Abstract
Several advances in molecular genetics and cardiac imaging of patients with hypertrophic cardiomyopathy (HCM) have been developed in recent years. The commercially available genetic testing is currently used (a) to identify affected relatives in families known to have HCM and (b) to differentiate HCM from metabolic storage disorders and other HCM phenocopies. Cardiovascular magnetic resonance has emerged as a useful tool to assess the magnitude and distribution of hypertrophy or mitral valve apparatus anatomy, which may have an impact on decision making regarding invasive management of HCM.
Introduction
HCM is characterized by a thickened but non-dilated left ventricle that is disproportionate to cardiac hemodynamic load and unexplained by other systemic conditions [1]. It is the most common monogenic cardiovascular disease, affecting 1 in 500 of the general population, and is the most common cause of sudden death in young people (including young athletes) and is an important cause of heart failure and stroke disability [2]. Since the first description over 50 years ago, there are still substantial gaps in our basic understanding of the molecular pathophysiology of HCM as well as the translation of basic insights into the diagnosis, prevention, and therapy of this disease and hence future researches in these areas are needed [3]. This brief review summarizes four major areas of advances: (a) major changes in clinical recommendation from HCM guidelines, (b) new insights from genetic testing for HCM, (c) new SCD (sudden cardiac death) prediction model, and (d) new insights from multimodality imaging.
New changes in hypertrophic cardiomyopathy guidelines
The recently updated 2011 American College of Cardiology Foundation (ACCF) and American Heart Association (AHA) guidelines discussed several new aspects of the management of HCM, including the use of genetic testing, septal reduction therapy, and the role of implantable cardioverter-defibrillator (ICD) to prevent SCD. The recommendations in the new guidelines are rooted primarily in expert consensus (Level C) given a lack of high levels of evidence regarding HCM provided by clinical trials. The key points of the new guidelines are summarized as follows.
Clinical genetic testing
The new guidelines point out that the relation between known HCM-related gene variants and clinical outcome has proved unreliable due to genetic and phenotypic heterogeneity. Therefore, genetic testing is not recommended (Class III) unless the index patient has a definitive pathogenic mutation. In Class I recommendation, the guidelines indicate that screening (clinical with or without genetic testing) is recommended for all first-degree relatives of patients with HCM and that “genetic testing for HCM and other genetic causes of unexplained cardiac hypertrophy is recommended in patients with an atypical clinical presentation of HCM or when another genetic condition is suspected to be the cause” [1].
Septal reduction therapy
The guidelines emphasize that the invasive correction of left ventricular outflow tract (LVOT) obstruction should be performed only by experienced operators in the context of a comprehensive HCM clinical program and only for the treatment of eligible patients with severe drug-refractory symptoms and a resting or provoked LVOT gradient of at least 50 mm Hg (Class I). Surgical septal myectomy is generally preferred when septal reduction is appropriate; alcohol septal ablation can be considered in patients with serious comorbidities or when surgery is contraindicated (Class IIa).
Implantable cardioverter-defibrillator therapy for prevention of sudden cardiac death
ICD therapy is recommended in patients with prior cardiac arrest, ventricular fibrillation, or hemodynamically significant ventricular tachycardia (VT) (Class I); in those with a history of SCD in a first-degree relative, marked LVH (left ventricular hypertrophy) (≥30 mm), or recent unexplained syncope (Class IIa); and in those with non-sustained VT or an abnormal systolic blood-pressure response (failure to increase ≥20 mm Hg or drop ≥20 mm Hg) to exercise (Class IIa or IIb, depending upon the presence of other risk modifiers, including marked LVOT obstruction). The new risk score for SCD risk prediction was recently developed (discussed below).
