Abstract
In fission yeast, the septation initiation network (SIN) is thought to promote cytokinesis by downstream activation of Rho1, a conserved GTPase that controls cell growth and division. Here we show that Etd1 and PP2A-Pab1, antagonistic regulators of SIN, are Rho1 regulators. Our genetic and biochemical studies indicate that a C-terminal region of Etd1 may activate Rho1 by directly binding it, whereas an N-terminal domain confers its ability to localize at the growing tips and the division site where Rho1 functions. In opposition to Etd1, our results indicate that PP2A-Pab1 inhibits Rho1. The SIN cascade is upstream-regulated by the Spg1 GTPase. In the absence of Etd1, activity of Spg1 drops down prematurely, thereby inactivating SIN. Interestingly, we find that ectopic activation of Rho1 restores Spg1 activity in Etd1-depleted cells. By using a cytokinesis block strategy, we show that Rho1 is essential to feedback-activate Spg1 during actomyosin ring constriction. Therefore, activation of Spg1 by Rho1, which in turn is regulated by Etd1, uncovers a novel feedback loop mechanism that ensures SIN activity while cytokinesis is progressing.
Keywords: cytokinesis, SIN, Etd1, PP2A-Pab1, Rho1, Spg1, Schizosaccharomyces pombe
FISSION yeast cells grow by elongating at their cell ends before dividing by forming a medially placed division septum during cytokinesis. New-born cells initiate growth in a monopolar manner from the old cell end that existed before division. During G2, cells initiate growth from the new end in a process known as NETO (new end take-off). These cells then grow in a bipolar mode until mitosis, when they stop growth and initiate cell division by septation (Mitchison and Nurse 1985; Hayles and Nurse 2001). Both polar growth and septation require proper cell membrane and cell-wall synthesis to ensure cell integrity. In Schizosaccharomyces pombe, the cell wall is composed of β-glucan, α-glucan, and mannoproteins. Synthesis of the major cell wall and primary septum polymer, 1,3-β-glucan, requires the activity of Drc1/Cps1/Bgs1 1,3-β-glucan synthase, which in turn is regulated by the guanosine triphosphate (GTP)-binding protein Rho1 (Garcia et al. 2006; Cortes et al. 2007). As with all GTPases, Rho1 acts as a binary switch, cycling between inactive GDP-bound and active GTP-bound conformational states (Yang et al. 2003). This transition is controlled by three types of proteins: GEFs (guanine exchange factors), which catalyze the exchange of GDP for GTP, rendering the protein active; GAPs (GTPase activating proteins), which enhance nucleotide hydrolysis, inactivating the GTPase; and guanine nucleotide dissociation inhibitors, which appear to block spontaneous activation (Garcia et al. 2006).
Upon the onset of mitosis, growth ceases at the cell ends and is re-established in the middle of the cell, where the actomyosin ring forms. At the end of mitosis, once the two sets of chromosomes have segregated, actomyosin ring constriction and septum formation is triggered by the septation initiation network (SIN) from the spindle pole body (SPB) (Krapp et al. 2004; Wolfe and Gould 2005). The nucleotide-binding state of Spg1, a small Ras GTPase, plays a central role in determining SIN activity. In interphase cells, Spg1 is in the inactive GDP-bound form. As the mitotic spindle forms during metaphase, active GTP-bound Spg1 accumulates at both SPBs until anaphase B, when Spg1-GTP is converted to the inactive (GDP-bound) form at the mother SPB (SPBm) (Schmidt et al. 1997; Cerutti and Simanis 1999). During anaphase B, the protein kinase Cdc7 is asymmetrically recruited to the daughter SPB (SPBd), followed by Sid1p–Cdc14 binding (Sohrmann et al. 1998). The Sid2–Mob1 protein kinase complex then transduces the SIN signal to the division site. This complex relocalizes from the SPB to the cell division site and likely triggers medial ring constriction and septation (Salimova et al. 2000; Hou et al. 2004). Multi-copy expression of Rho1 GTPase rescues sid2 mutants, suggesting that the SIN may promote proper cell-wall formation at the division site by stimulation of Drc1/Cps1/Bgs1 1,3-β-glucan synthase through Rho1 (Jin et al. 2006), although the mechanism remains unclear.
Etd1 is a key element of the SIN cascade (Daga et al. 2005). However, the role of this protein in cytokinesis is intriguing. Epistasis analysis suggests that Etd1 acts either upstream of Spg1 activation (Garcia-Cortes and Mccollum 2009) or as part of a feedback loop (Daga et al. 2005). In the budding yeast Saccharomyces cerevisiae, the pathway analogous to the SIN is known as MEN (mitotic exit network) (reviewed in Krapp and Simanis 2008). Contact between the activated SPB containing the Spg1 homolog, Tem1, and the putative Tem1-GEF Lte1 at the bud cortex has been proposed as a mechanism to ensure that mitotic exit occurs only after the spindle has been oriented correctly (Bardin et al. 2000). More recently, Lte1 has been shown to activate Tem1 by regulating the localization of the Bfa1 GAP at the SPBs, rather than by stimulating nucleotide exchange by Tem1 (Geymonat et al. 2009). The requirement of Etd1 for Spg1 activity and a weak sequence similarity between Etd1 and Lte1 suggests that these two proteins might be homologs (Garcia-Cortes and McCollum 2009).
Interestingly, Etd1 is the only SIN regulator found at both the growing tips and the medial septum (Daga et al. 2005), a localization largely coincident with that of Rho1 and its target Drc1/Cps1/Bgs1 1,3-β-glucan synthase (Cortes et al. 2002, 2005; Garcia et al. 2006). In addition, its assembly into the medial ring to initiate septation requires SIN activity. Therefore, Etd1 has also being proposed to link SIN with septum formation (Daga et al. 2005). Mutations in pab1, the gene encoding the protein phosphatase 2A (PP2A) regulatory subunit B, restore Spg1 activity and suppress the septation defects of etd1 mutants, indicating that PP2A-Pab1 and Etd1 are antagonistic regulators of cytokinesis (Lahoz et al. 2010). Here we report for the first time that Etd1 and PP2A-Pab1 act as antagonistic regulators of Rho1. Furthermore, we show that Rho1 GTPase positively regulates Spg1 GTPase, providing a novel feedback loop mechanism that interconnects SIN signaling with cytokinesis.
Materials and Methods
Media, strains, and general methods
The genotypes of the yeast strains used in this study are listed in Supporting Information, Table S1. Growth conditions in standard yeast extract with supplements (YES) or EMM medium and strain manipulations were carried out as previously described (Moreno et al. 1991). Experiments in liquid culture were carried out in EMM and supplemented as required with a starting cell density of 2–4 × 106 cells/ml, corresponding to midexponential-phase growth. For regulated nmt expression, previously described methods were used (Maundrell 1993). Conditional deficiency for etd1 was assessed by using the null etd1Δ allele (37°, permissive temperature) or the etd1Δ nmt81x-etd1 strain at any growth temperature, where etd1 expression is regulated by thiamine under the weak repressible nmt81x promoter (Daga et al. 2005). This latter strain was used to produce etd1Δ mutants with the desired genetic background, removing the nmt81x-etd1 marker by tetrad dissection. cdr1-191 block was achieved by temperature shift to 36° (Liu et al. 2000), except in Etd1-depletion experiments, in which 34° was used.
Previously reported sid2-as alleles were not sensitive to most of the small-molecule inhibitors, being only partially sensitive to 40 μm of 3-BrB-PP1 (Cipak et al. 2011). To create a new sid2-as mutant strain, we employed site-directed mutagenesis (QuikChangeII, Site Directed Mutagenesis Kit) and mutated the methionine M285 gatekeeper residue to a glycine (Gly) as described by Cipak et al. (2011). However, instead of using a hygromycin-resistant sid2-as construct in a sid2Δ/sid2+ heterozygous strain (Cipak et al. 2011), we PCR-amplified the sid2-as mutant allele, and the resulting DNA fragment was transformed in the sid2-150 strain. Growing colonies were selected at 35° (including revertants, suppressors, and carrying-sid2-as allele colonies). Correct integration of the mutant sid2-as allele was confirmed by colony PCR. The presence of the M285-to-Gly mutation was verified by sequencing of the PCR-amplified sid2-as allele from genomic DNA of the selected colony. The resulting sid2-as allele conferred sensitivity to 10 μm 1NMPP1 (Figure S1).
Genetic and cytological techniques were performed according to described methods (Moreno et al. 1991). Double mutants were constructed by tetrad dissection. Transformation was carried out using the lithium acetate transformation protocol (Moreno et al. 1991).
Microscopy
Cell wall and chromatin regions were, respectively, visualized by Calcofluor white (Sigma) and 4,6-diamidino-2-phenylindole (DAPI; Sigma). Cells were examined using a Leica fluorescence microscope equipped with a plan Apo ×100 lens. Confocal images were obtained with a confocal Leica SP2 microscope. For scanning electron microscopy, etd1-1 cells were collected by centrifugation at 4° and fixed with 2% glutaraldehyde in 0.1 M cacodilate buffer (pH 7.2) for 1 hr. Cells were washed three times (for 5 min) with the same buffer and post-fixed with 1% OsO4 for 1 hr. After fixation, cells were washed with distilled water and dehydrated in increasing concentrations of ethanol. Cells were collected in Millipore filters (0.2 μm), further dried with the critical point method, and gold sputter-coated. Observations were made using a Jeol JSM-840 scanning electron microscope operated at 10, 20, or 25 kv.
Live-cell imaging was performed under Deltavision wide-field microscope systems (Applied Precision, Issaquah, WA). For time-lapse experiments, exponentially growing cells were concentrated by centrifugation and suspended in 100 μl of EMM medium. Five-microliter aliquots of the cell suspension were placed in 35-mm glass-bottom culture dishes (P35-1.5-10-C; MatTek) with 5 μl of 1 mg/ml soybean lectin (L1395; Sigma-Aldrich) and immersed in 3 ml of medium. Time-lapse experiments were performed at the indicated temperature by acquiring images every 5 min as an image stack of 10 × 0.5 µm z-planes with 2 × 2 binning, with the exception of pab1-4 and pab1-4 etd1Δ where acquisition was performed by using 20 × 0.5 µm z-planes per image stack. Fluorescence intensity for Cdc7-Tomato or Mob1 GFP was measured with ImageJ software [National Institutes of Health (NIH), Bethesda, MD] by placing a circle around the corresponding SPB and measuring the maximal fluorescence (Daga et al. 2005). A nearby cytosolic region was used as background. Intensity and Ring diameter was also measured with ImageJ software (NIH, Bethesda, MD) by placing a line along the medial ring-localized Mob1-GFP.
Plasmids
Gene fragments were obtained by PCR amplification from either S. pombe genomic DNA or complementary DNA (cDNA) library, as appropriate. For overexpression/repression assays, nmt promoter-containing vectors pREP3X, pREP41X, and pREP81X were used (Forsburg 1993). The open reading frame of the desired gene or gene domain was amplified by PCR from the cDNA of the corresponding gene by using suitable primers and cloned into the cloning sites of the vector (Maundrell 1993). For moderate overexpression, the pREP41x vector was used (nmt41x-driven expression) (Forsburg 1993). Repression was achieved by adding thiamine, and nmt41x-repressed cells were analyzed 3 hr after the addition.
