Abstract
Range expansions are a ubiquitous phenomenon, leading to the spatial spread of genetic, ecological, and cultural traits. While some of these traits are advantageous (and hence selected), other, nonselected traits can also spread by hitchhiking on the wave of population expansion. This requires us to understand how the spread of a hitchhiking trait is coupled to the wave of advance of its host population. Here, we use a system of coupled Fisher-Kolmogorov-Petrovsky-Piskunov (F-KPP) equations to describe the spread of a horizontally transmitted hitchhiking trait within a population as it expands. We extend F-KPP wave theory to the system of coupled equations to predict how the hitchhiking trait spreads as a wave within the expanding population. We show that the speed of this trait wave is controlled by an intricate coupling between the tip of the population and trait waves. Our analysis yields a new speed selection mechanism for coupled waves of advance and reveals the existence of previously unexpected speed transitions.
Keywords: wave of advance, Fisher–KPP equation, traveling waves, horizontal transmission, range expansion
THE expansion of a population into a new spatial territory, known as a range expansion, is an important and fundamental process in evolution, ecology, and anthropology. Range expansions are driven by selection for advantageous traits, whether these be genetic, ecological, technological, or cultural. However, they typically lead to the concomitant spread of other traits, which may not be advantageous but spread by “hitchhiking” with the selected trait. Understanding spatial patterns of genetic, ecological, and cultural diversity requires us to understand how the spread of a hitchhiking trait is coupled to the wave of advance of its host population.
The wave of advance model was introduced by Fisher (1937) to describe the spread of an advantageous gene within a spatially extended population and was used independently by Kolmogorov et al. (1937) to describe general growth and diffusion processes and by Skellam (1951) in the ecological context of the invasion of new territory by a colonizing species. This model describes the advance of a population in space with the following equation, which we denote the Fisher-Kolmogorov-Petrovsky-Piskunov (F–KPP) equation,
| (1) |
where x denotes position, t denotes time, and u(x, t) is the population density. In this model, population expansion arises from a balance between diffusion of individuals in space (with diffusion constant D) and local growth (with maximum population density or carrying capacity K and linear growth rate αK). This equation has extremely broad biological relevance (Ammerman and Cavalli-Sforza 1971; Mollinson 1991; van den Bosch et al. 1992; Young and Bettinger 1995; Hethcote 2000; Murray 2004; Ackland et al. 2007; Barrett-Freeman et al. 2008; Rouzine et al. 2008; Greulich et al. 2012) and is also important in other fields, including applied mathematics (McKean 1975; Merkin and Needham 1989; Merkin et al. 1993; van Saarloos 2003), statistical physics (Derrida and Spohn 1988; Brunet and Derrida 1997), and computer science (Majumdar and Krapivsky 2002, 2003). The F–KPP equation predicts that the population advances as a traveling wave with a well-defined speed given by for a wide class of initial conditions. This wave speed is determined by a mathematically subtle speed selection principle that depends critically on the population dynamics at the very tip of the wave, as well as the initial condition, and that has been a topic of discussion for more than half a century (Kolmogorov et al. 1937; McKean 1975; Larson 1978; van Saarloos 2003).
Spatial expansion can have important effects on the genetic structure of a population. In particular, range expansions are often associated with genetic bottlenecks, in which the population of the newly colonized territory is descended from only a few “pioneer” individuals. Recent work (Klopfstein et al. 2006; Hallatschek et al. 2007; Excoffier and Ray 2008; Hallatschek and Nelson 2008, 2009; Excoffier et al. 2009; Korolev et al. 2010) has focused on the amplification of genetic drift at the low-density fronts of expanding populations, strikingly demonstrated in experiments with neutral, fluorescently labeled strains of bacteria and yeast (Hallatschek et al. 2007; Hallatschek and Nelson 2009). Range expansion may also favor the maintenance of cooperative traits, both by enrichment of cooperators at the front and by allowing them to outrun noncooperative cheats (Sen Datta et al. 2013).
Population expansions are often accompanied by the spread of nonselected traits that hitchhike with those that are advantageous. These traits may be genetic variants (alleles) that are vertically transmitted and can be gained and lost by mutation or recombination (Maynard Smith and Haigh 1974; Barton 1998, 2000; Etheridge et al. 2006). However, hitchhiking traits can also be infections, cultural variants, or genetic elements that are transmitted horizontally between individuals (Fagan et al. 2002; Bar-David et al. 2006; Ackland et al. 2007). Important examples include parasites carried by an invading population, which may have catastrophic consequences for the native species (Prenter et al. 2004; Bar-David et al. 2006) or in some cases be used as a means to control the invaders (Fagan et al. 2002), and horizontal gene transfer within spatially structured bacterial communities, which presents dangers for the spread of antibiotic resistance, but also opportunities for bioremediation (Molin and Tolker-Nielsen 2003; Fox et al. 2008).
The classic case of genetic hitchhiking for vertically transmitted alleles in well-mixed populations has been the topic of a large body of theory (Maynard Smith and Haigh 1974; Barton 1998, 2000; Etheridge et al. 2006). Recently, extensions of this work have shown that spatial structure can have nontrivial effects, typically decreasing the frequency of global selective sweeps that lead to hitchhiking but also possibly favoring hitchhiking by allowing local sweeps (Barton 2000; Barton et al. 2013). For horizontally transmitted traits, the focus of studies so far has mainly been on maintenance of directly selected (or neutral) traits within well-mixed (Lipsitch et al. 1995; Bergstrom et al. 2000) or spatially structured (Krone et al. 2007; Court et al. 2013) populations. The situation in which a horizontally transmitted trait spreads in a population by hitchhiking on selection for a different trait has been addressed in specific ecological and cultural contexts (Fagan et al. 2002; Bar-David et al. 2006; Ackland et al. 2007), but baseline theoretical results remain lacking.
In this article, we present a baseline model for the spread of a horizontally transmitted hitchhiking trait within the wave of advance of an expanding population. Extending Fisher–KPP wave theory to model the coupled waves of advance of the population and the hitchhiking trait, we uncover a new speed selection mechanism that leads to a nontrivial result (our Equation 7) for the speed of advance of the hitchhiking trait. We find that population expansion can significantly affect the speed at which a trait spreads within the population. Our analysis also reveals the existence of abrupt, initial-condition-dependent transitions in the speed of waves of invading traits.