New insights in genetic testing
Phenotype and genotype heterogeneity and complexity
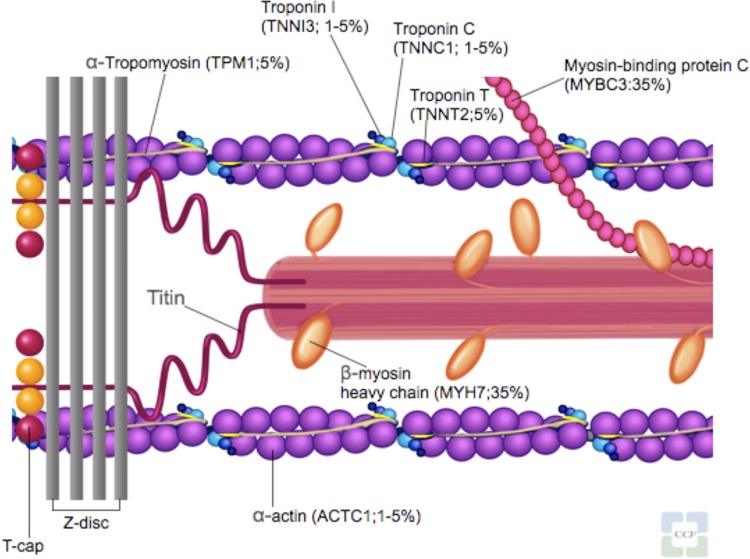
HCM is a primary autosomal-dominant disorder of the myocardium. It was initially called a “disease of the sarcomere” when the first identified genes of HCM were found to encode the contractile unit within the cardiac myocytes [4]. To date, over 1400 mutations in 20 sarcomere-related and myofilament-related genes have been identified in HCM [2,5] (Figure 1 and Table 1). However, not all of these genes listed as causing HCM are adequately proven and even within the proven HCM disease genes (MYH7, MYBPC3, TNNT2, TNNI3, TPN3, MYL2, MYL3, ACTC1, and CSRP3), the causality to individual alleles is still unknown [6]. Most HCM mutations (about 90%) are missense and unique to individual families [7]. Because of the age-related and incomplete penetrance (i.e. the proportion of mutation-positive subjects who express a clinically detectable phenotype), absence of disease at one evaluation cannot exclude subsequent development.
Figure 1. Sarcomere and myofilament diagram.
Sarcomere-related and myofilament-related genes known to cause hypertrophic cardiomyopathy.
Table 1. Hypertrophic cardiomyopathy-causing genes and associated phenotypes.
| Gene | Protein | Frequencies in patients with HCM | Associated phenotype |
|---|---|---|---|
| MYH7 | β-Myosin heavy chain | 25%-35% | Mild or severe HCM |
| MYBPC3 | Myosin-binding protein C (cardiac type) | 20%-30% | Expression similar to MYH7, late onset |
| TNNT2 | Troponin T (cardiac muscle) | 3%-5% | Mild hypertrophy, sudden death |
| TNNI3 | Troponin I (cardiac muscle) | <5% | Extreme intrafamilial heterogeneity, no sudden death without severe disease |
| TPM1 | Tropomyosin 1α | <5% | Variable prognosis, sudden death |
| MYL2 | Regulatory myosin light chain 2 (ventricular/cardiac-muscle isoform) | <5% | Skeletal myopathy |
| MYL3 | Essential myosin light chain 3 | Rare | Skeletal myopathy |
| ACTC | α-Cardiac actin 1 | Rare | Skeletal myopathy |
| TTN | Titin | Rare | Typical HCM |
| TNNC1 | Troponin C, slow skeletal and cardiac muscles | Rare | Typical HCM |
| MYH6 | α-Myosin heavy chain | Rare | Late onset |
| CSRP6 | Muscle LIM protein | Rare | Late onset, variable penetrance |
| MYLK2 | Myosin light chain kinase 2 | Rare | Early onset |
| LDB3 | LIM-binding domain 3 | Rare | Mainly sigmoidal HCM |
| TCAP | Telethonin | Rare | Typical HCM, variable penetrance |
| VCL | Vinculin/metavinculin | Rare | Obstructive midventricular hypertrophy |
| ACTN2 | α-Actinin 2 | Rare | Mainly sigmoidal HCM |
| PLN | Phospholamban | Rare | Typical HCM, variable penetrance |
| MYOZ2 | Myozenin 2 | Rare | Typical HCM |
| JPH2 | Junctophilin 2 | Rare | Typical HCM |
HCM, hypertrophic cardiomyopathy. Reprinted with permission from Nature Publishing Group [5].