Immunoprecipitation and in vitro kinase assay of Sid2
Cells of wt, pab1-4, etd1Δ, and pab1-4 etd1Δ strains in a chromosomally tagged sid2-myc genetic background were grown at 37° (permissive temperature for etd1Δ cells) and post-incubated for 3 hr at 25° (restrictive temperature for etd1Δ). A wild-type (sid2+) strain was used as a negative control. Cell pellets, obtained by centrifugation, were washed with stop buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3 pH 8.0) and frozen at −80°. Cells were lysed in NP40 buffer (1% NP40, 150 mM NaCl, 2 mM EDTA, 6 mM Na2HPO4, 4mM NaH2PO4, 100 μg/ml PMSF, 1 µg/ml pepstatin, 10 µg/ml leupeptin, 10 µg/ml aprotinin). Cells extracts were clarified by centrifuging for 10 min at 14,000 × g and 4°. Immunoprecipitations were performed by adding 1 μg of anti-Myc (9E10, Santa Cruz) to 50 μl of MagnaBind Goat anti-mouse immunoglobulin (Pierce 21354) prewashed with PBS as protocol describes. After 1 hr of incubation at 4° on a rotating wheel, the beads were washed once with MagnaBind, two times with PBS, and another two times with lysis buffer that included protease inhibitors. One milligram of total protein was added to 50 μl of MagnaBind with anti-Myc (α-Myc) in a final volume of 200 μl. After 3 hr of incubation on a rotating wheel at 4°, the beads were washed with lysis buffer.
For detection of protein kinase activity, the immune complex beads were washed three times with 100 μl of kinase assay buffer (10 mM Tris, pH 7.4, 10 mM MgCl2, 100 μg/ml PMSF, 1 µg/ml pepstatin, 10 µg/ml leupeptin, 10 µg/ml aprotinin) and then incubated for 30 min at 30° in 20 μl of kinase buffer with 10 μl of reaction cocktail that contains 9.45 μl of 1 mg/ml myelin basic protein (Sigma), 0.05 μl of 10 mM cold ATP, and 0.5 μl of [γ-32P]ATP (3000 Ci/mmol at 10 mCi/ml; Amersham Pharmacia). Reactions were stopped by addition of 20 μl of SDS-PAGE sample buffer. Samples were resolved on SDS-PAGE (15%), followed by imaging and quantification on a PhosphorImager. Relative kinase activity was determined by subtracting background kinase activity associated with beads incubated with lysates not expressing the tagged Sid2 kinase, followed by normalization for the relative levels of Sid2 protein as determined by immunobloting (Hou et al. 2004).
Measurement of 1,3-β-glucan synthase activity
To determine 1,3-β-d-glucan synthase activity in wt, etd1Δ, pab1-4, and etd1Δ pab1-4 strains, cells were grown at 37° and post-incubated for 3 hr at 25°. Cell extracts were prepared and 1,3-β-d-glucan synthase activity was assayed following described protocols (Arellano et al. 1996) with some modifications. Cells (3–4 × 109) were harvested, washed with 1 mM EDTA, and resuspended in 300 µl of 50 mM Tris–HCl, pH 8.0, in 1 mM EDTA. Lysis was achieved in a Fast-Prep as described above. The resulting homogenates were collected by adding 30 ml of the same buffer. Cell walls were removed by 5 min of centrifugation at 1000 × g. The resulting membrane pellet was resuspended in 0.5 ml of buffer containing 50 mM Tris–HCl, 1 mM EDTA, 1 mM β-mercaptoethanol, and 30% glycerol. The supernatant was centrifuged for 30 min at 50,000 × g. Protein concentration was determined by BCA kit quantification protein assay method (Pierce). The amount that catalyzes the incorporation of 1 µmol of substrate (UDP-D-glucose) per minute at 30° was considered the unit of activity.
Pull-down assay for GTP-bound Rho1 protein
To analyze levels of GTP-bound Rho1 protein in the absence of Etd1 and/or Pab1, previously described methods for S. pombe cells were employed (Calonge et al. 2003). Briefly, cells of wt, etd1Δ, pab1-4, and etd1Δ pab1-4 strains in a chromosomally tagged HA-rho1 genetic background were grown at 37° and post-incubated for 3 hr at 25°. To analyze the effect of Etd1 or Pab1 overexpression, HA-rho1 leu1-32 cells were transformed with either pREP41x-etd1 or pREP3x-pab1, and the transformed strains were incubated for 18 hr at 25° in the absence of thiamine. The amount of GTP-bound Rho proteins was analyzed in cell extracts by using the Rho-GTP pull-down assay (Ren et al. 1999). The total amount of Rho proteins in the extracts was determined by Western blotting using the anti-HA. GTP-bound Rho proteins were purified by binding to GST-rhotekin Rho-binding domain, previously obtained and purified from Escherichia coli (Reid et al. 1996). GTP-Rho1 levels were quantified by using the Image-J software (NIH).
Co-immunoprecipitation analysis of Etd1 and Rho1
Total protein extracts were prepared from 3–5 × 108 cells using HEPES buffer (Moreno et al. 1991). Cell extracts (2 mg of total protein) of the indicated Etd1-GFP HA-Rho1 strain were incubated with the GFP antibody and protein G-Sepharose beads for 2–4 hr at 4°. The beads were washed four times with 1 ml of HB buffer and resuspended in sample buffer. Immunoprecipitates were resolved on SDS-PAGE (12%) and blotted and probed with anti-HA (1:1000) (F-7, Santa Cruz) or anti-GFP (1:1000) antibody (gift from P. Nurse).
Results
High Rho1 activity suppresses lethality in Etd1-depleted cells
In fission yeast, the GTPase Rho1 is a key regulator that, among other functions, controls synthesis of the cell wall and primary septum polymers (Arellano et al. 1996, 1997; Sayers et al. 2000). Depletion of Rho1 impedes cell growth and leads to cell lysis (Arellano et al. 1996). The SIN is thought to regulate Rho1 during cytokinesis. Accordingly, SIN-deficient cells fail to produce a septum and undergo lysis as a terminal phenotype (Jin et al. 2006).
Little is known about the mechanisms that link the SIN pathway with Rho1 and the cell-wall synthesis activity, but the SIN regulator Etd1 could play a role in this link (Daga et al. 2005). As occurs with other sin mutants, Etd1-depleted cells undergo lysis, and lethality of these mutant cells can be rescued by adding an osmotic stabilizer (1.2 M sorbitol) to the medium (Figure 1A). To further study the influence of Etd1 in the synthesis of cell-wall polymers, we initially examined cell wall in etd1 mutant cells by using scanning electron microscopy. In agreement with the role of Etd1 in proper cell-wall formation, we observed that Etd1 depletion caused severe defects in cell-wall integrity (Figure 1B). Etd1 is the only SIN protein that localizes to the division site and to the cell tips (Daga et al. 2005), where Rho1 and the cell-wall biosynthesis machinery are known to function. In addition, Etd1 localization to the medial ring requires Sid2-Mob1 activity (Daga et al. 2005). Thus, we further analyzed whether Etd1 is involved in connecting Sid2-mediated SIN signaling with activation of Rho1 and the cell-wall synthesis machinery.
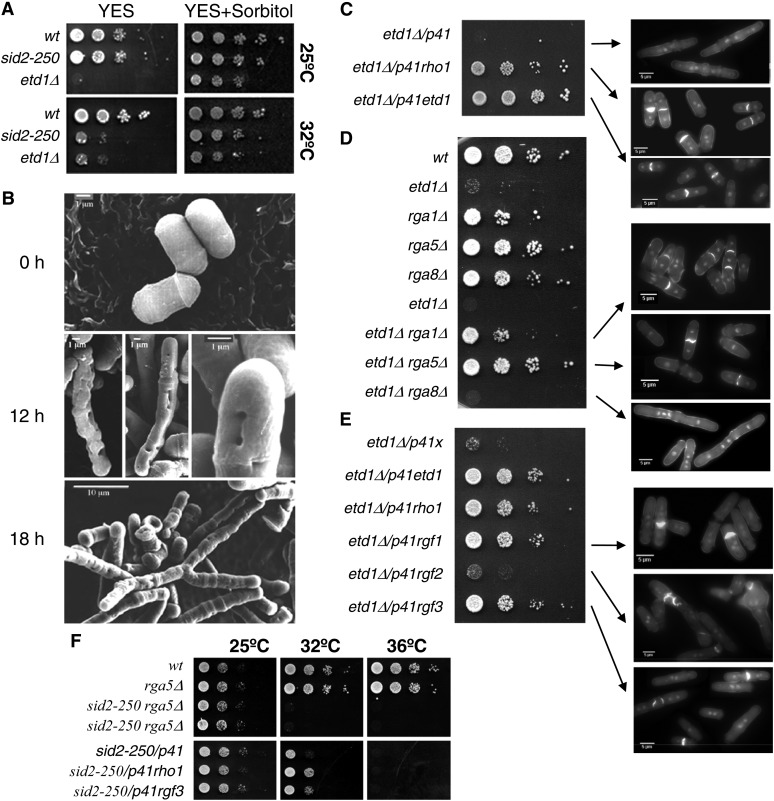
Figure 1.
Etd1-depleted cells are deficient for cell-wall integrity and septation, a phenotype suppressed by high Rho1 activity. (A) Growth assay (serial dilution drop tests on plates) of etd1Δcells in YES media (left panels) and YES with 1.2 M sorbitol (right panels). The wild-type and sid2-250 strains were used as controls. Cells were grown (at 37° for etd1Δ and wt and at 25° for sid2-250), and 10-fold dilutions (starting with 104 cells) were spotted and incubated at 25° or 32°. (B) Electron microscopy images of etd1Δ cells grown at 37° (permissive temperature for etd1Δ after 0-, 12-, and 18-hr incubations at 25° (restrictive temperature for etd1Δ). Etd1-deficient cells show severe defects in cell-wall integrity. (C) Growth assay (serial dilution drop tests on plates) of etd1Δ cells overexpressing Rho1 protein under nmt41x expression in the pREP41x vector (p41rho1, left). Cells were grown at 37°, and 10-fold dilutions (starting with 104 cells) were spotted and incubated at 30° in the absence of thiamine. The pREP41x-driven expression of Etd1 (p41etd1) and the empty pREP41x plasmid (p41) were used as positive and negative controls, respectively. DAPI- and Calcofluor-stained cells of these etd1Δ and etd1Δ/p41rho1 strains are shown (right panels). (D) Growth assay (serial dilution drop tests on plates) (left) and cell phenotype (DAPI and Calcofluor staining) (right panels) of etd1Δ rga1Δ, etd1Δ rga5Δ, and etd1Δ rga8Δ cells. Cells were grown at 37°, and 10-fold dilutions (starting with 104 cells) were spotted and incubated at 30°. Wild-type (wt) and single-deletion strains were used as control. (E) Growth assay (as described in C) and cell phenotype (DAPI and Calcofluor staining) (right panels of etd1Δ cells overexpressing Rgf1, Rgf2, or Rgf3 proteins under nmt41x expression in the pREP41x vector (p41rgf1, p41rgf2, and p41rgf3, respectivelly) (left). p41etd1 and p41rho1 were used as positive controls. The empty p41 plasmid was used as a negative control. (F) Growth assay of sid2-250 cells deleted for rga5 (single sid2-250 and rga5Δ mutant strains were used as controls) (top panels) or overexpressing rgf3 (p41rgf3) (bottom panels). The p41rho1 construct was used as a positive control. The empty p41 plasmid was used as a negative control. Cells were grown at 25°, and 10-fold dilutions (starting with 104 cells) were spotted and incubated at 25°, 32°, or 36° in the absence of thiamine in the case of overexpression assays (bottom panels).