Background: Fisher–KPP Wave Theory
We begin by reviewing the main results of F–KPP theory for expanding populations. The standard F–KPP equation (Equation 1) has traveling wave solutions of the form u(x, t) = U(x − vt). At large times the population asymptotically expands in a wave of constant shape that moves at a constant speed v. A detailed analysis (Kolmogorov et al. 1937; McKean 1975; Larson 1978) reveals that the wave speed v is governed by a simple selection principle that forms the basis of F–KPP wave theory: if the initial profile decays more steeply than , where λ* = v*/2D, then ; whereas if u(x, 0) decays less steeply than , then v > v*.
For future reference we review how this result can be understood in a simple way by examining the dynamics of the leading edge of the wave where u ≪ 1 (Figure 1). Then the linearized version of (1),
| (2) |
exhibits traveling wave solutions with possible velocities v ≥ v*. The full time-dependent solution of this linearized equation beginning from exponentially decaying initial conditions u(x, t = 0) = exp(−λx) for x > 0 can be constructed (van Saarloos 2003). The solution reveals two different, large-time asymptotic regimes (see supporting information, File S1): the front of the wave (Figure 1A) travels with the marginal speed v*; whereas the tip of the wave travels with a higher speed v(λ) = Dλ + αK/λ determined by the initial profile. If λ > λ* = v*/(2D), the crossover point between the two regimes moves faster than the front and asymptotically the wave travels with speed v*. However, if λ < λ*, the front catches up with the crossover point and the wave speed is then determined by v(λ) > v*. Thus for suitably steep initial conditions the marginal speed v* is selected. The fact that the behavior of the wave at its very tip is crucial in determining its speed has important implications for systems with noise or discrete cutoffs at the tip (Brunet and Derrida 1997, 2001; Hallatschek 2009, 2011).
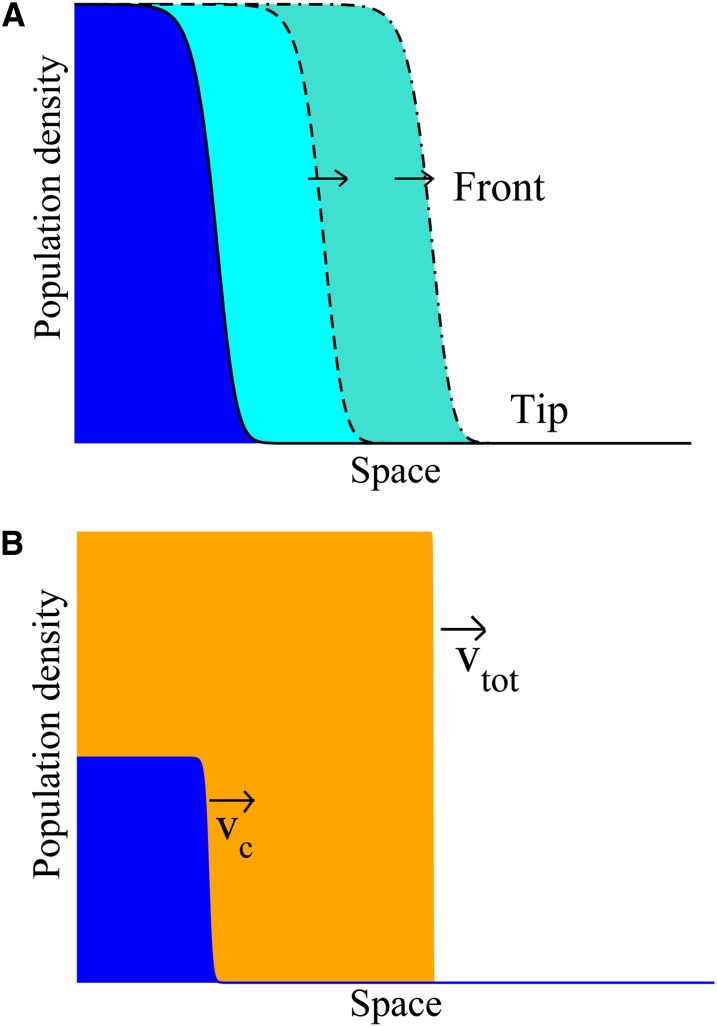
Figure 1.
Propagation of single and coupled F–KPP waves. (A) A population described by the F–KPP equation advances as a traveling wave. The speed of the main part of the wave (the “front”) depends critically on the details of the profile in the tip region, far ahead of the front. (B) The scenario investigated in this study. The subpopulations of individuals with and without the horizontally transmitted trait are shown in blue and orange, respectively. The total population advances as an F–KPP wave, with speed , while the subpopulation with the trait also advances as a wave, which lags behind the wave of the total population.
Hitchhiking on a Wave of Advance: Coupled F–KPP Equations
To model the spread of a horizontally transmitted hitchhiking trait within an expanding population, we extend the F–KPP framework to consider the dynamics of two subpopulations: subpopulation A, which carries the trait, and subpopulation B, which does not. Both subpopulations diffuse in space and replicate, competing for resources. To model horizontal transmission, we suppose that contact between individuals of types A and B can result in both individuals becoming type A. The trait is also lost at a constant rate. Because our aim is to present a baseline model, we assume that there are no fitness differences between the two subpopulations, i.e., that they have equal diffusion constants, growth rates, and carrying capacities. Our model is described by the following set of equations,
| (3) |
where NA(x, t) and NB(x, t) are the densities of the subpopulations with and without the trait; NT = NA + NB is the total population density; and D, K, and α are defined as before. The new parameters γ and β control the rates of horizontal transmission and loss of the trait, respectively; these processes exchange individuals between the two subpopulations.
There are three homogeneous density, steady-state solutions of Equations 3 that in terms of (NT, NA) read (0, 0) (no population), (K, 0) (population contains no trait carriers), and (K, K − β/γ) (coexistence of trait and nontrait subpopulations). The last solution is physical (i.e., has positive densities) only if γ ≥ β/K, i.e., if the rate of horizontal transmission is high enough that the trait can be sustained in the population. In addition to this condition on γ we also restrict the parameter values to γ < α so that
| (4) |
The range (4) corresponds to an intermediate transmission rate of the trait, relevant, for example, to the spread of chronic diseases in a colonizing population (Bar-David et al. 2006).
Summing the two Equations 3 gives a standard F–KPP equation,
| (5) |
and thus the total population advances as a wave, at constant speed , determined by the F–KPP speed selection principle (as long as the initial condition decays steeply enough).
