Abstract
We report discoveries of different haplotypes associated with the centromeres of three potato chromosomes, including haplotypes composed of long arrays of satellite repeats and haplotypes lacking the same repeats. These results are in favor of the hypothesis that satellite repeat-based centromeres may originate from neocentromeres that lack repeats.
Keywords: placeholder, Centromere, neocentromere, satellite repeat, centromere evolution
THE centromere is the chromosomal domain that guides the assembly of the kinetochore where microtubules attach during cell division. Centromeres are defined by the presence of a centromere-specific histone H3 variant, cenH3 (CENP-A in humans) (Henikoff et al. 2001). In many higher eukaryotes, centromeres are composed of long arrays of a single family of satellite repeat (Henikoff et al. 2001; Jiang et al. 2003). For example, human centromeres contain megabase-sized arrays of the 171-bp alpha satellite repeat (Willard and Waye 1987; Vafa and Sullivan 1997). Centromeric satellite repeats can evolve rapidly, which may result in different centromeric repeats among closely related species (Lee et al. 2005). Thus, a new centromeric repeat can sweep through the entire genome and rapidly replace the old repeat in all centromeres via poorly understood mechanisms. The cenH3/CENP-A nucleosomes in rice and humans are highly phased on the 155/171-bp monomers of the CentO and alpha satellite repeats, respectively, and show a distinct structure compared to canonical nucleosomes (Hasson et al. 2013; Zhang et al. 2013b). Therefore, satellite repeats are intrinsically more suitable for organizing centromeric nucleosomes than single- and low-copy DNA sequences.
Centromeres can be activated from noncentromeric regions by acquiring cenH3/CENP-A, resulting in “neocentromeres” (duSart et al. 1997). Neocentromere activation is usually associated with the loss of the original centromere (duSart et al. 1997), or with inactivation but retention of the original centromere on the same chromosome (Amor et al. 2004; Wang et al. 2014). The vast majority of neocentromeres, including several plant neocentromeres (Nasuda et al. 2005; Fu et al. 2013; Wang et al. 2014; Zhang et al. 2013a), do not contain long arrays of satellite repeats that are typically associated with centromeres. The neocentromeric DNA sequences are generally deficient in genes, but otherwise not distinct from average genome sequences (Marshall et al. 2008; Wang et al. 2014). A centromere can also “move” to a different position along the chromosome during evolution (Ventura et al. 2001; Han et al. 2009). Such “centromere reposition” events are likely mediated via neocentromere activation (Rocchi et al. 2012). Results from extensive neocentromere activation and centromere repositioning research have led to a hypothesis of centromere evolution: many existing centromeres may have originated as neocentromeres.
Mature centromeres are typically composed of long arrays of satellite repeats. Thus, an evolutionarily new centromere likely acquires a long array of satellite repeats during evolution. Such a repeat-based haplotype will eventually sweep through populations and replace the original neocentromere that lacks satellite repeats. According to this hypothesis, if a centromere is in the transition stage during evolution, we would find different haplotypes associated with the same centromere in populations, including the haplotype that contains mainly single- or low-copy DNA sequences and an additional haplotype(s) that acquired satellite repeats. However, no case of multiple haplotypes associated with the same centromere has been reported in literature.
Potato (Solanum tuberosum, 2n = 4x = 48) is an autotetraploid with a highly heterogeneous genome. The 12 potato centromeres show drastically different structure and DNA composition. Five potato centromeres (Cen4, Cen6, Cen10, Cen11, and Cen12) contain mainly single- and low-copy sequences. No satellite repeats were identified in these five centromeres. Thus, the structures of these five centromeres resemble neocentromeres (Gong et al. 2012). In contrast, six potato centromeres (Cen1, Cen2, Cen3, Cen5, Cen7, and Cen8) contain megabase-sized arrays of satellite repeats. The satellite repeats are specific to individual centromeres and appeared to completely occupy the cenH3-binding domains of the six potato centromeres (Gong et al. 2012). The fact that each potato centromere contains different DNA sequences provides unique opportunities to study evolution of DNA sequences associated with individual centromeres.