Overexpression of Rho1 partially rescues lethality of Sid2 mutants at low restrictive temperature (Jin et al. 2006). Interestingly, we observed that moderate overexpression of Rho1 under the nmt41x promoter in a plasmid (p41rho1) efficiently suppressed lethality of etd1Δ mutants (Figure 1C), suggesting that Etd1, as proposed for Sid2 (Jin et al. 2006), functions in downstream activation of Rho1. The rho1-596 is a hypomorphic thermo-sensitive allele with reduced Rho1 activity (Viana et al. 2013). It has been recently shown that the rho1-596 thermo-sensitive growth defect is completely rescued by overexpression of rho1+ from pREP41x. On the contrary, neither overexpression of the dominant negative allele rho1-T20N nor overexpression of the constitutively active allele rho1-G15V suppressed lethality of rho1-596 mutant cells, suggesting that GTP-GDP cycling is required for the suppression (Viana et al. 2013). Identical results were obtained for lethality suppression of etd1Δ cells with the expression of these rho1 alleles (Figure S2), suggesting that Etd1 might be closely related to the regulation of the Rho1 GTP-GDP cycle.
The activity of Rho GTPases is positively regulated by GEFs, in opposition to GAPs, which act as negative regulators. S. pombe contains seven genes bearing a Rho-GEF domain and nine genes encoding putative Rho-GAPs. Among them, Rgf1, Rgf2, and Rgf3 GEFs and Rga1, Rga5, and Rga8 GAPs are Rho1-specific regulators (Nakano et al. 2001; Yang et al. 2003; Mutoh et al. 2005; Garcia et al. 2006). To determine whether increasing Rho1 activity can suppress the lethality of Etd1 depletion, we next studied the effects of Rho1 hyper-activation in the etd1Δ strain by deletion of these Rho1 GAPs or by overexpression of these Rho1 GEFs. Neither overexpression of rgf2 nor deletion of rga8 rescued etd1Δ lethality (Figure 1, D and E), whereas deletion of rga1 and rga5 (Figure 1D) as well as overexpression of rgf1 and rgf3 (Figure 1E) efficiently suppressed septation and cell-wall defects of etd1Δ cells, leading to cell proliferation. Thus, Etd1 may act as an activator of Rho1 in specific Rho1 functions (those specifically inhibited by Rga1 and Rga5 and activated by Rgf1 and Rgf3). As shown in Figure 1E, ectopic expression of Rgf3 partially rescued lethality of sid2-250 cells at low restrictive temperature, and suppression was not observed in sid2-250 rga5Δ cells, suggesting for Etd1 a more specific role in Rho1 activation than for Sid2.
SIN-independent activation of cytokinesis by Rho1
SIN is required to trigger cytokinesis (Krapp and Simanis 2008). To gain further insight into the mechanism by which high Rho1 activity rescues lethality of etd1 mutants, we analyzed SIN signaling in the etd1Δ/p41rho1 strain by examining the cellular distribution of the Sid2-Mob1 complex, the last kinase of this pathway.
Expression of sid2-GFP is synthetically lethal in etd1-mutant alleles (Daga et al. 2005). Therefore, we used a mob1-GFP construct for the in vivo localization of the complex by time-lapse microscopy. As shown in Figure 2A, Mob1-GFP localized to both SPBs during mitosis and at the division site during septation in wild-type cells (Salimova et al. 2000; Hou et al. 2004). In etd1Δ cells, Mob1-GFP localized at the SPBs, but was not observed at the division site (Figure 2A), as previously reported (Daga et al. 2005). In the etd1Δ/p41rho1 strain, most of the cells completed the division cycle, in agreement with the ability of rho1 overexpression to rescue septation defects of etd1 mutant cells (Figure 1C). As expected, etd1Δ/p41rho1 cells entered cytokinesis but, unexpectedly, Mob1-GFP was not observed at the cleavage site (Figure 2A). This result indicates that high Rho1 activity is sufficient to trigger cytokinesis in Etd1-depleted cells, bypassing the requirement of Sid2-mediated SIN signaling. It is remarkable that binucleate cells can be observed at a high frequency (Figure 1, C–E), suggesting that mitosis and cytokinesis are not properly coupled in these cells.
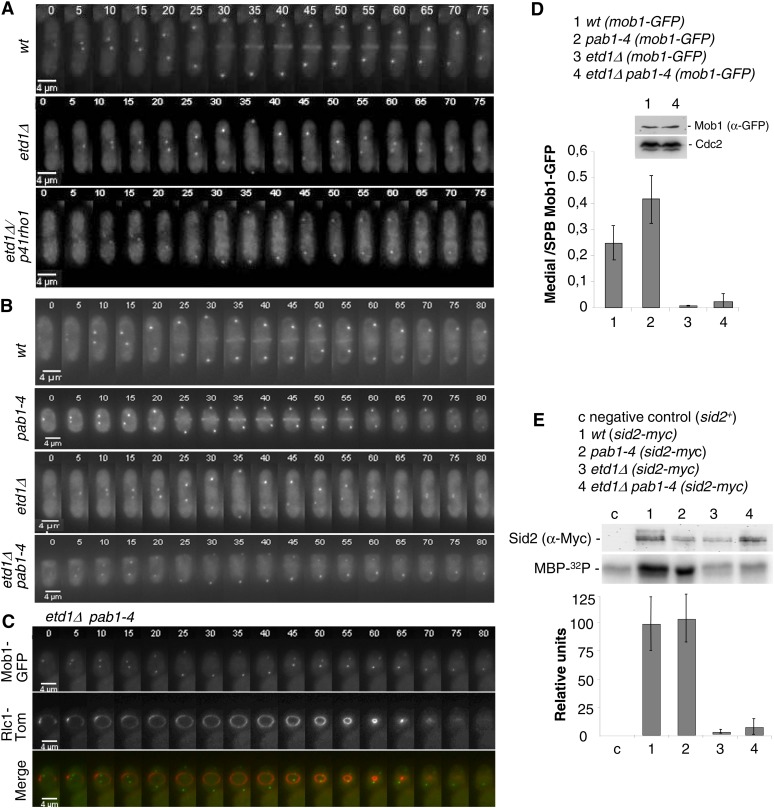
Figure 2.
Etd1 is required for Sid2-Mob1 kinase activity and proper localization at the division site. (A) Mob1-GFP was imaged in live wild-type cells (top), etd1Δ mutant cells (middle) and etd1Δ/p41rho1 cells (bottom) by time-lapse microscopy at 5-min intervals at 25°, restrictive conditions for etd1Δ mutants. (B) Mob1-GFP was imaged in live wild-type cells (top), etd1Δ mutant cells (middle), pab1-4 (middle), and etd1Δ pab1-4 cells (bottom) by time-lapse microscopy as described in A. (C) Mob1-GFP and the actomyosin ring marker Rlc1-Tom were co-expressed and imaged in these cells as above. (D) Mob1-GFP fluorescence intensity was quantified in five cells of each strain, and maximum values of medially located Mob1-GFP with respect to SPB-associated Mob1-GFP are represented. Levels of Mob1-GFP in etd1Δ pab1-4 and wild-type cells, determined by Western blot analysis, are shown. The amount of Cdc2 was tested as control. (E) Sid2 kinase activity in wt, etd1Δ, pab1-4, and etd1Δ pab1-4 strains (in a sid2-myc genetic background) was determined in cell extracts by 32P incorporation into MBP-32P. Cells were grown at 37° (permissive temperature for etd1Δ cells) and post-incubated for 3 hr at 25° (restrictive temperature for this strain). Total Sid2 protein (from the sid2-myc construct) was analyzed by Western blot (α-Myc), and a wild-type sid2+ strain was used as negative control. Kinase activity was quantified and represented (mean value was derived from three experiments). Error bars indicate SEM.
As occurs with Rho1 hyper-activation, the pab1-4 mutation (pab1 codes for the B-regulatory subunit of PP2A) efficiently rescues lethality in etd1Δ cells. However, pab1-4 restores Spg1 activity in these Etd1-depleted cells (Lahoz et al. 2010), suggesting that this mutant could counteract Etd1 deficiencies by recovering SIN activity rather than bypassing it. To study whether pab1-4 completely restored the SIN cascade in Etd1-depleted cells, we also examined the localization dynamics of the Sid2-Mob1 complex in the etd1Δ pab1-4 strain. As shown in Figure 2B, Mob1-GFP localized to both SPBs during mitosis in the pab1-4 mutant strain as in the wild type. According to previous results, this protein was not observed at the division site in etd1Δ cells. In the etd1Δ pab1-4 strain, cells entered cytokinesis but, to our surprise, Mob1-GFP failed to localize to the division site (Figure 2B). To substantiate this observation, we used the Rlc1-Tom construct to analyze initiation and progression of cytokinesis in these cells. By using this medial ring marker, we determined that the actomyosin ring undergoes normal constriction in etd1Δ pab1-4 cells, although Mob1-GFP fluorescence at the division site was undetectable in the dividing cells (Figure 2C). Fluorescent quantification of time-lapse microscopy experiments confirmed that the amount of Mob1-GFP that localizes at the septum was drastically reduced in this strain (Figure 2D). Western blot analysis determined, however, that the total amount of Mob1-GFP protein in etd1Δ pab1-4 cells was similar to that of wild-type cells (Figure 2D). This result suggests that the drastic reduction of Sid2-Mob1 localized at the medial ring is unlikely due to a reduction in total Sid2-Mob1 protein levels. Thus, although pab1-4 leads to Spg1 activation in the etd1Δ pab1-4 strain (Lahoz et al. 2010), we conclude that SIN signaling is not properly transduced by the Sid2-Mob1 complex to the division site in this strain. Furthermore, this result indicates that Etd1 is required for proper Sid2-Mob1 localization at the actomyosin ring during cytokinesis.
Interestingly, while the Sid2/Mob1 complex localizes symmetrically at both SPBs in wild-type cells (Spark et al. 1999) (Figure S3A), we observed that Mob1-GFP was asymmetrically recruited to the SPBs in etd1 mutant cells (Figure S3A). Therefore, Etd1 influences the association of Sid2-Mob1 at the division site but also the symmetric localization of this complex at the SPBs. Co-expression of Mob1-GFP and Cdc7-Tom in the etd1Δ pab1-4 strain allowed us to determine that Sid2-Mob1 was enriched at the new SPB (SPBd) in Etd1-depleted cells (Figure S3B).