To determine the qualitative phenomenology of our model we studied Equations 3 numerically (see File S1 for details). Our simulations (Figure 1B) reveal that the subpopulation of individuals carrying the trait also advances as a traveling wave (which we denote the “trait wave”), but it has a slower speed than that of the total population. Thus the trait spreads in space as the population advances, but it lags behind the advancing population. In this article, we analyze the speed of advance of this trait wave and show that it is controlled by an intricate coupling between the population densities of the two subpopulations at the very tips of the two waves.
An analysis of the linear stability (see File S1) of the three homogeneous fixed point solutions (NT, NA) reveals that under conditions (4), (0, 0) is unstable, (K, 0) is a saddle point (one stable and one unstable direction), and (K, K − β/γ) is stable. Thus the observed coupled traveling wave solutions correspond to solution (0, 0) being invaded by solution (K, 0) (population without trait) and, in turn, solution (K, 0) being invaded by solution (K, K − β/γ) (the coexisting state).
Spread of a trait in an established population
As a point of comparison, we begin with the well-studied case where a horizontally transmitted, neutral, trait invades an already established population. In this case, the total population NT is equal to the carrying capacity K throughout the domain. Setting NT = K in Equation 3 leads to a single F–KPP equation for the spread of the trait:
| (6) |
Thus, the trait invades an existing population as a traveling wave with amplitude K − β/γ and speed predicted by the F–KPP speed selection principle (assuming a sufficiently steep initial condition for subpopulation A). Figure 2 (top row) shows numerical simulation results for this scenario.
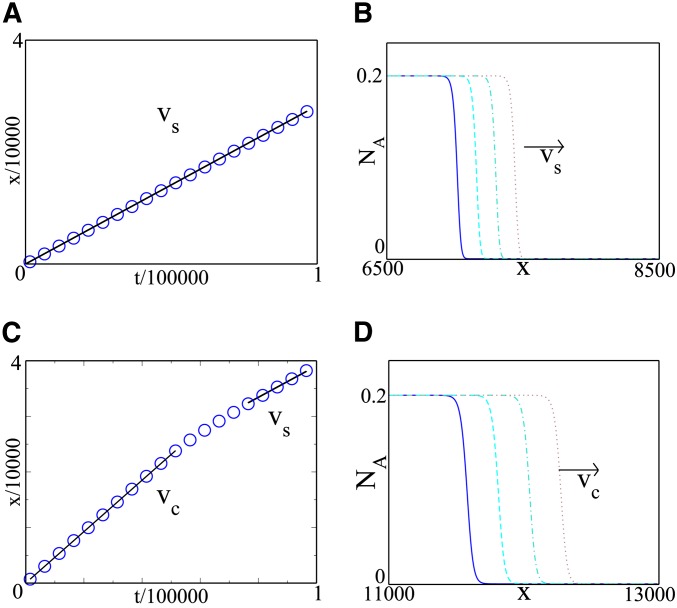
Figure 2.
Invasion of the trait wave into established and expanding populations. The top row (A,B) shows numerical simulation results (see File S1) for a trait that invades an already established population, i.e., with initial condition NB = NT = K for x > 0, while the bottom row (C,D) shows results for invasion of an expanding population, i.e., with initial condition NB = NT = 0 for x > 0. The simulation parameters are identical in the two simulations (K = 1, D = 1, α = 1, β = 0.08, and γ = 0.1). In each case, the right panel (B,D) shows wave profiles at several different times (the times plotted are the same in the top and bottom panels), while the left panel (A,C) tracks the position of the wave front as a function of time. The trait wave invades the established population as an F–KPP wave with speed vs. It also invades the expanding population as a wave, but with a faster speed vc, which eventually crosses over to vs.
The trait spreads faster in an expanding population
We now use our model to investigate what happens when a horizontally transmitted trait invades a population as it expands (Figure 1B). Figure 2 (bottom row) shows numerical simulation results starting with the spatial domain initially empty. After an initial transient (not visible in Figure 2), the trait advances as a traveling wave whose front moves at a speed vc that is greater than the speed vs at which it invades an already established population—but still smaller than the speed vtot of the total population wave. This rapid rate of invasion is maintained for a long time [many times the generation time, given by ∼(αK)−1]. At very long times, when the total population wave is very far ahead of the trait wave, the speed of the trait wave reverts to vs (Figure 2).
Distinctive features of the trait wave
Based on our understanding of the standard F–KPP wave, we expect the speed of the trait wave to be determined by the dynamics close to the tip. We therefore make a detailed study of this region. Figure 3A shows the profiles of the total population wave and the trait wave, during the time period when the front of the trait wave is moving forward at speed vc.
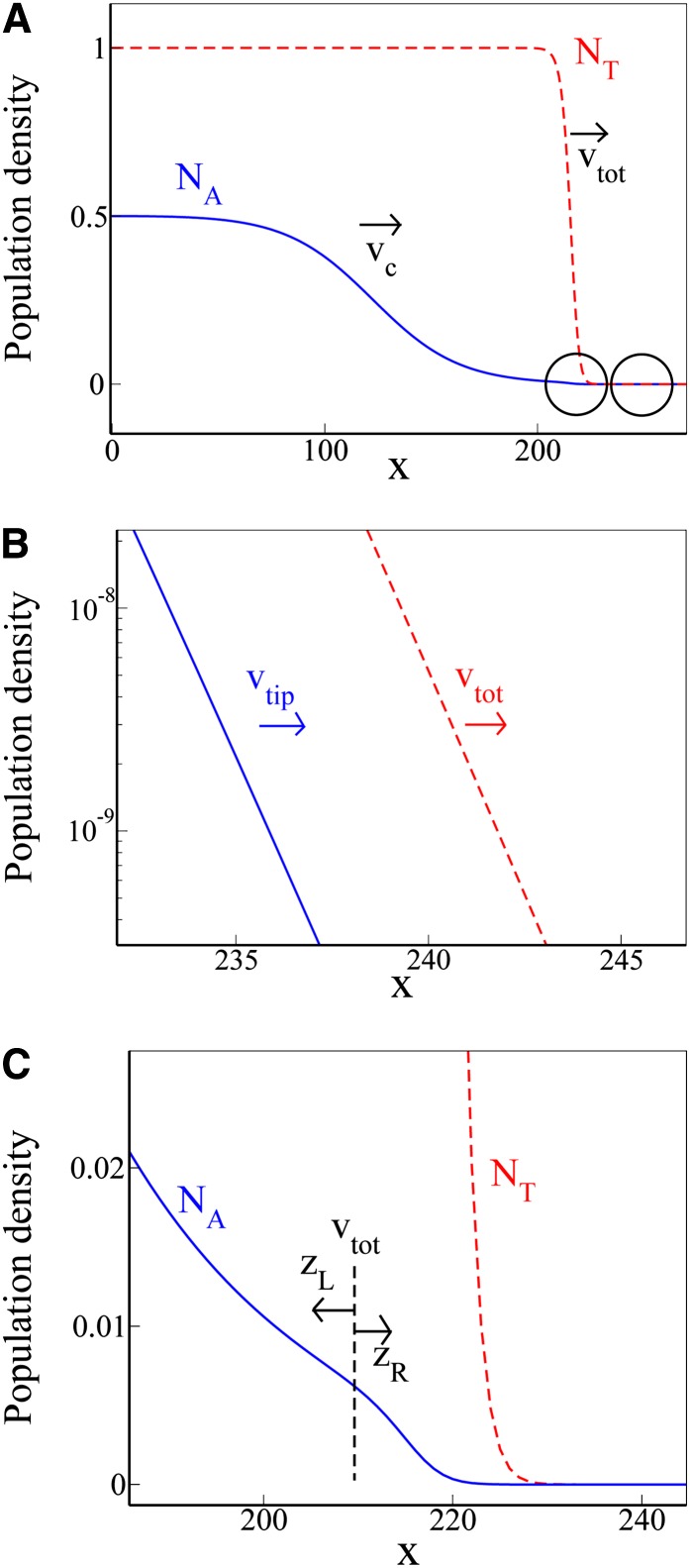
Figure 3.