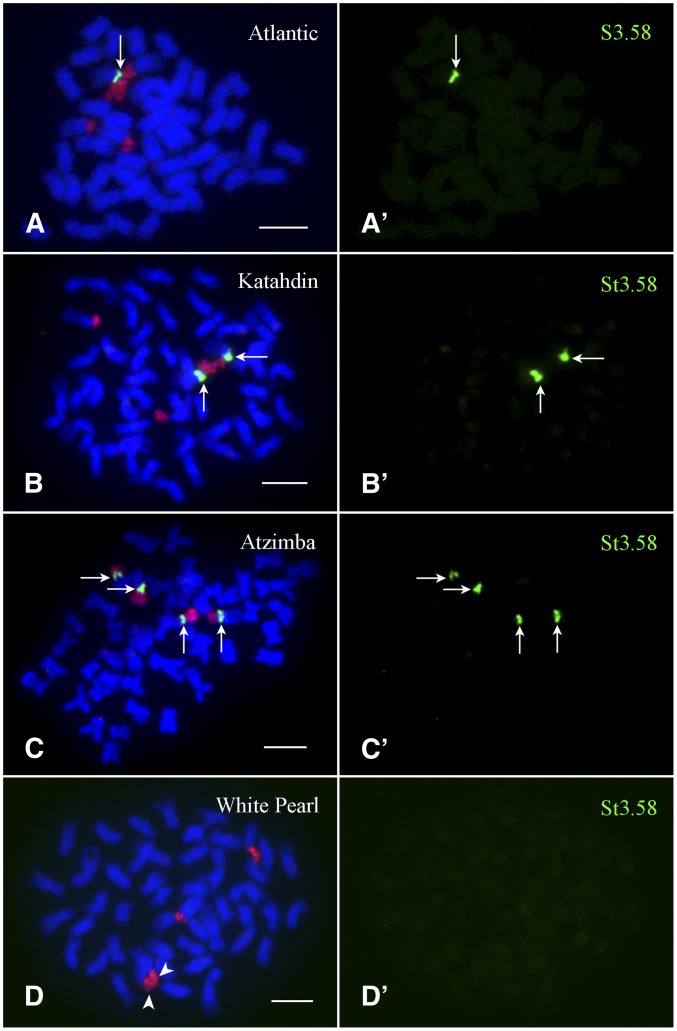
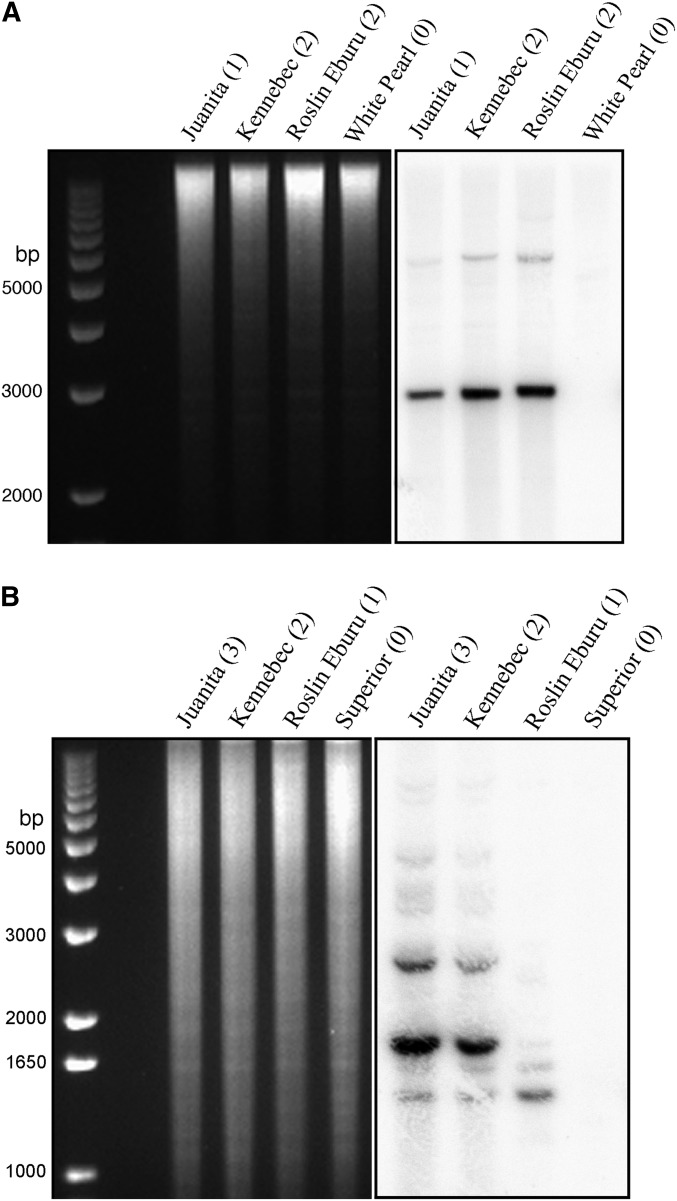
The potato centromeric repeats were isolated from DM 1-3 516 R44 (DM), a homozygous diploid clone (2n = 2x = 24) (Potato Genome Sequencing Consortium 2011). We were interested in the distribution of these repeats in cultivated potato genotypes because the autotetraploidy nature allows potato cultivars to maintain chromosomal variants that could be lethal to diploid species (Iovene et al. 2013). We conducted fluorescence in situ hybridization (FISH) for three repeats, repeat St24 for the centromere of chromosome 1 (Cen1), repeat St3.58 for Cen2, and repeat St57 for Cen7, in a set of 17 diverse potato cultivars developed in Europe and North America (Table 1). Each probe was expected to hybridize to all four copies of each homologous potato chromosome if the same centromere haplotype was fixed in populations. For example, repeat St3.58 is associated with Cen2 in DM. We expected that the St3.58 probe would hybridize to all four homologs of chromosome 2 in all cultivars. Surprisingly, the St3.58 probe hybridized to all four homologs of chromosome 2 in only two of the 17 cultivars examined (Table 1). Only one or two homologs of chromosome 2 contained the St3.58 repeat in 14 cultivars (Figure 1). One cultivar, White Pearl, did not contain this repeat (Figure 1D). Absence of the St3.58 repeat in White Pearl was also confirmed by