Although the amount of Sid2-Mob1 at the middle of the cell is extremely low during anaphase, hyper-activation of its kinase could provide sufficient Sid2-Mob1 function to trigger cytokinesis. To test this possibility, we examined Sid2-Mob1 kinase activity in the etd1Δ pab1-4 double-mutant strain. Chromosomally tagged Sid2-13Myc was immunoprecipitated with an anti-Myc antibody, and in vitro Sid2 kinase assays were performed with myelin basic protein (MBP) as a kinase substrate (Sparks et al. 1999). As shown in Figure 2E, Sid2 kinase activity was high in pab1-4 and wild-type cells. However, this kinase activity was extremely low both in etd1-mutant cells and in the etd1Δ pab1-4 double-mutant strain (Figure 2E). Given that actomyosin ring contraction and septation take place efficiently in etd1Δ pab1-4 cells, in which the Sid2-Mob1 complex fails to transduce SIN signaling to the medial ring (Figure 3, A and B), pab1-4 might directly activate Rho1, bypassing the requirement of Sid2-Mob1 and Etd1 to trigger cytokinesis and conferring on PP2A-Pab1 an inhibitory role in the regulation of Rho1.
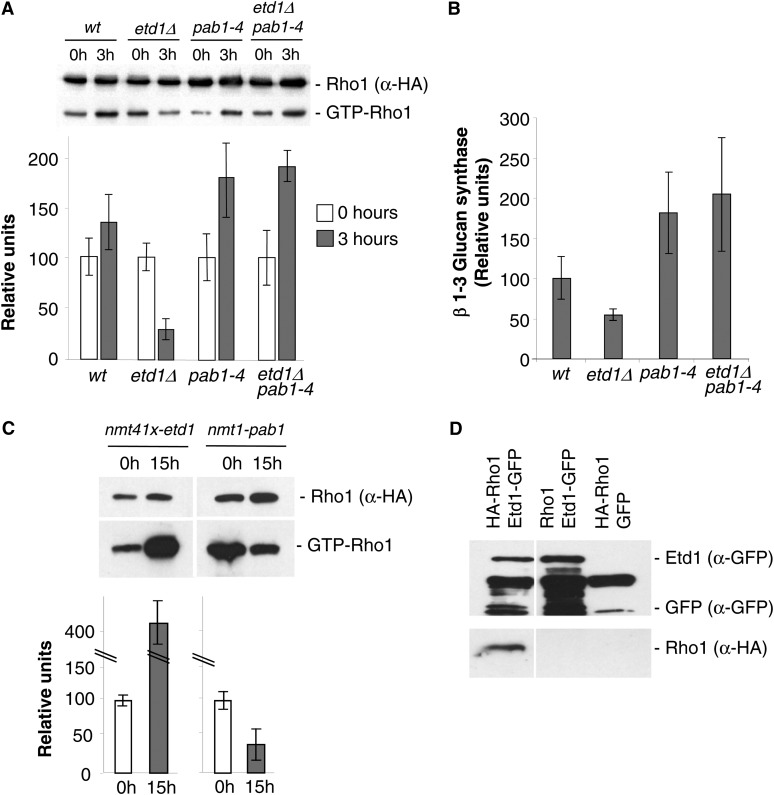
Figure 3.
Etd1 and PP2A-Pab1 are antagonistic regulators of Rho1 activity. (A) Determination of GTP-bound Rho1 (GTP-Rho1) levels vs. the total amount of Rho1 protein (α-HA) in extracts from wt, etd1Δ, pab1-4, and etd1Δ pab1-4 strains (in a HA-rho1 genetic background) at time 0 (incubated at 37°, the permissive temperature for etd1Δ cells) and at 3 hr post-incubation at the restrictive growth temperature for etd1Δ (25°). Quantification of relative GTP-Rho1 levels is shown (mean value derived from three experiments). Error bars indicate SEM. (B) 1,3-β-glucan synthesis activity was analyzed in cell extracts from the indicated strains and temperature (restrictive for the corresponding conditional mutant) (mean value derived from three experiments). Error bars indicate SEM. (C) Determination of GTP-bound Rho1 (Rho1p-GTP) levels as compared to the total amount of Rho1 protein (α-HA) in extracts from chromosomally tagged HA-rho1 strains overexpressing Etd1 (nmt41x-etd1 in plasmid pREP41x) or Pab1 (nmt1-pab1 in plasmid pREP3x) at 0 and 18 hr after nmt de-repression at 25° (see Materials and Methods for experimental details). Quantification of relative GTP-Rho1 levels is shown (mean value derived from three experiments). Error bars indicate SEM. (D) Physical association of Etd1 and Rho1. Protein extracts prepared from cells expressing Etd1-GFP (rho1+ genetic background), HA-Rho1 (with GFP expression as a control), or both were immunoprecipitated with anti-GFP antibodies; the immunoprecipitates were run on SDS–PAGE gels and probed with anti-GFP and anti-HA antibodies. Western blots of total extracts were also probed with anti-GFP and anti-HA antibodies to check the levels of tagged proteins.
Regulation of Rho1 activity by Etd1 and PP2A-Pab1
Our genetic results described above suggest that Etd1 and PP2A-Pab1 are antagonistic regulators of Rho1. This small GTPase controls 1,3-β-glucan synthesis, a major component of the cell wall and primary septum (Garcia et al. 2006). To check whether Etd1 and PP2A-Pab1 could regulate Rho1 GTPase activity, we analyzed levels of GTP-bound Rho1 (active) and measured 1,3-β-glucan synthase activity in etd1Δ, pab1-4 and the double etd1Δ pab1-4 mutant strains. Etd1 is essential at growth temperature <35° (Garcia-Cortes and McCollum 2009), and growth is significantly affected in pab1-4 cells at incubation temperatures <30° (Lahoz et al. 2010). Thus, wild-type, etd1Δ, pab1-4, and etd1Δ pab1-4 strains were grown at 37°, and the role of these proteins in Rho1 activity was determined by purifying GTP-Rho1 after a 3-hr shift to 25°. As shown in Figure 3A, the amount of GTP-bound Rho1 was drastically reduced in etd1Δ cells in comparison to the wild-type control [similar results were obtained in the etd1Δ nmt81xetd1 strain at 25° under expression and repression conditions for etd1 (data not shown)], suggesting for Etd1 a positive role in Rho1 activity. On the other hand, the increase in GTP-bound Rho1 observed in pab1-4 and etd1Δ pab1-4 cells after a 3-hr incubation at 25° suggests a role in Rho1 inhibition for PP2A-Pab1. Accordingly, we observed that 1,3-β-glucan synthase activity was consistently diminished in etd1Δ cells, while pab1-4 and etd1Δ pab1-4 cells showed twofold higher activity than the wild-type control (Figure 3B). Therefore, we conclude that Etd1 functions as a positive regulator of Rho1 and 1,3-β-glucan synthase while Pab1 plays an antagonistic role to Etd1, acting as an inhibitor of this GTPase.
To further test this possibility, we measured levels of GTP-bound Rho1 in cells overproducing Etd1. Very high levels of Etd1 expression, under the control of a de-repressed nmt1 promoter, cause severe cell lysis that makes a molecular analysis difficult (Garcia-Cortes and McCollum 2009). However, we found that moderate nmt41x-promoter-driven ectopic expression of Etd1 strongly increased levels of GTP-bound Rho1 (Figure 3C). Conversely, overexpression of Pab1 reduced the level of active Rho1-GTP (Figure 3C), in agreement with opposing roles for PP2A-Pab1 and Etd1 in regulating Rho1 activity.
To address whether Rho1 regulation by Etd1 and PP2A-Pab1 could be exerted directly, we tested for a possible physical interaction between Rho1 GTPase and Etd1 and Pab1 proteins. While we failed to detect association between Pab1 and Rho1, we did find that HA-Rho1 copurified with Etd1-GFP (Figure 3D). Similar results were obtained by using an Etd1-GST construct (data not shown). This physical interaction between Etd1 and Rho1 suggests that Etd1 might activate Rho1 through association with this GTPase. Given that physical interactions between Pab1 and Rho1 have not been proven so far (Lahoz et al. 2010), the PP2A-Pab1 protein phosphatase complex may inhibit Rho1 via its regulators.
Functional analysis of Etd1 protein domains
Etd1 has been proposed to directly activate the GTPase Spg1 (Garcia-Cortes and McCollum 2009); however, little is known about the molecular mechanism by which Etd1 promotes GTPase activation. In an attempt to uncover possible mechanisms for Etd1 activity, we constructed a number of etd1 deletions (etd1-D1 to etd1-D11) (outlined in Figure 4A) fused to GFP and studied their function and cell localization. To determine the domains essential for Etd1 function, corresponding truncated variants were expressed in etd1Δ cells for an in vivo complementation assay. To identify Etd1 domains required for its proper cell localization, these variants were expressed in wild-type cells.
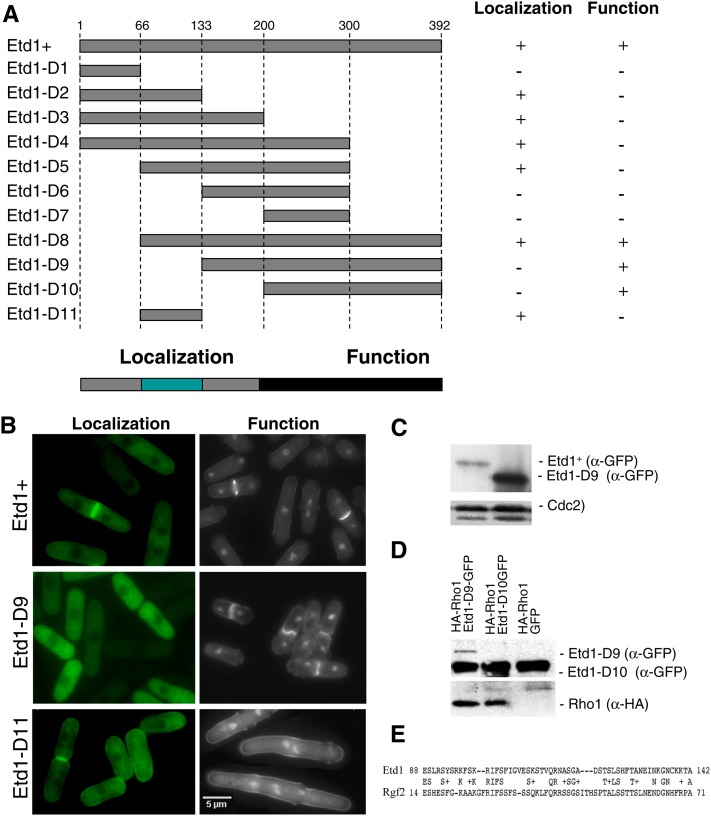
Figure 4.