Detailed features of the profile of the trait wave. (A) Profiles of the trait wave (blue) and the total population wave (red, dashed), during the time when the trait wave is advancing at speed νc. (B) Zoom-in on the region at the very tip of the two waves (indicated by the rightmost circle in A). Here both profiles decay exponentially (note the log scale on the vertical axis), and the tip of the trait wave advances at speed . (C) Zoom-in on the region of space corresponding to the front of the total population wave (indicated by the left circle in A), where the trait wave population density is still low. A distinct kink is observed in the trait wave profile NA(x), due to coupling with the total population wave. This kink advances at the speed of the front of the total population wave . The trait population density at the kink decreases in time, as the trait wave front falls behind the total population wave front. C also illustrates the change of coordinate system used to match the asymptotic solutions to the left and right of the kink.
We first zoom in on the region ahead of the total population wave, as indicated by the rightmost circle in Figure 3A. Figure 3B shows that the profiles of both the total population and the subpopulation with the trait decay exponentially (note the logarithmic scale on the vertical axis). In this tip region, we can make an analytical prediction for the speed vtip at which the tip of the trait wave advances. Because both NA and NB are very small, with NA ≪ NT, we can linearize Equation 3 to give ∂NA/∂t = D(∂2NA/∂x2) + NA(αK − β). Then the standard speed selection principle outlined earlier implies that the marginal speed is selected and NA(x) ∼ exp[−vtip(x − vtipt)/2D]. This prediction is verified by tracking the speed of the very tip of the trait wave in our numerical simulations. Our simulations also show that vtip > vc—thus, as for the standard F–KPP wave, the tip of the trait wave advances at a faster speed than its front.
Next, we inspect the trait wave farther back in its profile, at the point where it overlaps with the front of the total population wave. This point, shown by the leftmost circle in Figure 3A, lies well ahead of the front of the trait wave, so that the density of the subpopulation with the trait is still very small. Close inspection of our numerical simulations reveals that the trait wave profile has a distinctive “kink” that coincides with the front of the total population wave. This kink, shown in Figure 3C, advances with the front of the total population wave: i.e., its speed is and its position at time t is x* = vtott. As the trait wave front falls behind the total population wave front, the trait population density at the kink decreases.
To summarize, while the trait advances as a traveling wave with speed vc, the dynamics at its tip, where the population density is very small, are rather complex. The tip itself moves forward at speed , but behind the tip the trait wave profile has a kink created by coupling to the front of the total population wave; this kink advances at speed . The relative magnitudes of these speeds are vtot > vtip > vc.
Speed Selection Mechanism for the Trait Wave
We can obtain an analytic expression for the speed vc at which the trait invades an expanding population, by matching asymptotic expansions for the profile of the trait wave on either side of the kink.
Ahead of the kink, we define a coordinate zR = x − vtott, which measures the distance from the kink (Figure 3C). Following the result of our earlier analysis of the tip of the trait wave, we expect the profile in this region to decay as NA(zR, t) ∼ exp[−vtip(zR + (vtot − vtip)t)/(2D)].
Behind the kink, the trait wave advances at speed vc. We expect the trait wave profile to increase exponentially with distance behind the kink, so we write NA(zL, t) ∼ exp[−at + bzL], where zL = vtott − x measures the distance from the kink and a and b are unknown constants such that vc = vtot − a/b. By demanding that our two asymptotic expressions must match at the kink, i.e., NA(zR = 0, t) = NA(zL = 0, t), we can determine the constant a = vtip(vtot − vtip)/(2D). To find the remaining constant b, we linearize Equation 3 in the region behind the kink, where the total population is large (NT ≈ K), but the amplitude of the trait wave is still small (NA ≪ 1). This gives ∂NA/∂t = D(∂2NA/∂x2) + NA(γK − β). Substituting in our exponential ansatz for NA(zL, t) gives . For the inside of the square root to be positive, we need the second condition of (4), α > γ.
This calculation results in two solutions for the speed vc of the trait wave, corresponding to the positive and negative square roots in the expression for b. It turns out that the positive square root gives a speed vc that is greater than the speed of the tip vtip, so we discard that solution. Taking the negative square root, we arrive at the following expression for the speed of the trait wave,
| (7) |
which can be written in terms of the parameters of the model as
| (8) |
Figure 4 shows that this prediction is in excellent agreement with our simulation results.