Southern blot hybridization (Figure 2A). Based on the size and intensity of the FISH signals, the lengths of the St3.58 repeat array associated with different Cen2 in different cultivars appeared to be uniform and similar to those in DM (Gong et al. 2012). These results showed that the St3.58-containing Cen2 represents only one haplotype of this centromere in potato.
Table 1. Summary of FISH results of three centromeric repeats in tetraploid potato genotypes.
| Cultivar | Origin | St24 (Cen1) | St3.58 (Cen2) | St57 (Cen7) |
|---|---|---|---|---|
| Atlantic | United States | 2a | 1 | 1S, 2W |
| Atzimba | Mexico | 1 | 4 | 1S, 2W |
| Freedom Russet | United States | 3 | 2 | 1 |
| Hindenburg | Germany | 1 | 2 | 2S, 1W |
| Juanita | Mexico | 2S, 1Wb | 1 | 1 |
| Kalkaska | United States | 3 | 2 | 1 |
| Katahdin | United States | 3 | 2 | 1S, 2W |
| Kennebec | United States | 1S, 1W | 2 | 1 |
| Kerr’s Pink | United Kingdom | 2 | 1 | 2S, 1W |
| MegaChip | United States | 3 | 2 | 1S, 1W |
| Norkotah | United States | 2 | 2 | 1S, 2W |
| Ranger Russet | United States | 2 | 4 | 1S, 1W |
| Roslin Eburu | United Kingdom | 1 | 2 | 3 |
| Russet Burbank | United States | 4 | 2 | 1S, 2W |
| Snowden | United States | 1S, 1W | 2 | 1S, 1W |
| Superior | United States | 0 | 2 | 1 |
| White Pearl | United States | 1 | 0 | 1 |
The numbers indicate the number of homologs of the chromosome that hybridized to the centromeric repeat.