Functional analysis of different Etd1 domains. (A) The indicated PCR-derived fragments of the Etd1-coding region (Etd1-D1 to Etd1-D10) were expressed in the pREP41x-GFP plasmid. Position of amino-acid residues flanking each Etd1 fragment is indicated. For cell localization assays, the GFP-fused constructs were expressed in wild-type cells in the absence of thiamine at 25°. For the functional complementation test, these constructs were expressed in the etd1Δ strain. Plasmids containing etd1Δ cells were grown at 37°, and the ability to rescue growth and division in this strain was assessed in the absence of thiamine at 25°. Etd1-truncated variants were classified as functional (+) or nonfunctional (−) according to their cell localization and complementation properties. The fragment containing the entire Etd1-coding region was used as control. Essential regions required for proper Etd1 cell localization and function are indicated. (B) Cell localization assay and functional complementation test of Etd1+ (control), Etd1-D9, and Etd1-D11 fragments. GFP fluorescence of wild-type cells expressing Etd1-GFP, Etd1-D9-GTP, or Etd1-D11-GTP constructs (left panels). DAPI and Calcofluor staining of etd1Δ cells under the expression of these constructs (right panels). (C) Western blot analysis of Etd1+ and Etd1-D9 (GFP-tagged). Cdc2 protein was used as a control. (D) Physical association of functional truncated variants Etd1-D9 and Etd1-D10 (GFP-tagged) and Rho1 (HA-tagged). Protein extracts prepared from cells expressing HA-Rho1 and Etd1-D9-GFP, Etd1-D10-GFP or GFP (negative control) were immunoprecipitated with anti-GFP antibodies; the immunoprecipitates were run on SDS–PAGE gels and probed with anti-GFP and anti-HA antibodies. Western blots of total extracts were also probed with anti-GFP and anti-HA antibodies to check the levels of tagged proteins. (E) Sequence similarity between Etd1 and Rgf2 in a region that largely overlaps with the Etd1 localization domain.
As shown in Figure 4A, Etd1-D2, Etd1-D3, Etd1-D4, Etd1-D5, and Etd1-D8 GFP fusions localized indistinguishably from full-length Etd1 protein. This assay identified a 67-amino-acid domain ranging from residue 66 to 133 that is responsible for cell cortex and division site localization. This fragment (Etd1-D11) was sufficient for Etd1 localization (Figure 4A). The functional complementation assay determined that Etd1-D8, Etd1-D9, and Etd1-D10 fully suppressed growth and cytokinesis defects of Etd1-depleted cells, indicating that a carboxyl-terminal domain, between amino-acid residues 200 to 392, is essential for Etd1 function (Figure 4A). There is only one previously characterized loss-of-function allele (etd1-1) for this gene (Jimenez and Oballe 1994; Daga et al. 2005). We cloned the genomic fragment of etd1 from the etd1-1 mutant strain, and DNA sequence determined that this allele yields a C-terminal 88-amino-acid truncated protein, which confirms the relevance of the C-terminal domain in Etd1 function.
Interestingly, the localization fragment (Etd1-D11) was not functional, but it was sufficient to confer full-length Etd1+ localization (Figure 4A). Reciprocally, Etd1-D9 and Etd1-D10 were functionally active, but these variants did not accumulate at the cell cortex or medial ring. Instead, these truncated Etd1 proteins localized to the cytoplasm (see Figure 4B for Etd1-D9). This result demonstrates that the in vivo function of Etd1 at the cell cortex and the division site can be also executed from the cytoplasm. By Western blot analysis, we determined that levels of Etd1-D9 and Etd1-D10 were much higher than that of Etd1+ in similar expression conditions (see Figure 4C for Etd1-D9). Therefore, in wild-type cells, the localization domain is likely required to accumulate Etd1 at the growing tips and the division site, where Rho1 and its regulators localize. In the case of Etd1-D9- and Etd1-D10-expressing cells, their increased abundance may permit sufficient levels of cytoplasmic Etd1-D9 and Etd1-D10 to target Rho1 protein at the cell cortex and the medial ring.
Significantly, by using these GFP fusion constructs, we determined that Etd1 variants containing the functional C-terminal fragment physically associated with Rho1 (see in Figure 4D for Etd1-D9 and Etd1-D10), while such interaction was not observed in nonfunctional variants. Therefore, the Etd1-Rho1 interaction revealed by in vitro copurification assays might be relevant to the molecular mechanism by which Etd1 activates Rho1 in vivo.
Detailed sequence analysis reveals a weak similarity of Etd1 with Rgf1 and Rgf2 Rho1 GEFs at the Etd1 cell localization domain (see Figure 4E for Rgf2). No known motifs or significant similarities to proteins of known function were identified at the functional C-terminal region.
Feedback activation of Spg1 by Rho1
High Rho1 activity restores cytokinesis in Etd1-deficient cells (Figure 1C and Figure 2A). Given that Pab1 inhibits Rho1 (Figure 3), the pab1-4 mutation could rescue etd1Δ cells by increasing Rho1 activity. SIN-independent activation of cytokinesis is observed in both cases (Figure 2). Interestingly, pab1-4 is known to recover Spg1 activity in the etd1Δ strain (Lahoz et al. 2010). This observation suggests that PP2A-Pab1 could independently inhibit Spg1 and Rho1 GTPases. Alternatively, Spg1 activation by pab1-4 could be exerted through Rho1. To check this possibility, we also studied Spg1 GTPase activity in etd1Δ cells, either overexpressing Rho1 (the etd1Δ/p41rho1 strain) or lacking the Rga5 Rho1 inhibitor (the etd1Δ rga5Δ strain).
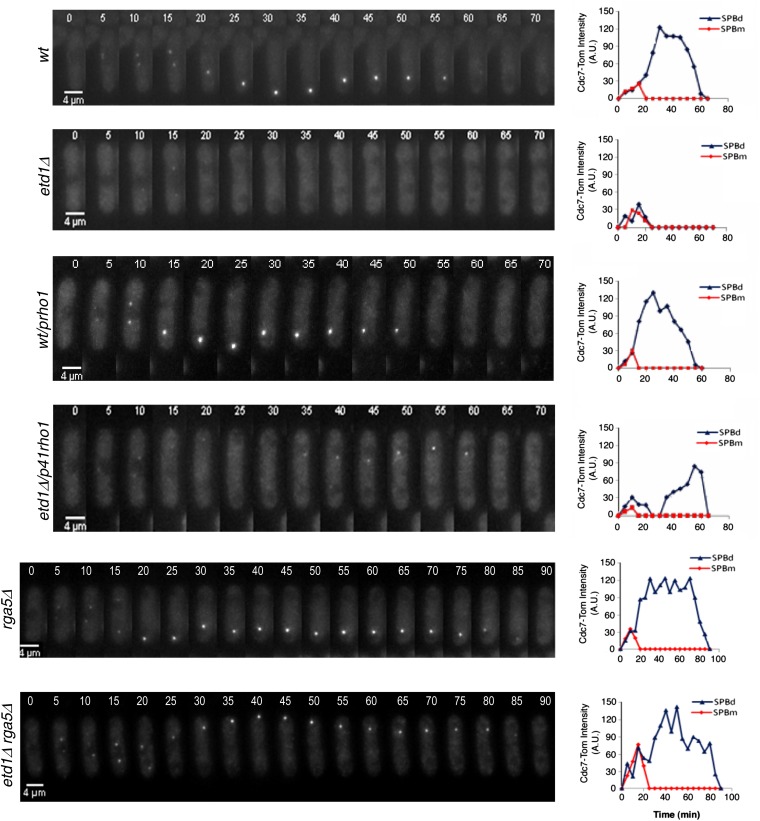
The Cdc7 protein kinase binds only to the active form of Spg1. Therefore, the Cdc7-Tom fusion protein is a useful marker for the in vivo analysis of Spg1 function in conjunction with time-lapse microscopy. As shown in Figure 5, Cdc7-Tom appeared at both SPBs at the initiation of mitosis in wild-type cells. As cells progressed through anaphase, Cdc7-Tom localized to only one of the SPBs (SPBd) until the completion of cell division, consistent with established results (Sohrmann et al. 1998). In Etd1-deficient cells, Cdc7-Tom also localized to both SPBs in early mitosis, but signal rapidly decayed early in anaphase (Figure 5), as reported previously (Daga et al. 2005; Garcia-Cortes and McCollum 2009). Interestingly, we found that high Rho1 activity, due to Rho1 overproduction or Rga5 depletion, led to the restoration of Cdc7-Tom SPB localization in etd1Δ mutant cells (Figure 5). Thus, high levels of Rho1 activity are sufficient for Spg1 activation in Etd1-depleted cells. Spg1 is thought to activate Rho1 through the SIN cascade (Jin et al. 2006). The feedback activation of Spg1 by Rho1 establishes an unexpected cross talk between these two GTPases that might serve to interconnect SIN and cytokinesis.
Figure 5.
Effects of high Rho1 activity on Spg1 activation in Etd1-depleted cells. Cdc7-Tom was imaged by time-lapse microscopy at 5-min intervals in living wild-type (wt), etd1Δ, wt/p41rho1 (nmt41x-driven expression of rho1 in plasmid pREP41x), etd1Δ/p41rho1, rga5Δ, and etd1Δ rga5Δ cells under restrictive conditions for the etd1Δ mutation (25°). Fluorescence intensity was quantified (arbitrary units) and represented for each SPB (SPBd and dSPBm).
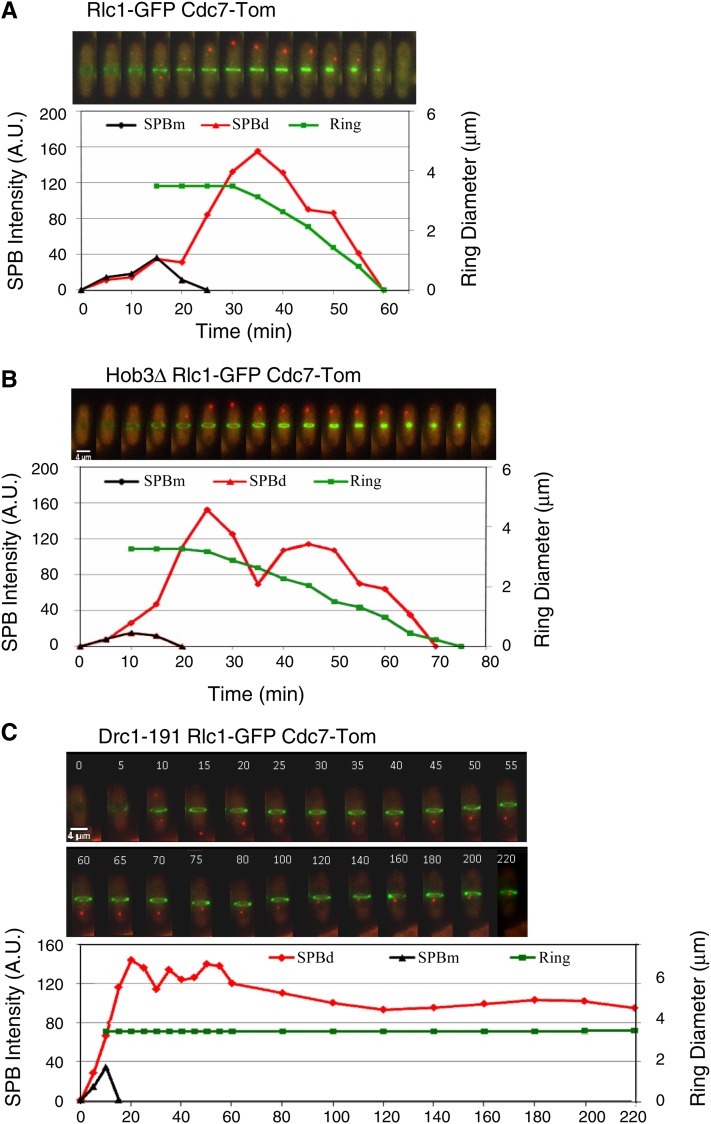
It is well established that cytokinesis is triggered by SIN (Krapp and Simanis 2008). To test whether SIN is feedback-connected with cytokinesis, we studied the correlation between the rate of actomyosin ring constriction (measured as the medial ring diameter during cytokinesis by using the Rlc1-GFP ring marker) and SIN activity (determined by fluorescent quantification of the Cdc7-Tom signal at the active SPB) in wild-type cells and in mutant cells with an altered rate of medial ring contraction. In wild-type cells under normal growth conditions, high Spg1 activity triggered ring constriction, and this activity decreased as the medial ring constricted (Garcia-Cortes and McCollum 2009) (Figure 6A). Hob3 is a BAR domain-containing protein required to recruit and activate Cdc42 at the cell division site. Interestingly, the actomyosin ring contraction is slower in hob3Δ than in wild-type cells (Coll et al. 2007). In agreement with a feedback loop model interconnecting SIN and cytokinesis, slowing down the rate of ring constriction by means of the hob3Δ mutation resulted in a proportional reduction in the rate of Spg1 inactivation (Figure 6B). Furthermore, the complete block of ring constriction in drc1-191 mutant cells (Liu et al. 2000) led to a permanent activation of Spg1 (Figure 6C). Hence, SIN activity and medial ring constriction are likely functionally interconnected events.