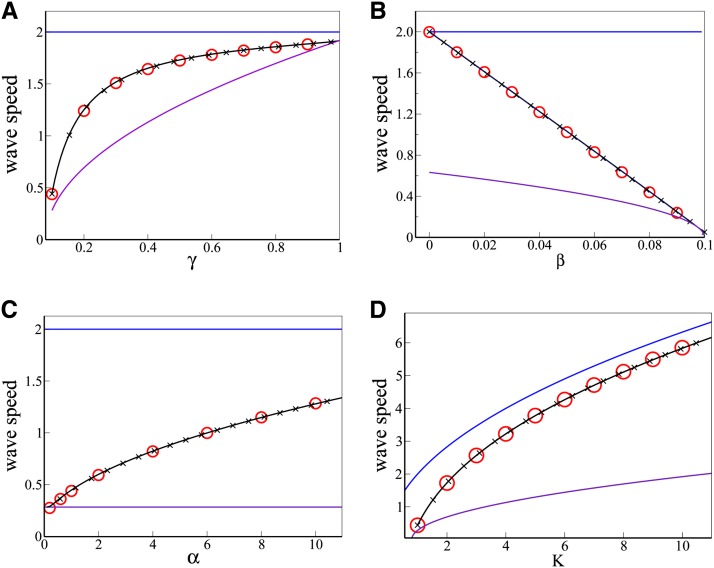
Figure 4.
Analytical prediction, and simulation results, for the speed vc of the trait wave as it invades an expanding population. (A) wave speed versus γ (B) wave speed versus β (C) wave speed versus α (D) wave speed versus K. The black lines show the analytical result, Equation 8, while the red circles show simulation results for the trait wave speed and the black crosses show the approximate result, Equation 9. Except where otherwise indicated on the horizontal axis, the parameters are D = α = K = 1.0, γ = 0.1, and β = 0.08. For comparison, the blue lines show the speed of the total population wave , while the purple lines show the speed of the trait wave as it invades an established population, .
The implications of our result can be understood by noting that in many scenarios we expect that β ≪ αK. This can occur either through a low trait loss rate β or through a high growth rate or carrying capacity (α or K). Expanding to first order in β/(αK) leads to a simple expression for the speed of invasion of the trait:
| (9) |
Equation 9 demonstrates that traits that are poorly transmitted (small γ) or easily lost (large β) will lag farther behind the main population wave. Importantly, our result also predicts that as the carrying capacity of the population increases, the relative amount by which the trait lags behind the main population wave should decrease. This implies that horizontally transmitted hitchhiking traits (such as parasites or horizontally transmitted genetic elements) should be found relatively closer to the advancing front in populations with a high carrying capacity, compared to those with a lower carrying capacity.
Our analysis also provides a simple prediction for how much the invasion by the trait is speeded up in an advancing population compared to an established one—i.e., the ratio vc/vs. Expanding for large K (K ≫ β/γ and γ < α) we find that
| (10) |
For large carrying capacity K, relation (10) reduces to vc/vs ≈ (α/γ)1/2. Thus we expect the spread of a trait in an expanding population to be significantly faster than in an existing population if the birth rate is high and the transmission rate of the trait is low.
Speed Transitions
Interestingly, our simulations also show that horizontally transmitted hitchhiking traits can undergo abrupt transitions in wave speed (see, e.g., Figure 2, bottom row). These transitions have their origin in the intricate coupling between the trait wave and the total population wave in their tip regions.
Slowing down transition due to wave decoupling
Although the trait wave initially advances at speed vc, Figure 2 shows that eventually it undergoes a slowing-down transition to final speed vs (the speed at which it would invade an established population). Careful inspection of our simulations reveals that this transition occurs when the kink in the trait wave profile (Figure 3C) overtakes its tip—defining the tip as the point at which the density of the trait becomes unresolvable in our numerical simulations. Such a transition is inevitable since the kink advances faster than the tip [the tip moves at while the kink advances at ]. When the kink overtakes the tip, the two waves become decoupled at the level of precision of our simulations, and the trait wave behaves as if it were invading an already established population. The time at which this transition happens will of course depend on the details of the initial conditions for the simulations and on the level of resolution of the tip. However, we can predict that this time will scale inversely with the difference in these two speeds, which, for β/(αK) ≪ 1, is ∼.
Initial condition-dependent speeding-up transition
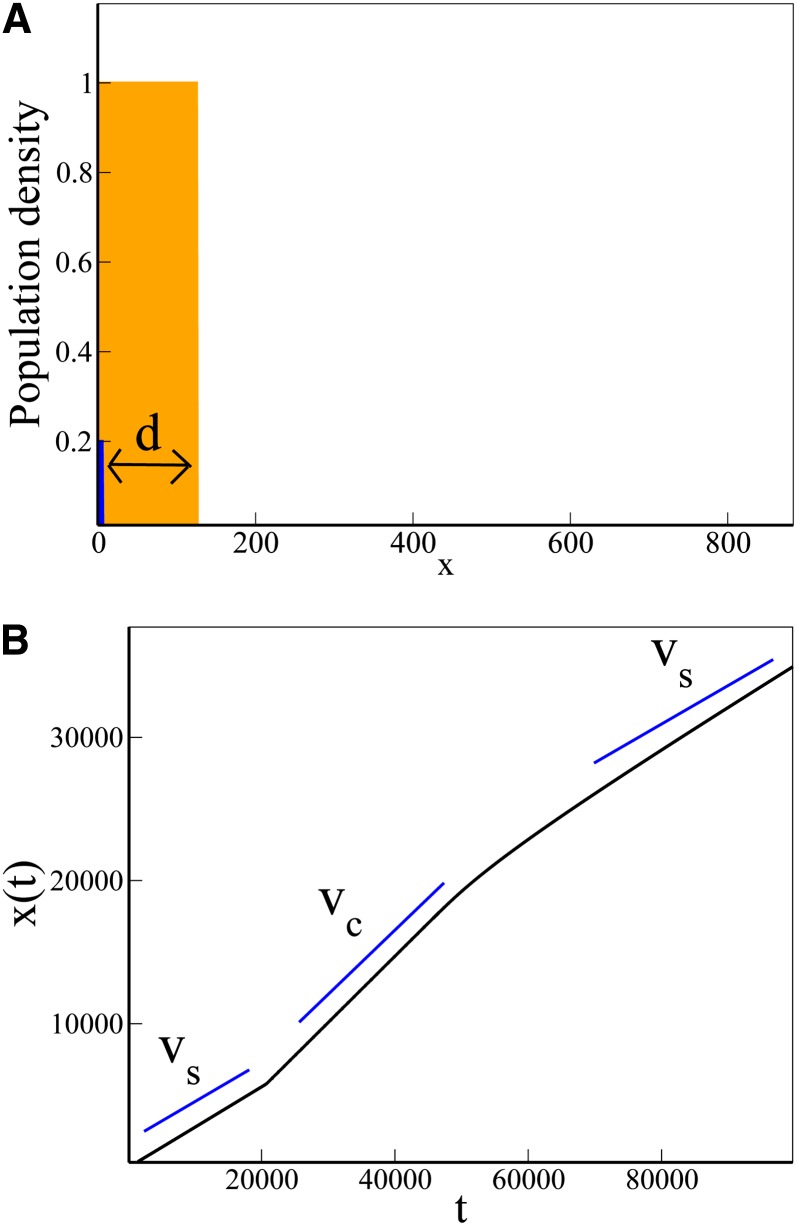
Our simulations also reveal that under some circumstances the trait wave can undergo abrupt speeding-up transitions. Figure 5 shows an example. Here, the spatial domain is partially colonized at the start of the simulation; the region 0 < x < d is occupied by individuals that do not carry the trait, while the rest of the domain is empty (Figure 5A). During the simulation, the population expands to fill the rest of the domain, while at the same time the trait invades the expanding population.