S, Strong signals; W, Weak signals. Numbers that are not followed by S or W refer to the numbers of strong or weak signals.
Figure 1.
FISH mapping of the Cen2-specific satellite repeat St3.58 in four potato cultivars. Potato chromosome 2 is identified by the 45S rDNA probe (red signals), which is located on the short arm of chromosome 2. (A) FISH signal from St3.58 (arrow) was detected on a single Cen2 in Atlantic. (A′) The FISH signal from St3.58 was digitally separated from A. (B) FISH signals from St3.58 (arrows) were detected on two of the four copies of Cen2 in Katahdin. (B′) The FISH signals from St3.58 were digitally separated from B. (C) FISH signals from St3.58 (arrows) were detected on all four copies of Cen2 in Atzimba. (C′) The FISH signals from St3.58 were digitally separated from C. (D) St3.58 did not hybridize to any of the four copies of Cen2 in White Pearl. The two arrowheads point to fused 45S rDNA signals from two chromosomes. (D′) The FISH signals from St3.58 were digitally separated from D. Plasmid 1331 was used to detect the St. 3.58 repeat. Bars, 5 µm.
Figure 2.
Southern blot hybridization analysis of the centromeric satellite repeats in different potato cultivars. (A) Southern blot hybridization analysis of the Cen2-specific satellite repeat St3.58 in four potato cultivars. The numbers in the parentheses indicate the copy number of chromosome 2 carrying St3.58 in each cultivar. Left: genomic DNAs from the four cultivars were digested by BamHI and separated by a 0.8% agarose gel. Right: blotted DNAs were hybridized to St3.58 (plasmid probe 1331). No hybridization was found to DNA of White Pearl. (B) Southern blot hybridization analysis of the Cen1-specific satellite repeat St24 in four potato cultivars. The numbers in parentheses indicate the copy number of chromosome 2 carrying St24 in each cultivar. Left: genomic DNAs from the four cultivars were digested by HindIII and separated by a 0.8% agarose gel. Right: blotted DNAs were hybridized to St24 (plasmid probe 1336). No hybridization was found to DNA of Superior. Southern blot hybridization followed published protocol (Stupar et al. 2002).
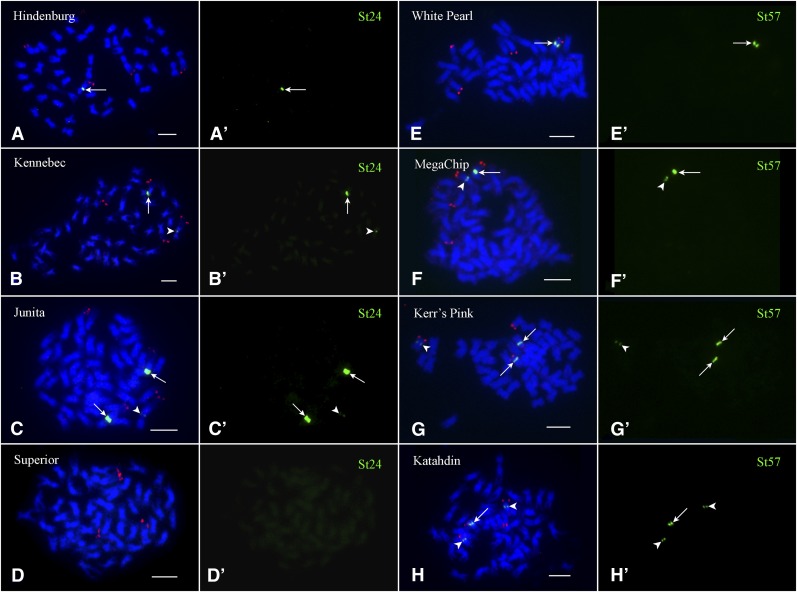
Repeat St24 is associated with Cen1 in DM (Gong et al. 2012). St24 hybridized to different numbers of chromosome 1 homologs among the 17 cultivars (Table 1). None of the four Cen1 in Superior contained St24, which was revealed by FISH (Figure 3D) and confirmed by Southern blot hybridization (Figure 2B). In addition, the sizes of the St24 repeat arrays appeared to vary significantly based on the size and intensity of the FISH signals associated with different Cen1 (Figure 3). Thus, expansion and/or contraction of the satellite repeat arrays may have occurred since the emergence of this repeat. Similar results were obtained with repeat St57 that was mapped to Cen7 in DM (Gong et al. 2012). St57 hybridized to only one to three copies of Cen7 among 17 cultivars (Figure 3 and Table 1). Different sizes and intensities of the FISH signals were also observed with the St57 repeat associated with different Cen7 (Figure 3).