Figure 6.
Kinetics of Spg1 activation and actomyosin ring contraction. Cdc7-Tom (Spg1 activity marker) and Rlc1-GFP (medial ring marker) were imaged by time-lapse microscopy at 5-min intervals in living (A) wild-type cells, (B) hob3Δ mutants (slows down medial ring constriction), and (C) drc1-191 mutants (blocks medial ring constriction) at 36° (restrictive temperature for drc1-191). Fluorescence intensity of Cdc7-Tom (arbitrary units) at each SPB (SPBd and SPBm) and medial ring diameter (μm) according to Rlc1-GFP signal are represented.
The thermo-sensitive drc1-191 mutation provides a unique tool to analyze components and mechanisms behind the proposed interconnection. At restrictive temperature, the Drc1-191 protein blocks actomyosin ring constriction (Liu et al. 2000), and the cell remains in a permanent medial ring-SPB signaling state (Figure 6C). This observation prompted us to test whether Sid2, Etd1, and/or Rho1 were required for this feedback connection.
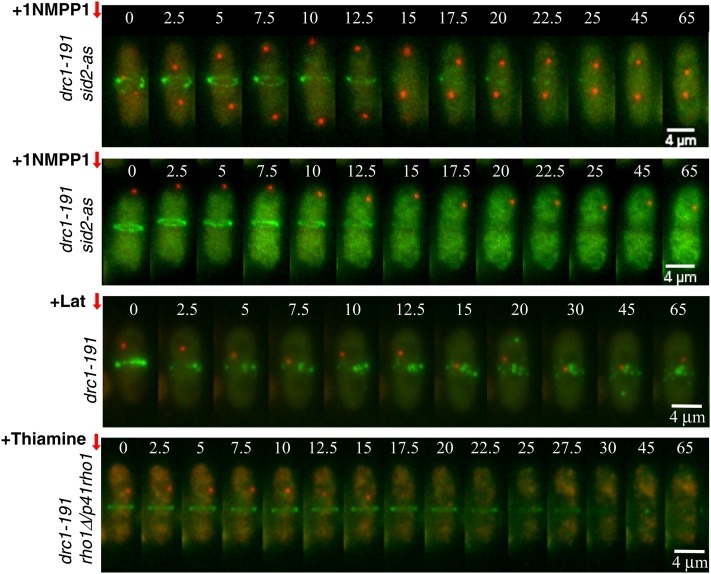
Sid2 is the last component of the SIN kinase cascade and localizes at both SPBs and the medial ring during cytokinesis. In fact, this kinase has been recently implicated in a novel feedback loop that hyper-activates SIN to generate SPB asymmetry (Feoktistova et al. 2012). Thus, Sid2 is a good candidate to feedback-signal medial ring constriction activity to SIN. To experimentally inactivate Sid2 in drc1-191 blocked cells, we first constructed a new conditional analog-sensitive sid2-as allele (Materials and Methods) and determined that controlled Sid2 inactivation (by adding 1NMPP1) in these sid2-as cells leads to a conventional sin− phenotype independently of the incubation temperature (see characterization of this strain in Figure S1).
To determine the effect of Sid2 inactivation in the proposed feedback loop, Cdc7-Tom and Rlc1-GFP were imaged by time-lapse microscopy in sid2-as drc1-191 cells, and Sid2-as was inactivated after shift to restrictive temperature. When Sid2 inactivation took place early in mitosis, before the generation of SPB asymmetry, Cdc7 SPB localization never became asymmetric as cells proceeded through anaphase (Figure 7). Interestingly, if Sid2 was inactivated during anaphase B, when asymmetric SPB location of active Spg1 is already generated, the asymmetry was maintained in these cells (Figure 7). These results indicate that Sid2 is required to generate the asymmetric activation of SIN as recently described (Feoktistova et al. 2012), but not to maintain it once asymmetry is established (Figure 7). In both cases, Sid2 inactivation resulted in disassembly of the actomyosin ring. However, Spg1 remained permanently active, indicating that Sid2 is required to maintain the actomyosin ring structure, although this observation demonstrates that it is not involved in the proposed cytokinesis-SIN interconnection.
Figure 7.
Involvement of Sid2, Etd1, and Rho1 in signaling Spg1 activation in drc1-191-blocked cells. Cdc7-Tom (Spg1 activity marker) and Rlc1-GFP (medial ring marker) were imaged by time-lapse microscopy at 2.5-min intervals in living drc1-191 sid2-as, drc1-191, and drc1-191 rho1D/p41rho1 mutants at 36°. Sid2-as inactivation (+10 μM 1NMPP1) before and after asymmetric localization of active Spg1, actomyosin ring disassembly (+100 µM latrunculin B), and repression of nmt41x-driven expression of rho1 in the p41rho1 plasmid (+ Thiamine) are indicated (Materials and Methods).
In Drc1-191-blocked cells, depolymerization of actomyosin ring structures by latrunculin treatment releases the mitotic arrest triggered by the cytokinesis checkpoint (Liu et al. 2000). However, as described above, medial ring disassembly by Sid2 inactivation had no effects in Spg1 activity (SPB-associated Cdc7) in Drc1-191-blocked cells. Identical results were obtained when the medial ring was disassembled by latrunculin treatment in these cells (Figure 7). Therefore, we conclude that while structural-dependent mechanisms are signaling mitotic arrest (Liu et al. 2000), trans-acting elements are likely involved in the feedback activation of SIN in Drc1-191 cells.
To determine the role of Rho1 in this activation, dynamics of Cdc7-Tom and Rlc1-GFP were analyzed by time-lapse microscopy at restrictive temperature for the drc1-191 mutation in drc1-191 rho1Δ/p41rho1 cells at the time that Rho1 was depleted by adding thiamine (Materials and Methods). Interestingly, Cdc7 signal disappeared from the active SPBd after Rho1 depletion, followed by medial ring disassembly (Figure 7). This result indicates that Spg1 requires Rho1 to maintain activity in drc1-191-blocked cells. Identical results were obtained when using the drc1-191 etd1Δ/p41etd1 strain (data not shown), according to the essential role of Etd1 in Rho1 activation proposed in this study. Hence, these results indicate that Rho1 is likely involved in a feedback loop mechanism that ensures SIN activity during progress of actomyosin ring constriction and septum formation. A working model is outlined in Figure 8.
Figure 8.
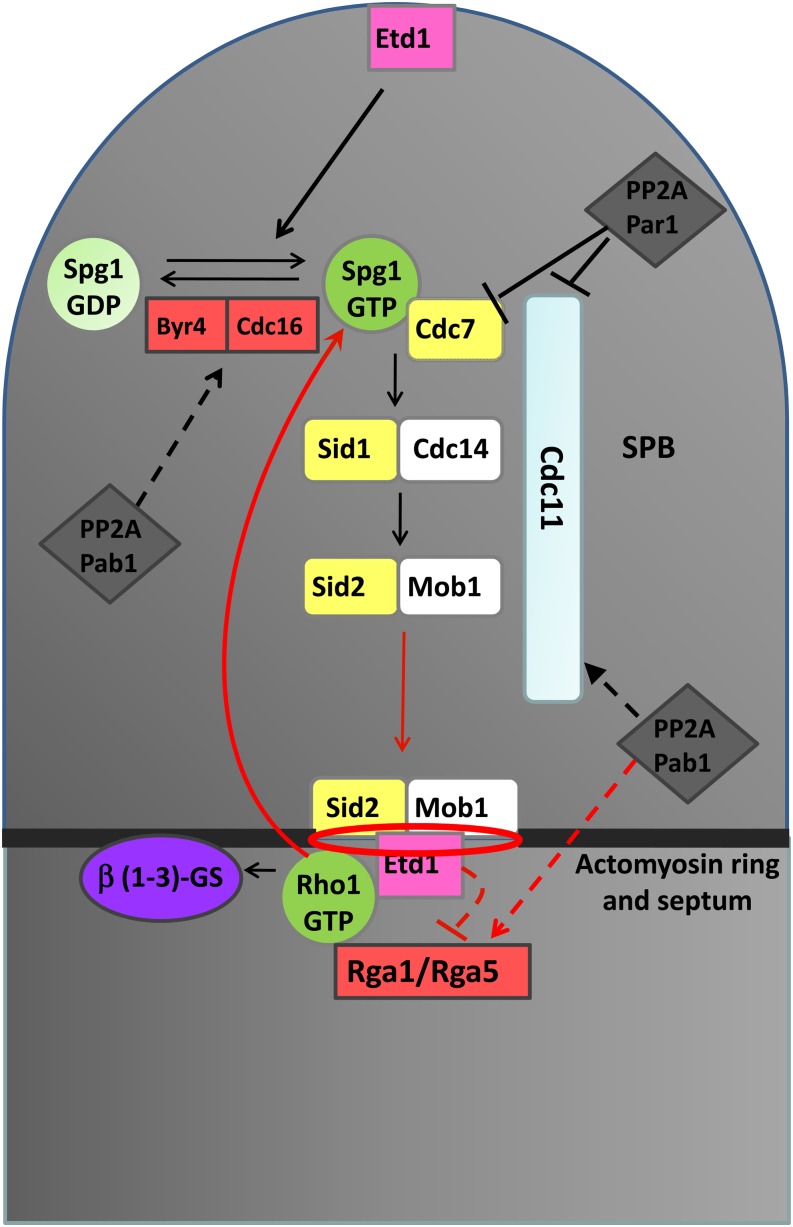
Schematic representation of mechanisms suggested for Spg1 and Rho1 coordination. The model includes the Cdc16 Spg1-specific GAP; Rga1 and Rga5 Rho1 GAPs; Etd1 and PP2A complexes regulating SIN; and the proposed feedback loop mechanism operating during cytokinesis. In red are the new pathways and regulatory mechanisms proposed in this study. Dashed lines represent hypothetical regulatory mechanisms.
Discussion
Etd1, a novel activator of Rho1
Rho1 is the key GTPase in the regulation of cell-wall and primary septum polymer synthesis (Garcia et al. 2006). Overproduction of Rho1 rescued lethality of the etd1Δ mutant strain (Figure 1C), pointing toward Rho1 as a target for Etd1 activity.
Among Rho1 regulators, depletion of Rho1 inhibitors such as Rga1 and Rga5 (Figure 1D), as well as ectopic expression of Rho1 activators Rgf1 and Rgf3 (Figure 1E) efficiently rescued growth and division defects of Etd1 mutants. Both Rga1 and Rgf1 are involved in Rho1-mediated regulation of cell-wall synthesis at the growing tips and septation (Nakano et al. 2001; Mutoh et al. 2005). Rga5 specifically regulates cell integrity (Calonge et al. 2003), while Rgf3 activates Rho1 functions to coordinate cell-wall synthesis with septation (Tajadura et al. 2004). Neither depletion of Rga8, a GAP connecting Cdc42/p21-activated kinase with Rho1 (Yang et al. 2003), nor overproduction of the Rho1 GEF Rgf2 involved in cell-wall synthesis during sporulation (Garcia et al. 2006), rescued etd1Δ lethality (Figure 1). Therefore, Etd1 is functionally redundant with the subset of Rho1 regulators that preserve cell integrity and control cell-wall synthesis and septation during vegetative growth.
These Rho1 regulators localize at the cell ends (Rga5), the septum (Rgf3), or both (Rga1 and Rgf1) (Calonge et al. 2003; Tajadura et al. 2004; Garcia et al. 2006). Consistent with a role in Rho1 activation, Etd1 localizes at the growing tips and the septum (Daga et al. 2005). Overall, our genetics and cellular localization analysis indicate that Etd1 is likely involved in the activation of Rho1 at the cell cortex and the medial ring to regulate cell-wall synthesis during cell growth and septation. Etd1-dependent levels of Rho1-GTP and its associated 1,3-β-glucan synthase activity (Figure 3) support the proposed role for Etd1 in Rho1 activation.
A molecular mechanism for the activation of Rho1 by Etd1
Etd1 positively regulates Rho1 activity (Figure 3), and its function is efficiently replaced by ectopic expression of established Rho1 GEFs such as rgf1 and rgf3 (Figure 1E) (Morrell-Falvey et al. 2005; Garcia et al. 2006). A certain degree of amino-acid sequence similarity to Rgf GEFs can be observed within the short stretch of Etd1 that confers proper cell localization (see Figure 4E for Rgf2). However, this similarity is unlikely to be involved in conferring GEF activity to Etd1 since this domain is not essential for Etd1 function (Figure 4A). In fact, the functional C-terminal region of Etd1 lacks motifs associated with GEF activity (Hoffman and Cerione 2002), and consistently, in vitro GEF activity has not been found for this protein (Garcia-Cortes and McCollum 2009). Thus, Etd1 is unlikely acting as a Rho1 GEF.
More recent experiments revealed that Lte1, a possible Etd1 budding yeast homolog (Garcia-Cortes and McCollum 2009), rather than acting as a GEF, contributes to the proper localization of MEN regulators such as the Bfa1 GAP (Geymonat et al. 2010) and the Kin4 kinase (Falk et al. 2011). Given that the removal of Rga1 or Rga5 Rho1 GAPs mimics Etd1 function (Figure 1D), we hypothesize that Etd1 binding to Rho1 (Figure 3D and Figure 4D) may activate this GTPase by competing for binding of its negative regulators. Additionally, Etd1 is required for the recruitment of the Sid2-Mob1 complex to the medial ring (Figure 2) and for maintaining this complex’s kinase activity during anaphase. We cannot discard that Sid2-Mob1 activity and its location at the ring could be interdependent features. However, independently of this observation, our results suggest that Etd1 plays a key role in connecting Sid2-mediated SIN signaling to the division site. The sid2-GFP construct is synthetically lethal in combination with etd1 mutant alleles (Daga et al. 2005), supporting the idea that Sid2-Mob1 might interact with Etd1 in this particular step. Showing whether the binding of Etd1 to Rho1 allows it to recruit Rho1 activators (for example, the Sid2-Mob1 kinase complex) and/or to compete with the binding of Rho1 inhibitors (likely Rga1 and Rga5) will be important questions to tackle in future studies.
Rho1 inhibition by PP2A-Pab1
PP2A is a heterotrimer composed of a catalytic subunit (C), a scaffold (A), and a regulatory or targeting subunit (B or B′) (Mayer-Jaekel and Hemmings 1994). Previous studies have indicated that the B′ subunit (Par1) is involved in the PP2A-mediated regulation of SIN components (Krapp et al. 2003). Par1 and the other B′ subunit known in S. pombe as Par2 inhibit SIN signaling upstream of the pathway at the level of the Spg1 GTPase and its associated Cdc7 kinase (Jiang and Hallberg 2001). The Pab1 B subunit is involved in SIN regulation at different levels (Lahoz et al. 2010). However, according to the results presented here, PP2A-Pab1 also inhibits septation downstream of the SIN cascade (Figure 2), directing PP2A activity toward inhibition of Rho1 (Figure 3).
It has been suggested that p190, a Rho-specific GAP, and the ECT2 GEF cooperate antagonistically to regulate the activity of RhoA (the Rho1 counterpart in higher eukaryotes) during cytokinesis in human cells (Mikawa et al. 2008). p190 GAP is inhibited by phosphorylation and activated by PP2A-mediated dephosphorylation (Mori et al. 2009). Similar results have been recently shown for Rho GAP activation in a number of different eukaryotic cells (Sopko et al. 2007; Zheng et al. 2007; Toure et al. 2008; McAvoy et al. 2009; Mori et al. 2009; Wolfe et al. 2009). Since rga5Δ and rga1Δ null mutants suppress lethality of Etd1-depleted cells, inhibition of Rho1 by PP2A-Pab1 could be exerted through dephosphorylation and activation of these Rho1 GAPs (see model in Figure 8). Accordingly, changes in Rho1-GTP levels and 1,3-β-glucan synthase activity caused by pab1-4 or by overexpressing pab1 (Figure 3) were very similar to changes described for rga5Δ and overexpression of rga5, respectively (Calonge et al. 2003). Phosphorylation studies of Rho1 GAPs will be very valuable in further addressing the molecular mechanism by which PP2A-Pab1 negatively regulates Rho1.
Coordination between SIN and cytokinesis
The suppression of lethality in Etd1-depleted cells by Rho1 hyper-activation (Figure 1 and Figure 2) revealed an unexpected feature. Hyper-activation of Rho1 in this strain by pab1-4, rga5Δ, or p41rho1 resulted in Spg1 activation (Lahoz et al. 2010) (Figure 5). We cannot discard the possibility that Rho1 regulators such as Pab1 or Rga5 could also target Spg1. However, Spg1 activation in etd1Δ/p41rho1 cells indicates that this GTPase is feedback activated by Rho1 itself during cytokinesis. The analysis of Spg1 activity during cytokinesis in wild-type cells and in mutant cells with a reduced rate of actomyosin ring constriction suggested that both events are interconnected during this phase (Figure 6). The use of the Drc1-191 mutant, which blocks the cell with a permanent medial ring-SIN signaling, allowed us to confirm that the feedback activation of SIN during cell division exists and that Rho1 and Etd1, but not Sid2, are involved in it (Figure 7). Thus, the feedback activation of Spg1 by Rho1 may serve to coordinate progress in cytokinesis with SIN activity. Interestingly, analog addition in Drc1-191 Sid2-as cells caused the complete disassembly of the persistent ring, indicating that Sid2 is required to maintain it (Figure 7). In the drc1-191 rho1Δ/p41rho1 strain, medial ring disassembly was also observed, but it took place only after Spg1 inactivation (Figure 7). This observation reinforces the existence of the proposed feedback loop: Spg1 inactivation by Rho1 depletion likely propitiates downstream Sid2 inactivation, which, in turn, results in actomyosin ring disassembly.
A relevant property of the SIN is that its activity is asymmetric on the two SPBs during anaphase. This asymmetry is thought to be important for correct regulation of SIN signaling (Johnson et al. 2012). Surprisingly, high Rho1 activity not only activated Spg1, but also restored asymmetric SPB activation in cells lacking Etd1 (Figure 5). Recruitment of Cdc7 to the new SPB by Sid2 phosphorylation of the scaffold protein Cdc11 is a key mechanism to break SPB activation symmetry (Feoktistova et al. 2012). Given that Sid2 kinase activity is extremely low in these cells (Figure 3), Spg1 activation by Rho1 may be involved in a Sid2-independent mechanism that generates and/or maintains SPB asymmetry.
The SIN is controlled at many levels to ensure that cytokinesis is executed once per cell cycle and only after cells segregate their DNA (Krapp and Simanis 2008). It has been recently shown that this network regulates proper spindle elongation and telophase nuclear positioning in mitosis (Mana-Capelli et al. 2012). Therefore, the feedback activation of SIN by Rho1 represents a new level of SIN regulation coupling late mitotic events with progression of cytokinesis. It is remarkable that in proliferating yeast cells that lack this SIN-cytokinesis coupling (Figure 2) nuclear division may eventually occur in the absence of cytokinesis, resulting in an increased incidence of binucleate cells (Figure 1, C–E). In mammalians, this phenotype is particularly relevant since it is frequently associated with cancer. Tetraploidy cells that stem from cytokinesis failure may give rise to more chromosome aberration, which promotes further aneuploidy and genomic instability and eventually leads to tumorigenesis (Holland and Cleveland 2009). Hence, our findings provide unexpected insights into a new mechanism, the Rho1-SIN cross talk, which may contribute to preventing cytokinesis failure during the mitotic cycle.
Etd1 is required for Spg1 activity at the daughter SPB (Daga et al. 2005; Garcia-Cortes and McCollum 2009). According to our results, Spg1 is likely activated by Etd1 through Rho1. However, Etd1 could also directly activate the Spg1 GTPase (Garcia-Cortes and McCollum 2009). Different GTPases may have common regulators to coordinate their functions. Lte1 is known to regulate Tem 1 GTPase at the SPB (Geymonat et al. 2010) and the small GTPases Ras and Bud1 at the cell cortex, a dual function that allows Lte1 to couple mitotic progression and morphological development in budding yeast (Geymonat et al. 2010). Thus, in addition to the feedback-loop mechanism proposed here, a parallel function of Etd1 in Spg1 and Rho1 regulation would help to coordinate ring constriction and SIN (Figure 8).