Figure 5.
Initial-condition-dependent speeding-up transition. (A) The starting point for a simulation in which the spatial domain is initially partially occupied by individuals that do not carry the trait. Here, the initial condition is NB = NT = K for 0 < x < d and NB = NT = 0 for x > d. The subpopulation density NB(x) is shown in orange; NA(x) is shown in blue. (B) tracks the position of the front of the trait wave as this simulation proceeds. Initially, the trait wave advances at speed (as for invasion of an established population). The wave then undergoes an abrupt transition to the faster wave speed vc (as given in Equation 8). Eventually, the waves become decoupled and the speed reverts to vs.
Figure 5B tracks the advance of the front of the trait wave in this simulation. Initially, the trait wave front advances at speed vs, as if it were invading a fully established population. However, the wave of invasion then makes an abrupt speeding-up transition to the faster speed vc. This transition from vs to vc has its origin in the evolution of the profile of the trait at the start of the simulation. This profile is initially sharp (in our simulations it is a step function). Starting from this steep initial condition, the trait wave develops a tip that, as in standard F–KPP wave theory, advances faster than the wave front (Sherratt 1998a; van Saarloos 2003). Initially this trait wave tip is far behind the front of the total population wave and the trait wave behaves as if it is invading a fully colonized environment, moving at speed vs. However, after some time the tip of the trait wave overtakes the front of the total population wave, and the waves become coupled. At this point, the profile of the trait wave develops a kink, and, following the speed selection mechanism outlined above, its front speed increases to vc. Figure 5B also shows that the wave speed eventually changes back to vs, in a slowing-down transition of the type discussed above.
Are the Observed Phenomena Biologically Relevant?
Our analysis predicts that a horizontally transmitted trait can invade an expanding population significantly faster than it would invade an already established population, but always at a speed slower than that of the expanding population front. Does this prediction still hold for parameter sets corresponding to real biological scenarios? To test this, we take as a model scenario the invasion of a semisolid agar matrix by an expanding population of nonmotile bacteria that undergo horizontal gene transfer by conjugation. This scenario mimicks the contamination of foodstuff by bacteria (Wimpenny et al. 1995; Wilson et al. 2002) and also relates to recent experiments on genetic segregation during range expansion (Hallatschek et al. 2007; Hallatschek and Nelson 2009). The diffusion constant of a nonmotile bacterium such as Escherichia coli in liquid medium is D ≈ 1μm2/sec (Berg 1983), and the carrying capacity in rich medium is K ≈ 1010 cells/ml (which is equivalent to 10−2 cells/μm3). E. coli has a doubling time in rich medium of ∼30 min, so that αK = (ln 2)/30 per minute (or alternatively α = 3.85 × 10−2 μm3⋅cell−1⋅sec−1). The rate of transfer of genetic material (plasmids) by conjugation has been estimated for some strains of E. coli as γ = 10−12 ml⋅cell−1⋅hr−1 (Simonsen et al. 1990) (or equivalently γ = 2.8 × 10−4 μm3⋅cell−1⋅sec−1). Bacterial plasmid loss rates are highly variable (Summers 2009). We choose the rate of loss of the horizontally transmitted trait β = 0.5γK, such that in a steady-state, homogeneous population, half the cells are trait carriers.
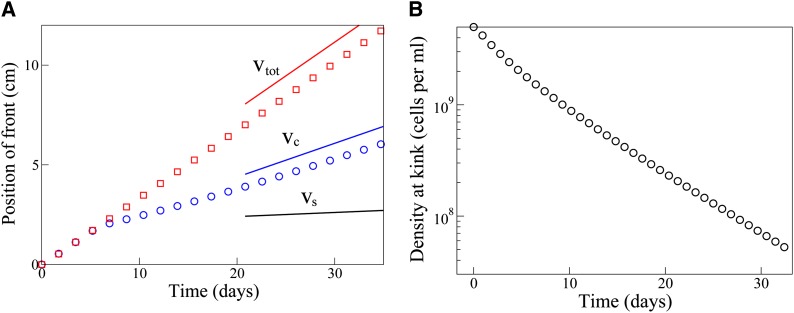
Figure 6A shows the results of numerical simulations, using these parameter values: after an initial transient period, the population advances as two waves, with the wave of trait carriers (blue circles) lagging behind that of the nontrait carriers (red squares). The speed at which the wave of trait-carrying cells invades the advancing population is indeed well predicted by our analytical expression for vc, Equation 8, which, for this parameter set, is greater than the speed vs at which the trait wave would invade an already established population, by a factor of ∼8. Figure 6B shows the population density of trait carriers, at the position of the front of the total population wave—i.e., the height of the kink in the trait wave (see Figure 3C). This decreases in time as the trait wave falls behind the total population wave. However, over the time period of our simulation, the population density at the kink remains significant.
Figure 6.
Numerical simulation results for a parameter set representing the invasion of an agar matrix by a population of nonmotile E. coli bacteria undergoing horizontal gene transfer by plasmid conjugation. The parameters used were D = 1 μm2/sec, K = 1010 cells/ml (equivalent to K = 10−2 cells/μm3), α = 3.85 × 10−2 μm3⋅cell−1⋅sec−1, γ = 2.8 × 10−4 μm3⋅cell−1⋅sec−1, and β = 0.5γK. (A) Trajectories of the front positions of the wave of cells without the trait (red squares) and the trait wave (blue circles). Initially, the spatial domain is empty except at its very edge where the two populations take their steady-state values. At early times, the two waves travel together as the traveling wave profiles are established. Thereafter, the trait wave invades the expanding population at the speed vc predicted by our theory, which is significantly faster than the speed vs with which it would invade an established population (vc, vs, and vtot are indicated by the blue, black, and red solid lines, respectively). (B) Population density of the trait wave at the kink, i.e., at the position of the front of the total population wave (see Figure 3C). The trait population density at the kink decreases in time approximately exponentially as the trait wave falls behind the total population wave, but remains significant over the timescale of our simulation (note the log scale on the vertical axis). For reasons of computational speed, these simulations were carried out using standard Euler integration rather than the operator splitting method used in our other calculations (see File S1).
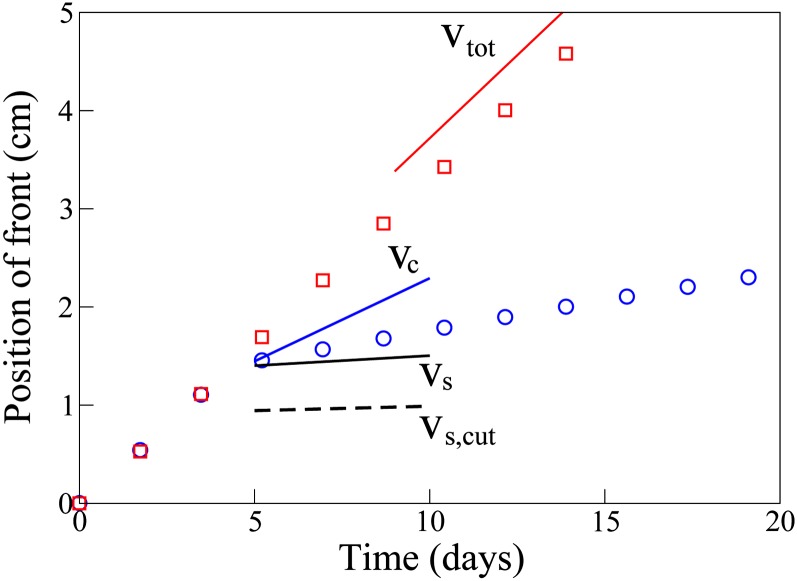
In real populations, individuals are of course discrete entities; this leads to a finite lower cutoff for the population density. The fact that, in our continuous simulations, the population density at the kink remains significant for long times (Figure 6B) suggests that our results may be robust to the effects of such a cutoff. To investigate this in more detail, we repeated the simulations of Figure 6, but introducing a cutoff for both the trait-carrying and the non-trait-carrying subpopulations, at a population density of 1 cell/ml. The cutoff is implemented by setting a density to zero when it becomes smaller than the threshold 1 cell/ml. Figure 7 shows the resulting trajectories for the waves of the two subpopulations. As well as the theoretical speeds vc, vs, and vtot, we also indicate the speed vs,cut at which the trait population invades an already established population, in simulations with the cutoff. Measuring the speed of the trait wave at time 7 days, we find that the trait invades the expanding population 7.2 times faster than it invades the established population. The key prediction of our theory thus still holds in the presence of the cutoff. Figure 7 also shows, however, that the actual magnitude of the trait wave speed is lower than vc in the simulations with the cutoff—suggesting that corrections to our theory will be need to account quantitatively for the effects of population discreteness.
Figure 7.
The effect of a discrete cutoff, for simulations of the invasion of an agar matrix by nonmotile E. coli bacteria undergoing horizontal gene transfer by plasmid conjugation. The parameters used were as in Figure 6, but with the addition of a cutoff in population density at 1 cell/ml = 10−12 cells/μm3. In our simulations, subpopulation densities smaller than this value were set to zero. Shown are the trajectories of the front positions of the wave of cells without the trait (red squares) and the trait wave (blue circles). Initially, the spatial domain is empty except at its very edge where the two populations take their steady-state values. After the initial transient (during which the steady-state wave profiles are established), the trait wave invades the expanding population at a speed faster than the speed vs for invasion of an established population but slower than the speed vc given by (7). Also indicated is the speed νs,cut at which the trait invades an established population in simulations with the same cutoff; this corresponds closely with vs. As in Figure 6, these simulations were carried out using standard Euler integration.
Discussion
The spread of a population in space, and the accompanying spread of genetic, ecological, and cultural traits, is a ubiquitous biological phenomenon; correspondingly, the speed selection principle of F–KPP wave theory, which describes the spatial advance of population waves, is one of the most important results in mathematical biology. In this article, we have presented a baseline model, consisting of coupled F–KPP waves, to describe the spread of a horizontally transmitted trait within an advancing population. Our results reveal a new selection mechanism that controls the speed at which the trait wave invades the expanding population. We have derived an analytic expression (Equation 7) for the speed of the trait wave, which reduces to a simple form in the biologically relevant case where the carrying capacity is large. We find that under some circumstances, the trait can invade an expanding population significantly faster than it would spread in a population that is already established. The mechanism underlying this speedup is that the front of the total population wave creates a kink in the tip region trait wave profile, which couples the two waves at their tips; matching asymptotic solutions on either side of the kink leads to the speed selection mechanism. We also reveal the possibility of abrupt transitions in speed in coupled waves of advance. For a parameter set corresponding to invasion of a semisolid matrix by a population of E. coli bacteria undergoing horizontal gene transfer by conjugation, our theory and simulations suggest that invasion of an expanding population by a horizontally transmitted gene may occur about eight times faster than invasion of an already established population. While this is a crude model [for example, in reality the rate of conjugation may depend on the growth rate (Merkey et al. 2011)], it does indicate that our results may indeed be biologically relevant.
An important consideration is the robustness of our results to noise. In real populations, stochastic fluctuations due to births and deaths of individual organisms are inevitable and can play an important role at the tips of F–KPP waves where population densities are low (Brunet and Derrida 1997, 2001; Hallatschek 2009). For standard F–KPP waves, finite size effects at the tip of the wave are known to cause a significant correction to the wave speed (Brunet and Derrida 1997). In our model, the kink in the trait wave (which plays a key role in determining its speed) occurs behind the tip, but in the region where the population density of individuals with the trait is still low. In our simulations in Figure 6, the trait population density at the kink remains significant over long times, suggesting that the kink, and the consequent coupling between the waves, would probably survive in the presence of weak noise. This conclusion is also borne out by the fact that we still observe faster invasion of an expanding population than an established one when we include a discrete cutoff (Figure 7). However, the cutoff does significantly affect the speed of the trait wave; likewise in the presence of noise, we would expect the correction of the total population wave speed to carry through into the speed of the trait wave.