Figure 3.
FISH mapping of the satellite repeats St24 and St57 in tetraploid potato genotypes. (A–D) FISH mapping of the Cen1-specific satellite repeat St24 in four potato cultivars, Hindenburg, Kennebec, Juanita, and Superior, respectively. Potato chromosome 1 was identified by bacterial artificial chromosome clone 96H03 (red signals). Arrows point to strong FISH signals and arrowheads point to weak FISH signals. (A′–D′) The FISH signals from St24 were digitally separated from A to D, respectively. (E–H) FISH mapping of the Cen7-specific satellite repeat St57 in four potato cultivars, White Pearl, MegaChip, Kerr’s Pink, and Katahdin, respectively. Potato chromosome 7 was identified by bacterial artificial chromosome clone 186I02 (red signals). Arrows point to strong FISH signals and arrowheads point to weak FISH signals. (E′–H′) The FISH signals from St57 were digitally separated from E to H, respectively. Plasmids 1336 and 1338 were used to detect repeats St24 and St57, respectively. Bars, 5 µm.
Repeat St3.58 was found only in cultivated potato, not in any wild Solanum species, suggesting that this repeat emerged after potato diverged from its most closely related wild relative (Gong et al. 2012). The highly similar size and intensity of the FISH signals associated with Cen2 in different potato cultivars (Figure 1) support that the St3.58 repeat array likely emerged very recently and has not undergone obvious expansion and/or contraction events. Repeat St24 was detected only in Solanum verrucosum, a wild species that is most closely related to potato and was proposed as the progenitor of cultivated potato, but was not detected in any other Solanum species that are more distantly related to potato (Gong et al. 2012). In addition, the St24 repeat in S. verrucosum is not associated with Cen1, but with another centromere (Gong et al. 2012). These results show that the St3.58 and St24 repeats in potato cultivars originated in S. tuberosum and were not introgressed from wild species during breeding. Repeat St57, however, was detected in Cen7 of S. verrucosum. Therefore, we cannot exclude the possibility that this repeat was introgressed from S. verrucosum during potato breeding.
There are two possibilities for the DNA compositions of the Cen1, Cen2, and Cen7 haplotypes that lack the St24, St3.58, and St57 repeats, respectively. These haplotypes may consist of single- and low-copy sequences or contain different satellite repeats. Megabase-sized satellite repeat arrays located in a single genomic location are rare in the literature. Thus, it would be unlikely that two different satellite repeat arrays have invaded the same centromeres and both have been maintained in the natural populations. We predict that the Cen1, Cen2, and Cen7 haplotypes that lack the St24, St3.58, and St57 repeats, respectively, may resemble the five DM centromeres (Cen4, Cen6, Cen10, Cen11, Cen12) that contain exclusively single- and low-copy sequences.
