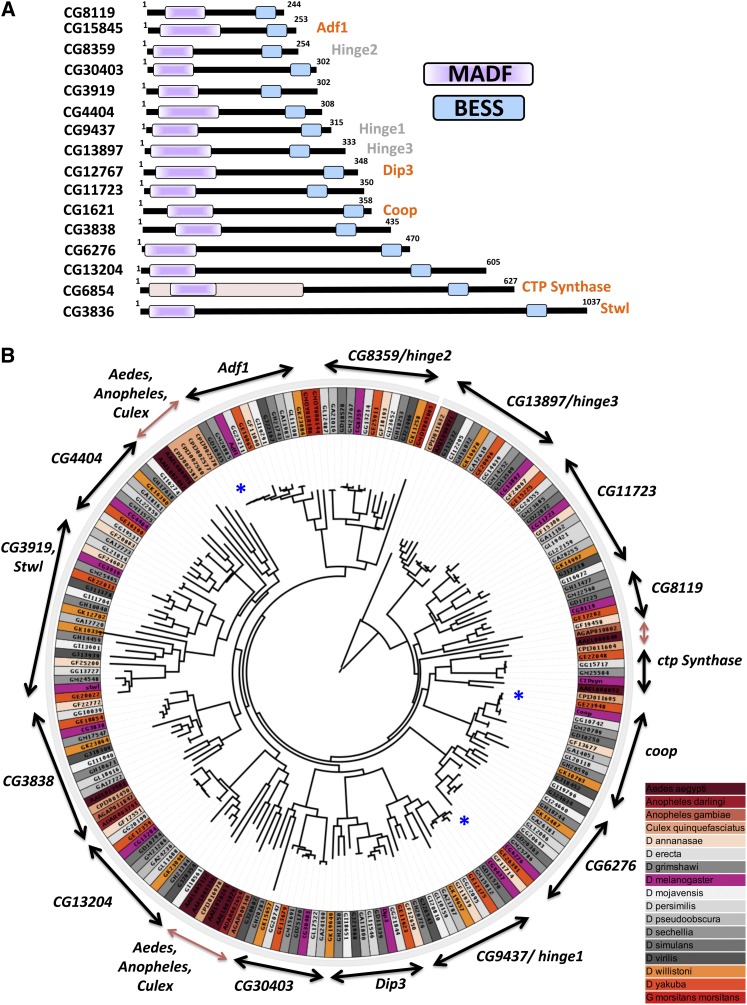
Figure 1.
The D. melanogaster MADF-BESS family codes for 16 members that are a consequence of gene duplication. (A) MADF-BESS family in D. melanogaster consists of 16 proteins coded by individual genes. The 16 proteins, represented in the figure, contain an N-terminal DNA-binding Myb/SANT like in Adf (MADF) domain and a C-terminal BEAF-32, Stonewall, Su (var) 3-7 homology (BESS) domain. MADF and BESS domains tend to be found together in a single polypeptide chain. Proteins labeled in orange have been characterized, while those in gray are the subject of this study and been named hinge genes based on their loss-of-function phenotype. (B) Maximum-likelihood phylogenetic tree of all MADF-BESS genes in dipterans. Branch lengths are proportional to mean substitutions per site. Orthologs of the 16 MADF-BESS genes cluster separately with the corresponding D. melanogaster gene (labeled on two-sided arrows), indicating that the duplication in the family occurred before the divergence of drosophilids. MADF-BESS genes from Culex, Anopheles, and Glossina morsitans have fewer orthologs and cluster separately (brown arrow) for the most part, indicating that, in their case, expansion in MADF-BESS was independent from that in drosophilids. The genes for different sequenced dipterans are color-coded to bring out this feature. The blue asterisk marks the genes that show short branch lengths and thus minimal sequence divergence.