To fully study stochastic effects within a reproducing population one would have to introduce an individual-based computer simulation model. Several stochastic systems designed to model the effects of noise on the F–KPP equation have been considered (Breuer et al. 1995; Riordan et al. 1995; Brunet and Derrida 1997, 2001); however, these do not directly simulate a reproducing population. Instead a reaction diffusion scheme of the type A + A ⇋ A is generally used, and it may be shown that these systems are related by a duality property to the stochastic F–KPP equation (Doering et al. 2003). Large-scale individual-based studies of population dynamics described by stochastic Fisher waves have, to our knowledge, rarely been attempted. One example of an individual-based stochastic algorithm is that used to simulate the population dynamics of sedimenting bacteria (Barrett-Freeman et al. 2008). The algorithm is defined on discrete spatial sites, labeled by i, each carrying an integer number ni of individuals. The results were shown to agree qualitatively with a F–KPP equation (with an additional advective term to model gravity). It would be important to make a systematic study of the effects of noise via individual-based algorithms such as the one just described, for the system of coupled F–KPP equations that we have studied in the present work. We hope to pursue this direction in the future.
Coupled systems of reaction–diffusion equations provide a rich source of interesting dynamical behavior, from models for infection dynamics, through spatially coupled autocatalytic chemical reaction systems (Merkin et al. 1993), to advancing fronts of oscillatory predator–prey systems (Sherratt 1998b; Sherratt et al. 2009). From a mathematical perspective, for systems of coupled F–KPP waves, the phenomenon of “anomalous spreading,” in which coupling between two populations influences the wave speeds, has been recognized in several examples of systems of coupled F–KPP equations (Weinberger et al. 2007; Holzer and Scheel 2012; Holzer 2012). These examples are more complex than the model studied here, in that the diffusion and growth parameters for the two populations are not identical, and the coupling terms are not symmetric. Our study therefore provides a baseline for understanding speed selection in coupled F–KPP waves in general. The asymptotic matching approach presented here should prove useful in understanding these more complex models. As a first step toward introducing the effects of selection for or against the horizontally transmitted trait, we have simulated a version of our model in which we allow the growth rate α to differ between the two subpopulations. We find that for small growth-rate differences the qualitative results described here remain unchanged (see File S1). Another interesting extension would be to the case where the external environment is spatially heterogeneous; here the range of the “host” population is limited, but may be extended by mutation (Holt and Gomulkiewicz 1997; Kirkpatrick and Barton 1997; Waclaw et al. 2010; Greulich et al. 2012) or, potentially, by selection for horizontally transmitted traits.
In conclusion, understanding existing spatial patterns of genetic, ecological, or cultural traits and predicting and controlling the consequences of future population expansions are important goals for both evolution and ecology. Many of these expansions involve the hitchhiking of horizontally transmitted traits. The model presented here, while clearly simplistic, reveals important phenomena associated with the spread of traits within expanding population waves, and with coupled systems of F–KPP waves in general, and should provide a basis on which to build more complex and detailed models.
Supplementary Material
Acknowledgments
The authors thank Davide Marenduzzo for advice on simulation methodology and Jonathan Sherratt for helpful discussions. J.V.-O. was funded by a University of Edinburgh College Studentship. R.J.A. was funded by a Royal Society University Research Fellowship and by a Royal Society research grant. This work was funded in part by the Engineering and Physical Sciences Research Council under grant EP/J007404/1.
Footnotes
Communicating editor: J. Hermisson
Literature Cited
- Ackland G. J., Signitzer M., Stratford K., Cohen M. H., 2007. Cultural hitchhiking on the wave of advance of beneficial technologies. Proc. Natl. Acad. Sci. USA 104: 8714–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman A. J., Cavalli-Sforza L. L., 1971. Measuring the rate of spread of early farming in Europe. Man 6: 674–688. [Google Scholar]
- Barrett-Freeman C., Evans M. R., Marenduzzo D., Poon W. C. K., 2008. Nonequilibrium phase transition in the sedimentation of reproducing particles. Phys. Rev. Lett. 101: 100602. [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Barton N. H., 2000. Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-David S., Lloyd-Smith J. O., Getz W. M., 2006. Dynamics and management of infectious disease in colonizing populations. Ecology 87: 1215–1224. [DOI] [PubMed] [Google Scholar]
- Barton N. H., Etheridge A. M., Kelleher J., Véber A., 2013. Genetic hitchhiking in spatially extended populations. Theor. Popul. Biol. 87: 75–89. [DOI] [PubMed] [Google Scholar]
- Berg H. C., 1983. Random Walks in Biology. Princeton University Press, Princeton, NJ. [Google Scholar]
- Bergstrom C. T., Lipsitch M., Levin B. R., 2000. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155: 1505–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer H.-P., Huber W., Petruccione F., 1995. The macroscopic limit in a stochastic reaction-diffusion process. Europhys. Lett. 30: 69. [Google Scholar]
- Brunet E., Derrida B., 1997. Shift in the velocity of a front due to a cutoff. Phys. Rev. E 56: 2597–2604. [Google Scholar]
- Brunet E., Derrida B., 2001. Effect of microscopic noise on front propagation. J. Stat. Phys. 103: 269–282. [Google Scholar]
- Court S. J., Blythe R. A., Allen R. J., 2013. Parasites on parasites: coupled fluctuations in stacked contact processes. EPL 101: 50001. [Google Scholar]
- Derrida B., Spohn H., 1988. Polymers on disordered trees, spin glasses and traveling waves. J. Stat. Phys. 51: 817–840. [Google Scholar]
- Doering C. R., Mueller C., Smereka P., 2003. Interacting particles, the stochastic Fisher-Kolmogorov-Petrovsky-Piscounov equation, and duality. Physica A 325: 243–259. [Google Scholar]
- Etheridge A., Pfaffelhuber P., Wakolbinger A., 2006. An approximate sampling formula under genetic hitchhiking. Ann. Appl. Probab. 16: 685–729. [Google Scholar]
- Excoffier L., Ray N., 2008. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 23: 347–351. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Foll M., Petit R. J., 2009. Genetic consequences of range expansions. Rev. Ecol. Evol. Syst. 40: 481–501. [Google Scholar]
- Fagan W. F., Lewis M. A., