.9500 |
| C15—C16 | 1.3912 (10) | C27—H27 | 0.9500 |
| C21—C22 | 1.3825 (10) | C28—H28 | 0.9500 |
| C21—C29 | 1.4286 (9) | C1A—C2A | 1.3906 (15) |
| C22—C23 | 1.4131 (11) | C1A—C3A | 1.3891 (14) |
| C23—C24 | 1.3708 (11) | C2A—C3Ai | 1.3876 (14) |
| C24—C30 | 1.4191 (11) | C1A—H1A | 0.9500 |
| C25—C26 | 1.3724 (12) | C2A—H2A | 0.9500 |
| C25—C30 | 1.4189 (11) | C3A—H3A | 0.9500 |
| C26—C27 | 1.4120 (13) | ||
| C2—N1—C8 | 106.49 (5) | C5—C4—H4 | 121.00 |
| C2—N1—C11 | 126.57 (6) | C9—C4—H4 | 121.00 |
| C8—N1—C11 | 126.72 (6) | C4—C5—H5 | 119.00 |
| C2—N3—C9 | 104.76 (6) | C6—C5—H5 | 119.00 |
| N1—C2—N3 | 113.12 (6) | C5—C6—H6 | 119.00 |
| N1—C2—C21 | 120.27 (6) | C7—C6—H6 | 119.00 |
| N3—C2—C21 | 126.61 (6) | C6—C7—H7 | 122.00 |
| C5—C4—C9 | 118.02 (7) | C8—C7—H7 | 122.00 |
| C4—C5—C6 | 121.39 (7) | C11—C12—H12 | 120.00 |
| C5—C6—C7 | 121.50 (7) | C13—C12—H12 | 120.00 |
| C6—C7—C8 | 116.58 (7) | C12—C13—H13 | 120.00 |
| N1—C8—C7 | 131.99 (6) | C14—C13—H13 | 120.00 |
| N1—C8—C9 | 105.10 (6) | C13—C14—H14 | 120.00 |
| C7—C8—C9 | 122.85 (6) | C15—C14—H14 | 120.00 |
| N3—C9—C4 | 129.80 (6) | C14—C15—H15 | 120.00 |
| N3—C9—C8 | 110.53 (6) | C16—C15—H15 | 120.00 |
| C4—C9—C8 | 119.65 (7) | C11—C16—H16 | 120.00 |
| N1—C11—C12 | 119.52 (6) | C15—C16—H16 | 120.00 |
| N1—C11—C16 | 119.59 (6) | C21—C22—H22 | 120.00 |
| C12—C11—C16 | 120.89 (6) | C23—C22—H22 | 120.00 |
| C11—C12—C13 | 119.26 (7) | C22—C23—H23 | 120.00 |
| C12—C13—C14 | 120.16 (7) | C24—C23—H23 | 120.00 |
| C13—C14—C15 | 120.30 (7) | C23—C24—H24 | 120.00 |
| C14—C15—C16 | 120.01 (7) | C30—C24—H24 | 120.00 |
| C11—C16—C15 | 119.37 (7) | C26—C25—H25 | 120.00 |
| C2—C21—C22 | 119.43 (6) | C30—C25—H25 | 120.00 |
| C2—C21—C29 | 120.29 (6) | C25—C26—H26 | 120.00 |
| C22—C21—C29 | 120.13 (6) | C27—C26—H26 | 120.00 |
| C21—C22—C23 | 120.80 (7) | C26—C27—H27 | 120.00 |
| C22—C23—C24 | 120.00 (7) | C28—C27—H27 | 120.00 |
| C23—C24—C30 | 120.97 (7) | C27—C28—H28 | 120.00 |
| C26—C25—C30 | 120.75 (7) | C29—C28—H28 | 120.00 |
| C25—C26—C27 | 119.94 (8) | C2A—C1A—C3A | 120.00 (9) |
| C26—C27—C28 | 120.78 (7) | C1A—C2A—C3Ai | 119.81 (9) |
| C27—C28—C29 | 120.63 (7) | C1A—C3A—C2Ai | 120.19 (9) |
| C21—C29—C28 | 122.80 (6) | C2A—C1A—H1A | 120.00 |
| C21—C29—C30 | 118.72 (6) | C3A—C1A—H1A | 120.00 |
| C28—C29—C30 | 118.48 (6) | C1A—C2A—H2A | 120.00 |
| C24—C30—C25 | 121.24 (7) | C3Ai—C2A—H2A | 120.00 |
| C24—C30—C29 | 119.33 (6) | C1A—C3A—H3A | 120.00 |
| C25—C30—C29 | 119.42 (6) | C2Ai—C3A—H3A | 120.00 |
| C8—N1—C2—N3 | −0.31 (8) | C16—C11—C12—C13 | −0.28 (11) |
| C8—N1—C2—C21 | 178.80 (6) | N1—C11—C16—C15 | −178.15 (7) |
| C11—N1—C2—N3 | −175.10 (6) | C12—C11—C16—C15 | 1.18 (11) |
| C11—N1—C2—C21 | 4.01 (10) | C11—C12—C13—C14 | −0.76 (11) |
| C2—N1—C8—C7 | −177.27 (8) | C12—C13—C14—C15 | 0.90 (12) |
| C2—N1—C8—C9 | −0.10 (7) | C13—C14—C15—C16 | 0.00 (12) |
| C11—N1—C8—C7 | −2.50 (12) | C14—C15—C16—C11 | −1.03 (12) |
| C11—N1—C8—C9 | 174.68 (6) | C2—C21—C22—C23 | −174.88 (7) |
| C2—N1—C11—C12 | −120.91 (8) | C29—C21—C22—C23 | 0.72 (11) |
| C2—N1—C11—C16 | 58.42 (10) | C2—C21—C29—C28 | −5.79 (10) |
| C8—N1—C11—C12 | 65.33 (9) | C2—C21—C29—C30 | 173.63 (6) |
| C8—N1—C11—C16 | −115.34 (8) | C22—C21—C29—C28 | 178.65 (7) |
| C9—N3—C2—N1 | 0.57 (8) | C22—C21—C29—C30 | −1.92 (10) |
| C9—N3—C2—C21 | −178.47 (7) | C21—C22—C23—C24 | 1.44 (12) |
| C2—N3—C9—C4 | 177.86 (7) | C22—C23—C24—C30 | −2.34 (12) |
| C2—N3—C9—C8 | −0.62 (8) | C23—C24—C30—C25 | −177.62 (8) |
| N1—C2—C21—C22 | 60.23 (9) | C23—C24—C30—C29 | 1.09 (11) |
| N1—C2—C21—C29 | −115.36 (7) | C30—C25—C26—C27 | 0.42 (13) |
| N3—C2—C21—C22 | −120.80 (8) | C26—C25—C30—C24 | 178.77 (8) |
| N3—C2—C21—C29 | 63.62 (10) | C26—C25—C30—C29 | 0.05 (13) |
| C9—C4—C5—C6 | 0.43 (11) | C25—C26—C27—C28 | −0.17 (13) |
| C5—C4—C9—N3 | −178.03 (7) | C26—C27—C28—C29 | −0.58 (13) |
| C5—C4—C9—C8 | 0.34 (10) | C27—C28—C29—C21 | −179.54 (7) |
| C4—C5—C6—C7 | −0.89 (11) | C27—C28—C29—C30 | 1.04 (11) |
| C5—C6—C7—C8 | 0.51 (11) | C21—C29—C30—C24 | 1.04 (10) |
| C6—C7—C8—N1 | 177.03 (7) | C21—C29—C30—C25 | 179.78 (7) |
| C6—C7—C8—C9 | 0.28 (11) | C28—C29—C30—C24 | −179.51 (7) |
| N1—C8—C9—N3 | 0.45 (8) | C28—C29—C30—C25 | −0.77 (10) |
| N1—C8—C9—C4 | −178.22 (6) | C3A—C1A—C2A—C3Ai | −0.10 (13) |
| C7—C8—C9—N3 | 177.95 (7) | C2A—C1A—C3A—C2Ai | 0.10 (14) |
| C7—C8—C9—C4 | −0.71 (11) | C1A—C2A—C3Ai—C1Ai | 0.10 (13) |
| N1—C11—C12—C13 | 179.04 (7) |
Symmetry code: (i) −x+1, −y, −z+1.
Hydrogen-bond geometry (Å, º)
Cg1, Cg2, Cg3, Cg4 and Cg8 are the centroids of the N1/C2/N3/C9/C8 imidazole ring, the C4–C9 fused benzene ring, the C11–C16 phenyl ring, the C21–C24,C30/C29 fused benzene ring and the C1A,C2A,C3A',C1A',C2A',C3A benzene ring, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C28—H28···N3 | 0.95 | 2.61 | 3.2113 (10) | 121 |
| C7—H7···Cg4ii | 0.95 | 2.75 | 3.6019 (8) | 150 |
| C15—H15···Cg8iii | 0.95 | 2.99 | 3.6981 (9) | 132 |
| C15—H15···Cg8iv | 0.95 | 2.99 | 3.6981 (9) | 132 |
| C22—H22···Cg1v | 0.95 | 2.91 | 3.6478 (8) | 136 |
| C24—H24···Cg3vi | 0.95 | 2.76 | 3.4888 (9) | 134 |
| C26—H26···Cg2iv | 0.95 | 2.87 | 3.5801 (9) | 133 |
| C27—H27···Cg1iv | 0.95 | 2.97 | 3.7258 (8) | 137 |
Symmetry codes: (ii) x+1, y, z; (iii) x, y+1, z; (iv) −x+1, −y+1, −z+1; (v) −x+1, −y+1, −z; (vi) x−1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: JJ2179).
References
- Agilent (2012). CrysAlis PRO Agilent Technologies, Yarnton, England.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Brown, K. L. (2005). Chem. Rev. 105, 2075–2150. [DOI] [PubMed]
- Clark, R. C. & Reid, J. S. (1995). Acta Cryst. A51, 887–897.
- Cross, E. M., White, K. M., Moshrefzadeh, R. S. & Francis, C. V. (1995). Macromolecules, 28, 2526–2532.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Jayamoorthy, K., Mohandas, T., Sakthivel, P. & Jayabharathi, J. (2013). Acta Cryst. E69, o244. [DOI] [PMC free article] [PubMed]
- Jayamoorthy, K., Rosepriya, S., Thiruvalluvar, A., Jayabharathi, J. & Butcher, R. J. (2012). Acta Cryst. E68, o2708. [DOI] [PMC free article] [PubMed]
- Rosepriya, S., Thiruvalluvar, A., Jayamoorthy, K., Jayabharathi, J. & Linden, A. (2011). Acta Cryst. E67, o3519. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spasov, A. A., Yozhitsa, I. N., Bugaeva, L. I. & Anisimova, V. A. (1999). Pharm. Chem. J. 33, 232–243.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681303331X/jj2179sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681303331X/jj2179Isup2.hkl
Supporting information file. DOI: 10.1107/S160053681303331X/jj2179Isup3.cdx
Supporting information file. DOI: 10.1107/S160053681303331X/jj2179Isup4.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report