In summary, we demonstrate that potato Cen1, Cen2, and Cen7 have been invaded by megabase-sized satellite repeat arrays (Gong et al. 2012). However, the satellite repeat-based haplotypes of these three centromeres have not swept potato populations, but have coexisted with their ancestral haplotypes. These results are in favor of the hypothesis that that repeat-based centromeres might originate from repeat-lack neocentromeres and that the repeat-lack and repeat-based haplotypes of the same centromeres can coexist in natural populations until the repeat-based haplotypes eventually sweep through populations.
Acknowledgments
We thank Shelley Jansky and Richard Veilleux for their valuable comments on the manuscript. This work was partially supported by grant DBI-0923640 from the National Science Foundation to J.J.
Footnotes
Communicating editor: A. Houben
Literature Cited
- Amor D. J., Bentley K., Ryan J., Perry J., Wong L., et al. , 2004. Human centromere repositioning “in progress.” Proc. Natl. Acad. Sci. USA 101: 6542–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- duSart D., Cancilla M. R., Earle E., Mao J. I., Saffery R., et al. , 1997. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 16: 144–153. [DOI] [PubMed] [Google Scholar]
- Fu S. L., Lv Z. L., Gao Z., Wu H. J., Pang J. L., et al. , 2013. De novo centromere formation on a chromosome fragment in maize. Proc. Natl. Acad. Sci. USA 110: 6033–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z. Y., Wu Y. F., Koblížková A., Torres G. A., Wang K., et al. , 2012. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24: 3559–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. H., Zhang Z. H., Liu C. X., Liu J. H., Huang S. W., et al. , 2009. Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc. Natl. Acad. Sci. USA 106: 14937–14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D., Panchenko T., Salimian K. J., Salman M. U., Sekulic N., et al. , 2013. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 20: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H. S., 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Iovene M., Zhang T., Lou Q. F., Buell C. R., Jiang J. M., 2013. Copy number variation in potato: an asexually propagated autotetraploid species. Plant J. 75: 80–89. [DOI] [PubMed] [Google Scholar]
- Jiang J. M., Birchler J. A., Parrott W. A., Dawe R. K., 2003. A molecular view of plant centromeres. Trends Plant Sci. 8: 570–575. [DOI] [PubMed] [Google Scholar]
- Lee H. R., Zhang W. L., Langdon T., Jin W. W., Yan H. H., et al. , 2005. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102: 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O. J., Chueh A. C., Wong L. H., Choo K. H. A., 2008. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82: 261–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasuda S., Hudakova S., Schubert I., Houben A., Endo T. R., 2005. Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA 102: 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium , 2011. Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. [DOI] [PubMed] [Google Scholar]
- Rocchi M., Archidiacono N., Schempp W., Capozzi O., Stanyon R., 2012. Centromere repositioning in mammals. Heredity 108: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R. M., Song J. Q., Tek A. L., Cheng Z. K., Dong F. G., et al. , 2002. Highly condensed potato pericentromeric heterochromatin contains rDNA-related tandem repeats. Genetics 162: 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O., Sullivan K. F., 1997. Chromatin containing CENP-A and satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7: 897–900. [DOI] [PubMed] [Google Scholar]
- Ventura M., Archidiacono N., Rocchi M., 2001. Centromere emergence in evolution. Genome Res. 11: 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wu Y. F., Zhang W. L., Dawe R. K., Jiang J. M., 2014. Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Res. DOI: .10.1101/gr.160887.160113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard H. F., Waye J. S., 1987. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 3: 192–198. [Google Scholar]
- Zhang B., Lv Z. L., Pang J. L., Liu Y. L., Guo X., et al. , 2013a Formation of a functional maize centromere after loss of centromeric sequences and gain of ectopic sequences. Plant Cell 25: 1979–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Talbert P. B., Zhang W. L., Wu Y. F., Yang Z. J., et al. , 2013b The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proc. Natl. Acad. Sci. USA 110: E4875–E4883. [DOI] [PMC free article] [PubMed] [Google Scholar]