Rho1/RhoA as well as its regulators and effectors are important players for ensuring successful cytokinesis. Our results strongly suggest that Etd1 and the PP2A-Pab1 complex are novel regulators of the universal Rho1 GTPase. The function of Etd1 in positively regulating Rho1 and the existence of a feedback mechanism connecting Rho1 and SIN may contribute to our understanding of how eukaryotic cells coordinate mitosis, cell growth, and cell division.
Supplementary Material
Acknowledgments
We thank Paul Nurse and Pilar Perez for providing strains and plasmids. We also thank Borja Fernandez, Viesturs Simanis, John R. Pearson, and Ann Yonetani for useful comments and discussions. This work was supported by grants BFU2009-13565 and BFU2010-21310 to J.J. and R.R.D., respectively, from the Ministerio de Ciencia e Innovación of the Spanish Government.
Footnotes
Communicating editor: D. Lew
Literature Cited
- Arellano M., Duran A., Perez P., 1996. Rho 1 GTPase activates the (1–3)beta-D-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 15: 4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Arellano M., Duran A., Perez P., 1997. Localisation of the Schizosaccharomyces pombe rho1p GTPase and its involvement in the organisation of the actin cytoskeleton. J. Cell Sci. 110: 2547–2555. [DOI] [PubMed] [Google Scholar]
- Bardin A. J., Visintin R., Amon A., 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102: 21–31. [DOI] [PubMed] [Google Scholar]
- Calonge T. M., Arellano M., Coll P. M., Perez P., 2003. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 47: 507–518. [DOI] [PubMed] [Google Scholar]
- Cerutti L., Simanis V., 1999. Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J. Cell Sci. 112: 2313–2321. [DOI] [PubMed] [Google Scholar]
- Cipak L., Zhang C., Kovacikova I., Rumpf C., Miadokova E., et al. , 2011. Generation of a set of conditional analog-sensitive alleles of essential protein kinases in the fission yeast Schizosaccharomyces pombe. Cell Cycle 10: 3527–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll P. M., Rincon S. A., Izquierdo R. A., Perez P., 2007. Hob3p, the fission yeast ortholog of human BIN3, localizes Cdc42p to the division site and regulates cytokinesis. EMBO J. 26: 1865–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J. C., Ishiguro J., Duran A., Ribas J. C., 2002. Localization of the (1,3)beta-D-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J. Cell Sci. 115: 4081–4096. [DOI] [PubMed] [Google Scholar]
- Cortes J. C., Carnero E., Ishiguro J., Sanchez Y., Duran A., et al. , 2005. The novel fission yeast (1,3)beta-D-glucan synthase catalytic subunit Bgs4p is essential during both cytokinesis and polarized growth. J. Cell Sci. 118: 157–174. [DOI] [PubMed] [Google Scholar]
- Cortes J. C., Konomi M., Martins I. M., Muñoz J., Moreno M. B., et al. , 2007. The (1,3)beta-D-glucan synthase subunit Bgs1p is responsible for the fission yeast primary septum formation. Mol. Microbiol. 65: 201–217. [DOI] [PubMed] [Google Scholar]
- Daga R. R., Lahoz A., Munoz M. J., Moreno S., Jimenez J., 2005. Etd1p is a novel protein that links the SIN cascade with cytokinesis. EMBO J. 24: 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J. E., Chan L. Y., Amon A., 2011. Lte1 promotes mitotic exit by controlling the localization of the spindle position checkpoint kinase Kin4. Proc. Natl. Acad. Sci. USA 108: 12584–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova A., Morrell-Falvey J., Chen J. S., Singh N. S., Balasubramanian M. K., et al. , 2012. The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol. Biol. Cell 23: 1636–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21: 2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P., Tajadura V., Garcia I., Sanchez Y., 2006. Role of Rho GTPases and Rho-GEFs in the regulation of cell shape and integrity in fission yeast. Yeast 23: 1031–1043. [DOI] [PubMed] [Google Scholar]
- Garcia-Cortes J. C., McCollum D., 2009. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J. Cell Biol. 186: 739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., de Bettignies G., Sedgwick S. G., 2009. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J. Cell Biol. 187: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Jensen S., Sedgwick S. G., 2010. Phosphorylation of Lte1 by Cdk prevents polarized growth during mitotic arrest in S. cerevisiae. J. Cell Biol. 191: 1097–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J., Nurse P., 2001. A journey into space. Nat. Rev. Mol. Cell Biol. 2: 647–656. [DOI] [PubMed] [Google Scholar]
- Hoffman G. R., Cerione R. A., 2002. Signaling to the Rho GTPases, networking with the DH domain. FEBS Lett. 513: 85–91. [DOI] [PubMed] [Google Scholar]
- Holland A. J., Cleveland D. W., 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M. C., Guertin D. A., McCollum D., 2004. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol. Cell. Biol. 24: 3262–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Hallberg R. L., 2001. Correct regulation of the septation initiation network in Schizosaccharomyces pombe requires the activities of par1 and par2. Genetics 158: 1413–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J., Oballe J., 1994. Ethanol-hypersensitive and ethanol-dependent cdc- mutants in Schizosaccharomyces pombe. Mol. Gen. Genet. 245: 86–95. [DOI] [PubMed] [Google Scholar]
- Jin Q. W., Zhou. A. Bimbo M., Balasubramanian M. K., McCollum D., 2006. A role for the septation initiation network in septum assembly revealed by genetic analysis of sid2–250 suppressors. Genetics 172: 2101–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. E., McCollum D., Gould K. L., 2012. Polar opposites: fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton (Hoboken) 69: 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A., Simanis V., 2008. An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36: 411–415. [DOI] [PubMed] [Google Scholar]
- Krapp A., Cano E., Simanis V., 2003. Mitotic hyperphosphorylation of the fission yeast SIN scaffold protein cdc11p is regulated by the protein kinase cdc7p. Curr. Biol. 13: 168–172. [DOI] [PubMed] [Google Scholar]
- Krapp A., Gulli M. P., Simanis V., 2004. SIN and the art of splitting the fission yeast cell. Curr. Biol. 14: R722–R730. [DOI] [PubMed] [Google Scholar]
- Lahoz A., Alcaide-Gavilan M., Daga R. R., Jimenez J., 2010. Antagonistic roles of PP2A-Pab1 and Etd1 in the control of cytokinesis in fission yeast. Genetics 186: 1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang H., Balasubramanian M. K., 2000. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J. Cell Sci. 113: 1223–1230. [DOI] [PubMed] [Google Scholar]
- Mana-Capelli S., McLean J. R., Chen C. T., Gould K. L., McCollum D., 2012. The kinesin-14 Klp2 is negatively regulated by the SIN for proper spindle elongation and telophase nuclear positioning. Mol. Biol. Cell 23: 4592–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K., 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130. [DOI] [PubMed] [Google Scholar]
- Mayer-Jaekel R. E., Hemmings B. A., 1994. Protein phosphatase 2A-a ‘menage a trois’. Trends Cell Biol. 4: 287–291. [DOI] [PubMed] [Google Scholar]
- McAvoy T., Zhou M. M., Greengard P., Nairn A. C., 2009. Phosphorylation of Rap1GAP, a striatally enriched protein, by protein kinase A controls Rap1 activity and dendritic spine morphology. Proc. Natl. Acad. Sci. USA 106: 3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa M., Su L., Parsons S. J., 2008. Opposing roles of p190RhoGAP and Ect2 RhoGEF in regulating cytokinesis. Cell Cycle 7: 2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison J. M., Nurse P., 1985. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75: 357–376. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Mori K., Amano M., Takefuji M., Kato K., Morita Y., et al. , 2009. Rho-kinase contributes to sustained RhoA activation through phosphorylation of p190A RhoGAP. J. Biol. Chem. 284: 5067–5076. [DOI] [PubMed] [Google Scholar]
- Morrell-Falvey J. L., Ren L., Feoktistova A., Haese G. D., Gould K. L., 2005. Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J. Cell Sci. 118: 5563–5573. [DOI] [PubMed] [Google Scholar]
- Mutoh T., Nakano K., Mabuchi I., 2005. Rho1-GEFs Rgf1 and Rgf2 are involved in formation of cell wall and septum, while Rgf3 is involved in cytokinesis in fission yeast. Genes Cells 10: 1189–1202. [DOI] [PubMed] [Google Scholar]
- Nakano K., Mutoh T., Mabuchi I., 2001. Characterization of GTPase-activating proteins for the function of the Rho-family small GTPases in the fission yeast Schizosaccharomyces pombe. Genes Cells 6: 1031–1042. [DOI] [PubMed] [Google Scholar]
- Reid T., Furuyashiki T., Ishizaki T., Watanabe G., Watanabe N., et al. , 1996. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, rhophilin in the rho-binding domain. J. Biol. Chem. 271: 13556–13560. [DOI] [PubMed] [Google Scholar]
- Ren X. D., Kiosses W. B., Schwartz M. A., 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18: 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimova E., Sohrmann M., Fournier N., Simanis V., 2000. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signaling the onset of septum formation. J. Cell Sci. 113: 1695–1704. [DOI] [PubMed] [Google Scholar]
- Sayers L. G., Katayama S., Nakano K., Mellor H., Mabuchi I., et al. , 2000. Rho-dependence of Schizosaccharomyces pombe Pck2. Genes Cells 5: 17–27. [DOI] [PubMed] [Google Scholar]
- Schmidt S., Sohrmann Hofmann M., K., A. Woollard, and V. Simanis, 1997. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 11: 1519–1534. [DOI] [PubMed] [Google Scholar]
- Sohrmann M., Schmidt S., Hagan I., Simanis V., 1998. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 12: 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Smith J. C., Figeys D., Andrews B. J., 2007. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 26: 4487–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks C. A., Morphew M., McCollum D., 1999. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol. 146: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajadura V., García B., García I., García P., Sánchez Y., 2004. Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall beta-glucan biosynthesis through the GTPase Rho1p. J. Cell Sci. 117: 6163–6174. [DOI] [PubMed] [Google Scholar]
- Toure A., Mzali R., Liot C., Seguin L., Morin L., et al. , 2008. Phosphoregulation of MgcRacGAP in mitosis involves Aurora B and Cdk1 protein kinases and the PP2A phosphatase. FEBS Lett. 582: 1182–1188. [DOI] [PubMed] [Google Scholar]
- Viana R. A., Pinar M., Soto T., Coll P. M., Cansado J., et al. , 2013. Negative functional interaction between cell integrity MAPK pathway and Rho1 GTPase in fission yeast. Genetics 195: 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L., 2005. Split decisions, coordinating cytokinesis in yeast. Trends Cell Biol. 15: 10–18. [DOI] [PubMed] [Google Scholar]
- Wolfe B. A., Takaki T., Petronczki M., Glotzer M., 2009. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7: e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Qyang Y., Bartholomeusz G., Zhou X., Marcus S., 2003. The novel Rho GTPase-activating protein family protein, Rga8, provides a potential link between Cdc42/p21-activated kinase and Rho signaling pathways in the fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 278: 48821–48830. [DOI] [PubMed] [Google Scholar]
- Zheng W., Chen J., Liu W., Zheng S., Zhou J., et al. , 2007. A Rho3 homolog is essential for appressorium development and pathogenicity of Magnaporthe grisea. Eukaryot. Cell 6: 2240–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.