Challenges in routine genetic testing
Cascade screening of family members of the proband is important [7]. Although HCM proband genetic testing has been commercially available since 2003 [8] and was shown to be a cost-effective strategy when added to conventional HCM management [9], there are still a number of limitations. Specifically, the pathogenic mutations can only be identified in 50%-60% of clinically affected probands [10,11], as all genes causing HCM have not yet been identified, and are absent from testing panels [8,12]. A recent study using high-throughput sequencing (HTS) technology showed that the frequency of non-sarcomeric variants (genes encoding Z-disc or calcium-handling proteins) was similar to the control population and hence raised the suspicion of whether the previously published pathogenic mutations in RYR2, ANK2, CAV3, and SCN5A may be only potential phenotype modifiers in HCM [13]. Additionally, DNA-based testing frequently identifies a “variant of uncertain significance” (VUS), the nucleotide change for which there is not sufficient data to determine if it causing disease or is a benign variant [1,12]. These ambiguous variants usually have unknown clinical implications for family screening and may create confusion in genetic testing interpretation.
Genotype-positive phenotype-negative hypertrophic cardiomyopathy
As more families undergo genetic testing for HCM, a new preclinical population (genotype-positive, phenotype-negative) is growing. These apparently healthy individuals carry the family's HCM-causing variant, yet do not have evidence of hypertrophy. The guidelines, however, recommended electrocardiogram (ECG), transthoracic echocardiogram (TTE), and clinical assessment in these patients at periodic intervals (12 to 18 months in children and adolescents and about every 5 years in adults), based on the patient's age and change in clinical status [1]. At the moment, the sudden death risk and disease progression of this population are still unknown and hence the decision about prophylactic ICD or disqualification from competitive sport participation is usually resolved on a case-by-case basis [2,14].
Phenocopies
The term phenocopy refers to apparently similar disorders with different causes [4]. Several genetic cardiomyopathies and metabolic diseases may present with left ventricular hypertrophy and mimic HCM (Table 2). Differentiation between sarcomeric HCM and its phenocopies is important, given differences in inheritance pattern, natural history, and management. For example, LAMP2 cardiomyopathy (Danon disease) is associated with a lethal clinical course within the first three decades that requires early recognition to consider prophylactic heart transplantation [15]. Fabry disease may benefit from recombinant α-galactosidase A enzyme replacement [16]. Phenocopy should be suspected when an atypical feature of classic HCM or multi-organ involvement is present, e.g. Wolff-Parkinson-White pattern in PRKAG2 and LAMP2, greatly increased precordial voltage and massive left ventricular hypertrophy (LVH) in patients with LAMP2 mutation, symmetric LVH and posterobasal late gadolinium enhancement in Fabry disease or multisystemic disease (central and peripheral nervous system and eyes) in mitochondrial cardiomyopathies [8].
Table 2. Hypertrophic cardiomyopathy phenocopies.
| Gene | Protein | Frequency in patients with HCM phenocopy diseases | Associated phenotype |
|---|---|---|---|
| PRKAG2 | AMP-activated protein kinase γ2 regulatory subunit | <1% | Wolff-Parkinson-White syndrome, conduction disease |
| GLA | α-Galactosidase A | <5% | Anderson-Fabry disease |
| GAA | Acid α-1,4-glucosidase | Rare | Pompe disease |
| AGL | Amylo-1,6-glucosidase | Rare | Forbes disease |
| LAMP2 | Lysosomal-associated membrane protein 2 | Rare | Danon disease |
| Various mitochondrial genes (e.g., MTTG, MTTI) | Protein-encoding mitochondrial ribosomal and transfer RNA | Rare | Mitochondrial cytopathy (MELAS, MERRF, LHON) |
| PTPN11 | Protein tyrosine phosphatase, non-receptor type 11, SHP-2 | Rare | LEOPARD syndrome, Noonan syndrome |
| FRDA | Frataxin | Rare | Friedreich's ataxia |
| KRAS | v-Ki-ras2 Kristen rate sarcoma viral oncogene homolog | Rare | Noonan syndrome |
| SOS1 | Son of sevenless homolog 1 | Rare | Noonan syndrome |
Abbreviations: LHON, Leber's hereditary optic neuropathy; MELAS, mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes; MERRF, myoclonus epilepsy associated with ragged-red fibers. Reprinted with permission from Nature Publishing Group [5].
New sudden cardiac death prediction model
The validation study of current strategies for identifying patients with HCM who are at high risk of SCD suggested that these algorithms overestimate risk, resulting in unnecessary prophylactic ICD implantation in a substantial number of patients [17]. These inappropriate ICD implantations are concerning because approximately one third of patients who had ICD will experience either an inappropriate shock or implant complication within 5 years [18]. A new, validated SCD risk prediction model was recently published [19]. The investigators used data from a retrospective, longitudinal cohort study of 3675 patients evaluated at one of six large centers in Europe. The study included only adults (>16 years) without metabolic diseases or syndromic causes of HCM and without a history of ventricular fibrillation (VF) or VT. Patients were followed up for a median of 5.7 years. Age, family history of SCD, left atrial diameter, maximal left ventricular wall thickness, maximal LVOT gradient, non-sustained VT, and unexplained syncope were significantly associated with SCD in the study cohort. This model suggested that for every 16 ICDs implanted in patients with at least 4% 5-year SCD risk, potentially one patient will be saved from SCD at 5 years. Clinicians should be cautious in its use as the authors pointed out that “the model should only be used in patients with similar characteristics to the study cohort” [19]. Some high-risk patient subgroups (those with prior SCD) were not included, and treatments such as surgical myectomy or septal ablation and beta-blockers were not considered (a risk calculator will become available online at www.HCMRisk.org).
Multimodality imaging in hypertrophic cardiomyopathy clinical decision-making
Echocardiography is particularly important in its ability to demonstrate dynamic outflow tract obstruction (both unprovoked or perturbated) as well as to define clinically relevant abnormalities in cardiac (as well as valvular) structure and function. Echocardiography may underestimate the degree of ventricular hypertrophy, especially when it is confined to the anterolateral wall [20], apex [21], and posterior septum [21]. Cardiac magnetic resonance (CMR) provides superior spatial resolution and complete visualization of the entire left ventricular myocardium. Therefore, CMR can clarify the diagnosis and phenotype of HCM, particularly with regard to distribution of LVH and mechanism of LVOT obstruction. However, to date, there has been no definitive evidence so far suggesting that adding CMR to echocardiography would affect the outcomes in the management of HCM.
Mitral valve and subvalvular apparatus
Coexisting intrinsic mitral valve (MV) abnormalities are common in patients with HCM, and approximately 14% of patients undergoing surgery for HCM require concomitant mitral valve surgery [22]. The anterior and posterior MV leaflet lengths were greater in patients with HCM compared with those of the control group [23]. Additionally, mitral annular calcification, degenerative changes, myxomatous changes, chordal rupture, and fibrosis of the mitral leaflets have been reported in HCM [24]. The differentiation between mitral regurgitation (MR) secondary to systolic anterior motion (SAM) versus MR from intrinsic MV disease is important given the different management (i.e. septal reduction therapy alone in the case of SAM-causing MR or concomitant MV surgery in the case of intrinsic MV disease). The presence of severe MR without SAM and the central or anterior-directed MR jet suggest intrinsic MV disease. Papillary muscle in HCM has been described to be abnormal in number, position, morphology, mobility, insertion, or a combination of these abnormalities. In patients whose papillary muscle is hypermobile and causes LVOT obstruction, papillary muscle reorientation surgery has been shown to improve symptoms and LVOT obstruction [25].
Myocardial crypt
The area of myocardial cleft, crypt, or crevices had been described in the basal and mid posterior left ventricular wall in HCM carriers who have not yet developed hypertrophy [26]. Basal ventricular septal crypts have also been reported in genotype-negative non-obstructive HCM patients. The morphology of septal crypts appeared to be deeper compared to the inferoseptal crypts. However, the clinical significance of these septal crypts is still unclear [27].
Role of fibrosis in hypertrophic cardiomyopathy
Foci of late gadolinium enhancement (LGE) in HCM usually occur in areas of maximal hypertrophy or right ventricle-left ventricle insertion points and have been correlated with the area of interstitial expansion due to myocardial fibrosis [28,29]. Patients with HCM with evidence of LGE on CMR imaging tend to have more markers of risk of SCD, such as non-sustained ventricular tachycardia (NSVT), on Holter monitoring than patients without LGE [30]. However, available data are currently insufficient to consider LGE as an independent risk marker of SCD in HCM.
T1 mapping in hypertrophic cardiomyopathy
LGE imaging has become the gold standard technique for imaging focal myocardial fibrosis in non-ischemic cardiomyopathy [31] but not diffuse fibrosis, as there are no reference regions of normal myocardium [32]. The recent development technique of T1 mapping, typically performed after gadolinium-based contrast agent administration, has proved useful in demonstrating expanded extracellular volume in HCM and showed good correlation with fibrosis on histopathology [33], a technique known as “non-invasive myocardial biopsy”. Given the prognostic data available in HCM for the evaluation of focal fibrosis with LGE, it is likely that T1 mapping may provide additional prognostic value in this disease as it reflects both focal and diffuse fibrosis [34].
Conclusions
Several issues in the management of HCM patients had changed since the updates of the 2011 guideline. Despite new advances in our pathophysiological understanding of HCM in recent years, their impact on clinical management strategies has been limited. Genetic testing can be a helpful tool to identify individuals at risk for developing disease or to clarify ambiguous diagnoses but needs to be approached and interpreted properly. The new SCD risk prediction model has been developed and validated. The emerging role of CMR was convincing in defining cardiac anatomy and physiology, but again the outcomes studies are still needed.
Abbreviations
- CMR
cardiac magnetic resonance
- HCM
hypertrophic cardiomyopathy
- ICD
implantable cardioverter-defibrillator
- LGE
late gadolinium enhancement
- LVH
left ventricular hypertrophy
- LVOT
left ventricular outflow tract
- MR
mitral regurgitation
- MV
mitral valve
- SAM
systolic anterior motion
- SCD
sudden cardiac death
- VF
ventricular fibrillation
- VT
ventricular tachycardia
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/m/6/12
References
- 1.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, Naidu SS, Nishimura RA, Ommen SR, Rakowski H, Seidman CE, Towbin JA, Udelson JE, Yancy CW. 2011 ACCF/AHA Guideline for the Diagnosis and Treatment of Hypertrophic Cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Developed in collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol. 2011;58:e212–60. doi: 10.1016/j.jacc.2011.06.011. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240410
- 2.Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381:242–55. doi: 10.1016/S0140-6736(13)60922-8. [DOI] [PubMed] [Google Scholar]
- 3.Force T, Bonow RO, Houser SR, Solaro RJ, Hershberger RE, Adhikari B, Anderson ME, Boineau R, Byrne BJ, Cappola TP, Kalluri R, LeWinter MM, Maron MS, Molkentin JD, Ommen SR, Regnier M, Tang WHW, Tian R, Konstam MA, Maron BJ, Seidman CE. Research priorities in hypertrophic cardiomyopathy: report of a Working Group of the National Heart, Lung, and Blood Institute. Circulation. 2010;122:1130–3. doi: 10.1161/CIRCULATIONAHA.110.950089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watkins H, Ashrafian H, Redwood C. Inherited cardiomyopathies. N Engl J Med. 2011;364:1643–56. doi: 10.1056/NEJMra0902923. [DOI] [PubMed] [Google Scholar]
- 5.Keren A, Syrris P, McKenna WJ. Hypertrophic cardiomyopathy: the genetic determinants of clinical disease expression. Nat Clin Pract Cardiovasc Med. 2008;5:158–68. doi: 10.1038/ncpcardio1349. [DOI] [PubMed] [Google Scholar]
- 6.Watkins H. Assigning a causal role to genetic variants in hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2013;6:2–4. doi: 10.1161/CIRCGENETICS.111.000032. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240411
- 7.Wang L, Seidman JG, Seidman CE. Narrative review: harnessing molecular genetics for the diagnosis and management of hypertrophic cardiomyopathy. Ann Intern Med. 2010;152:513–20. doi: 10.7326/0003-4819-152-8-201004200-00008. W181. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718240412
- 8.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60:705–15. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240413
- 9.Ingles J, McGaughran J, Scuffham PA, Atherton J, Semsarian C. A cost-effectiveness model of genetic testing for the evaluation of families with hypertrophic cardiomyopathy. Heart. 2012;98:625–30. doi: 10.1136/heartjnl-2011-300368. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240414
- 10.Landstrom AP, Ackerman MJ. Mutation type is not clinically useful in predicting prognosis in hypertrophic cardiomyopathy. Circulation. 2010;122:2441–9. doi: 10.1161/CIRCULATIONAHA.110.954446. discussion 2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bos JM, Towbin JA, Ackerman MJ. Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54:201–11. doi: 10.1016/j.jacc.2009.02.075. [DOI] [PubMed] [Google Scholar]
- 12.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Le Marec H, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, Zipes DP. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8:1308–39. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 13.Lopes LR, Zekavati A, Syrris P, Hubank M, Giambartolomei C, Dalageorgou C, Jenkins S, McKenna W, Plagnol V, Elliott PM. Genetic complexity in hypertrophic cardiomyopathy revealed by high-throughput sequencing. J Med Genet. 2013;50:228–39. doi: 10.1136/jmedgenet-2012-101270. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718240415
- 14.Maron BJ, Yeates L, Semsarian C. Clinical challenges of genotype positive (+)-phenotype negative (-) family members in hypertrophic cardiomyopathy. Am J Cardiol. 2011;107:604–8. doi: 10.1016/j.amjcard.2010.10.022. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240417
- 15.Maron BJ, Roberts WC, Arad M, Haas TS, Spirito P, Wright GB, Almquist AK, Baffa JM, Saul JP, Ho CY, Seidman J, Seidman CE. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301:1253–9. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718240418
- 16.Weidemann F, Niemann M, Breunig F, Herrmann S, Beer M, Störk S, Voelker W, Ertl G, Wanner C, Strotmann J. Long-term effects of enzyme replacement therapy on fabry cardiomyopathy: evidence for a better outcome with early treatment. Circulation. 2009;119:524–9. doi: 10.1161/CIRCULATIONAHA.108.794529. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240419
- 17.O'Mahony C, Tome-Esteban M, Lambiase PD, Pantazis A, Dickie S, McKenna WJ, Elliott PM. A validation study of the 2003 American College of Cardiology/European Society of Cardiology and 2011 American College of Cardiology Foundation/American Heart Association risk stratification and treatment algorithms for sudden cardiac death in patients with hypertrophic cardiomyopathy. Heart. 2013;99:534–41. doi: 10.1136/heartjnl-2012-303271. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240420
- 18.Mearns BM. Cardiomyopathy: New SCD risk prediction model. Nat Rev Cardiol. 2013;10:680. doi: 10.1038/nrcardio.2013.169. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240421
- 19.O'Mahony C, Jichi F, Pavlou M, Monserrat L, Anastasakis A, Rapezzi C, Biagini E, Gimeno JR, Limongelli G, McKenna WJ, Omar RZ, Elliott PM. A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM Risk-SCD) Eur Heart J. 2013 doi: 10.1093/eurheartj/eht439. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718144431
- 20.Maron MS, Lesser JR, Maron BJ. Management implications of massive left ventricular hypertrophy in hypertrophic cardiomyopathy significantly underestimated by echocardiography but identified by cardiovascular magnetic resonance. Am J Cardiol. 2010;105:1842–3. doi: 10.1016/j.amjcard.2010.01.367. [DOI] [PubMed] [Google Scholar]
- 21.Maron MS, Maron BJ, Harrigan C, Buros J, Gibson CM, Olivotto I, Biller L, Lesser JR, Udelson JE, Manning WJ, Appelbaum E. Hypertrophic cardiomyopathy phenotype revisited after 50 years with cardiovascular magnetic resonance. J Am Coll Cardiol. 2009;54:220–8. doi: 10.1016/j.jacc.2009.05.006. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240422
