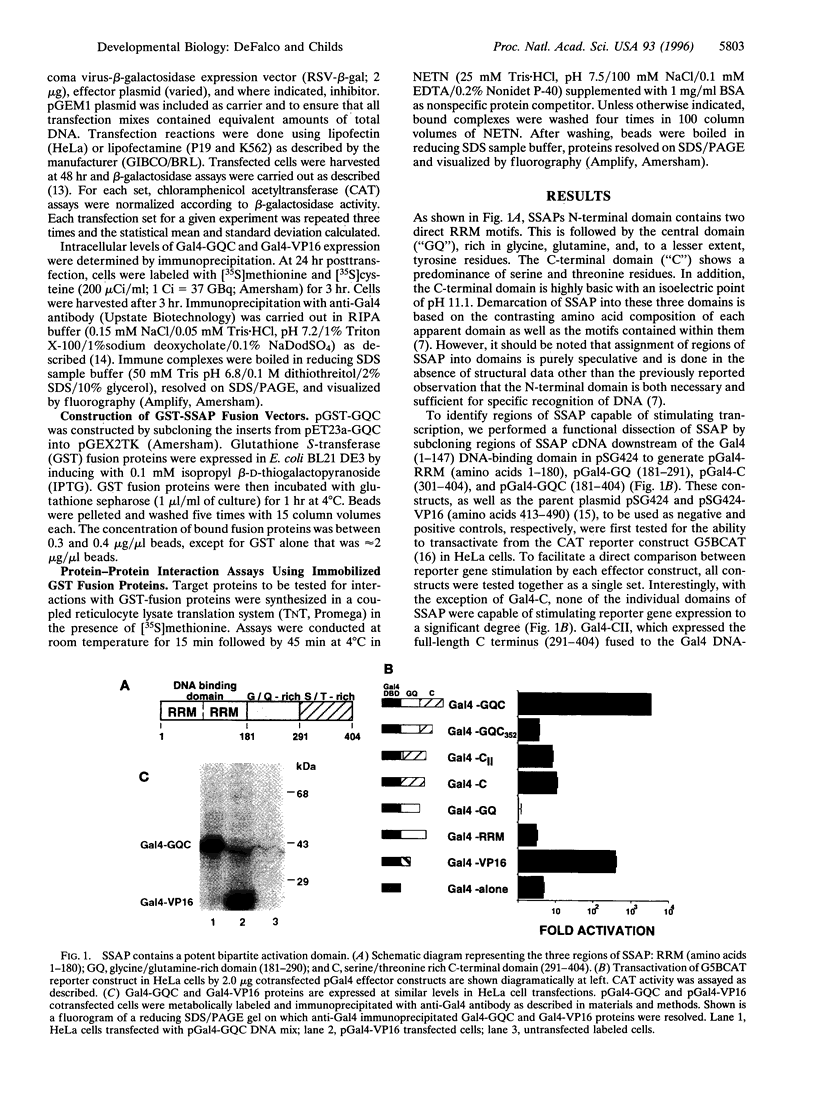
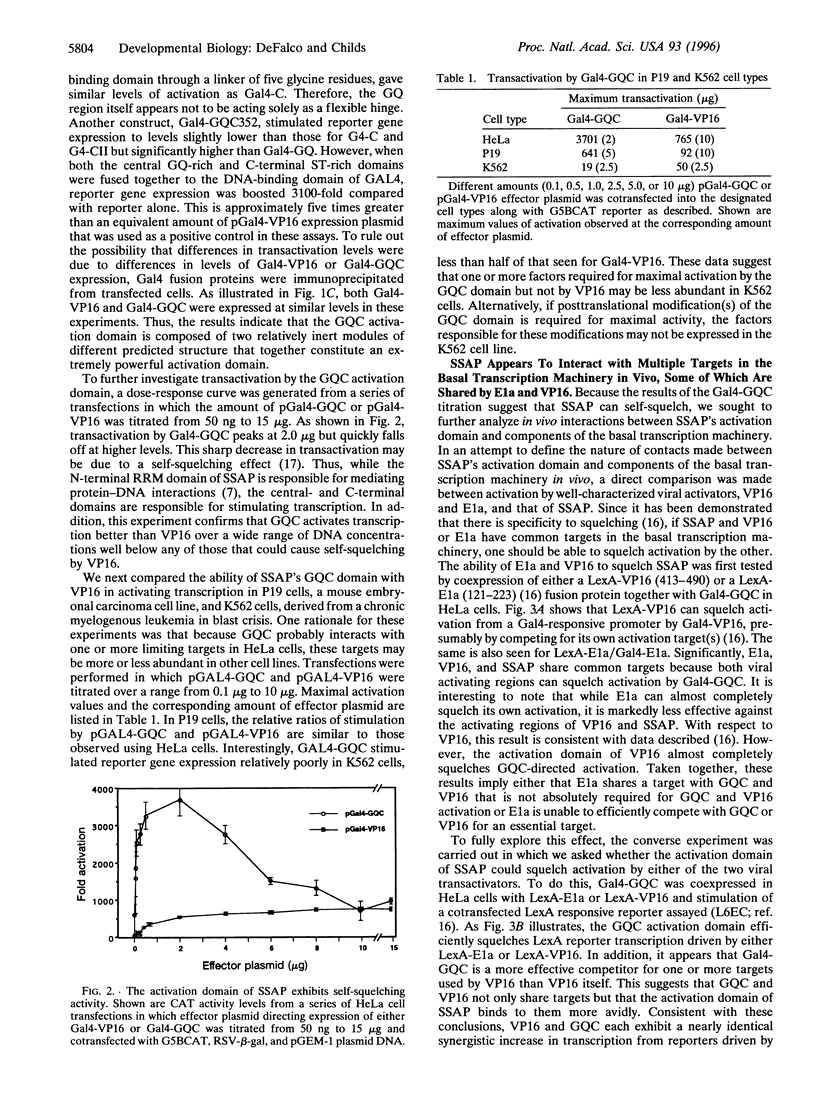
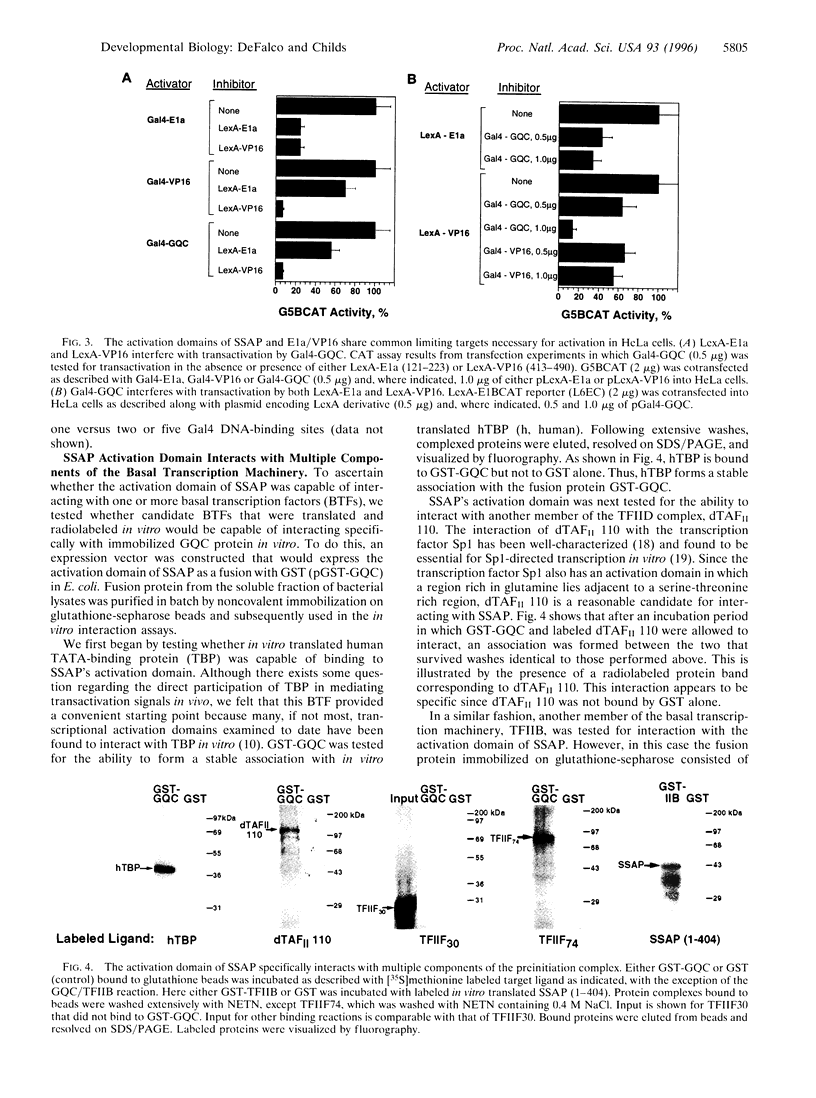
Abstract
Stage specific activator protein (SSAP) is a member of a newly discovered class of transcription factors that contain motifs more commonly found in RNA-binding proteins. Previously, we have shown that SSAP specifically binds to its recognition sequence in both the double strand and the single strand form and that this DNA-binding activity is localized to the N-terminal RNA recognition motif domain. Three copies of this recognition sequence constitute an enhancer element that is directly responsible for directing the transcriptional activation of the sea urchin late histone H1 gene at the midblastula stage of embryogenesis. Here we show that the remainder of the SSAP polypeptide constitutes an extremely potent bipartite transcription activation domain that can function in a variety of mammalian cell lines. This activity is as much as 3 to 5 times stronger than VP16 at activating transcription and requires a large stretch of amino acids that contain glutamine-glycine rich and serine-threonine-basic amino acid rich regions. We present evidence that SSAP's activation domain shares targets that are also necessary for activation by E1a and VP16. Finally, SSAP's activation domain is found to participate in specific interactions in vitro with the basal transcription factors TATA-binding protein, TFIIB, TFIIF74, and dTAF(II) 110.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blair W. S., Bogerd H. P., Madore S. J., Cullen B. R. Mutational analysis of the transcription activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol Cell Biol. 1994 Nov;14(11):7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Chen J. L., Attardi L. D., Verrijzer C. P., Yokomori K., Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994 Oct 7;79(1):93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- Choy B., Green M. R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993 Dec 9;366(6455):531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- Colgan J., Manley J. L. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 1992 Feb;6(2):304–315. doi: 10.1101/gad.6.2.304. [DOI] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- DeAngelo D. J., DeFalco J., Childs G. Purification and characterization of the stage-specific embryonic enhancer-binding protein SSAP-1. Mol Cell Biol. 1993 Mar;13(3):1746–1758. doi: 10.1128/mcb.13.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelo D. J., DeFalco J., Rybacki L., Childs G. The embryonic enhancer-binding protein SSAP contains a novel DNA-binding domain which has homology to several RNA-binding proteins. Mol Cell Biol. 1995 Mar;15(3):1254–1264. doi: 10.1128/mcb.15.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson L., Capone J. P. Purification and characterization of the carboxyl-terminal transactivation domain of Vmw65 from herpes simplex virus type 1. J Biol Chem. 1992 Jan 25;267(3):1411–1414. [PubMed] [Google Scholar]
- Finkelstein A., Kostrub C. F., Li J., Chavez D. P., Wang B. Q., Fang S. M., Greenblatt J., Burton Z. F. A cDNA encoding RAP74, a general initiation factor for transcription by RNA polymerase II. Nature. 1992 Jan 30;355(6359):464–467. doi: 10.1038/355464a0. [DOI] [PubMed] [Google Scholar]
- Gerber H. P., Seipel K., Georgiev O., Höfferer M., Hug M., Rusconi S., Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994 Feb 11;263(5148):808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- Gill G., Pascal E., Tseng Z. H., Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988 Aug 25;334(6184):721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Ha I., Roberts S., Maldonado E., Sun X., Kim L. U., Green M., Reinberg D. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 1993 Jun;7(6):1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- Hardwick J. M., Tse L., Applegren N., Nicholas J., Veliuona M. A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992 Sep;66(9):5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot V., Demma M., Prywes R. Interaction with RAP74 subunit of TFIIF is required for transcriptional activation by serum response factor. Nature. 1995 Feb 16;373(6515):632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- Kim T. K., Roeder R. G. Proline-rich activator CTF1 targets the TFIIB assembly step during transcriptional activation. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J. A., Lai Z. C., Childs G. J. Isolation, characterization, and expression of the gene encoding the late histone subtype H1-gamma of the sea urchin Strongylocentrotus purpuratus. Mol Cell Biol. 1987 Jan;7(1):478–485. doi: 10.1128/mcb.7.1.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z. C., Childs G. Characterization of the structure and transcriptional patterns of the gene encoding the late histone subtype H1-beta of the sea urchin Strongylocentrotus purpuratus. Mol Cell Biol. 1988 Apr;8(4):1842–1844. doi: 10.1128/mcb.8.4.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z. C., Maxson R., Childs G. Both basal and ontogenic promoter elements affect the timing and level of expression of a sea urchin H1 gene during early embryogenesis. Genes Dev. 1988 Feb;2(2):173–183. doi: 10.1101/gad.2.2.173. [DOI] [PubMed] [Google Scholar]
- Lieber T., Weisser K., Childs G. Analysis of histone gene expression in adult tissues of the sea urchins Strongylocentrotus purpuratus and Lytechinus pictus: tissue-specific expression of sperm histone genes. Mol Cell Biol. 1986 Jul;6(7):2602–2612. doi: 10.1128/mcb.6.7.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991 Mar 8;64(5):971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Lillie J. W., Green M. R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990 Jul 12;346(6280):147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- Nelson H. C. Structure and function of DNA-binding proteins. Curr Opin Genet Dev. 1995 Apr;5(2):180–189. doi: 10.1016/0959-437x(95)80006-9. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Regier J. L., Shen F., Triezenberg S. J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989 Sep 25;17(18):7539–7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundseth R., Hansen U. Activation of RNA polymerase II transcription by the specific DNA-binding protein LSF. Increased rate of binding of the basal promoter factor TFIIB. J Biol Chem. 1992 Apr 15;267(11):7845–7855. [PubMed] [Google Scholar]
- Sutherland J. A., Cook A., Bannister A. J., Kouzarides T. Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev. 1992 Sep;6(9):1810–1819. doi: 10.1101/gad.6.9.1810. [DOI] [PubMed] [Google Scholar]
- Tan S., Aso T., Conaway R. C., Conaway J. W. Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J Biol Chem. 1994 Oct 14;269(41):25684–25691. [PubMed] [Google Scholar]
- Tanaka M., Clouston W. M., Herr W. The Oct-2 glutamine-rich and proline-rich activation domains can synergize with each other or duplicates of themselves to activate transcription. Mol Cell Biol. 1994 Sep;14(9):6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora L., White J., Brou C., Tasset D., Webster N., Scheer E., Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989 Nov 3;59(3):477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- Triezenberg S. J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995 Apr;5(2):190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]