- 22.Kaple RK, Murphy RT, DiPaola LM, Houghtaling PL, Lever HM, Lytle BW, Blackstone EH, Smedira NG. Mitral valve abnormalities in hypertrophic cardiomyopathy: echocardiographic features and surgical outcomes. Ann Thorac Surg. 2008;85:1527–35. doi: 10.1016/j.athoracsur.2008.01.061. 1535.e1-2. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240423
- 23.Maron MS, Olivotto I, Harrigan C, Appelbaum E, Gibson CM, Lesser JR, Haas TS, Udelson JE, Manning WJ, Maron BJ. Mitral valve abnormalities identified by cardiovascular magnetic resonance represent a primary phenotypic expression of hypertrophic cardiomyopathy. Circulation. 2011;124:40–7. doi: 10.1161/CIRCULATIONAHA.110.985812. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240424
- 24.Cavalcante JL, Barboza JS, Lever HM. Diversity of mitral valve abnormalities in obstructive hypertrophic cardiomyopathy. Prog Cardiovasc Dis. 2012;54:517–22. doi: 10.1016/j.pcad.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Kwon DH, Smedira NG, Thamilarasan M, Lytle BW, Lever H, Desai MY. Characteristics and surgical outcomes of symptomatic patients with hypertrophic cardiomyopathy with abnormal papillary muscle morphology undergoing papillary muscle reorientation. J Thorac Cardiovasc Surg. 2010;140:317–24. doi: 10.1016/j.jtcvs.2009.10.045. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240425
- 26.Germans T, Wilde AAM, Dijkmans PA, Chai W, Kamp O, Pinto YM, van Rossum AC. Structural abnormalities of the inferoseptal left ventricular wall detected by cardiac magnetic resonance imaging in carriers of hypertrophic cardiomyopathy mutations. J Am Coll Cardiol. 2006;48:2518–23. doi: 10.1016/j.jacc.2006.08.036. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240426
- 27.Maron BJ, Lindberg J, Lesser JR. Ventricular septal crypt in hypertrophic cardiomyopathy. Eur Heart J. 2010;31:1923. doi: 10.1093/eurheartj/ehq140. [DOI] [PubMed] [Google Scholar]
- 28.Maron MS. Clinical utility of cardiovascular magnetic resonance in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson. 2012;14:13. doi: 10.1186/1532-429X-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718240429
- 29.Maron MS, Maron BJ, Harrigan C, Buros J, Gibson CM, Olivotto I, Biller L, Lesser JR, Udelson JE, Manning WJ, Appelbaum E. Hypertrophic cardiomyopathy phenotype revisited after 50 years with cardiovascular magnetic resonance. J Am Coll Cardiol. 2009;54:220–8. doi: 10.1016/j.jacc.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Adabag AS, Maron BJ, Appelbaum E, Harrigan CJ, Buros JL, Gibson CM, Lesser JR, Hanna CA, Udelson JE, Manning WJ, Maron MS. Occurrence and frequency of arrhythmias in hypertrophic cardiomyopathy in relation to delayed enhancement on cardiovascular magnetic resonance. J Am Coll Cardiol. 2008;51:1369–74. doi: 10.1016/j.jacc.2007.11.071. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1157477
- 31.Assomull RG, Prasad SK, Lyne J, Smith G, Burman ED, Khan M, Sheppard MN, Poole-Wilson PA, Pennell DJ. Cardiovascular magnetic resonance, fibrosis, and prognosis in dilated cardiomyopathy. J Am Coll Cardiol. 2006;48:1977–85. doi: 10.1016/j.jacc.2006.07.049. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1052861
- 32.Salerno M, Kramer CM. Advances in parametric mapping with CMR imaging. JACC Cardiovasc Imaging. 2013;6:806–22. doi: 10.1016/j.jcmg.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flett AS, Hayward MP, Ashworth MT, Hansen MS, Taylor AM, Elliott PM, McGregor C, Moon JC. Equilibrium contrast cardiovascular magnetic resonance for the measurement of diffuse myocardial fibrosis: preliminary validation in humans. Circulation. 2010;122:138–44. doi: 10.1161/CIRCULATIONAHA.109.930636. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718240430
- 34.Green JJ, Berger JS, Kramer CM, Salerno M. Prognostic value of late gadolinium enhancement in clinical outcomes for hypertrophic cardiomyopathy. JACC Cardiovasc Imaging. 2012;5:370–7. doi: 10.1016/j.jcmg.2011.11.021. [DOI] [PubMed] [Google Scholar]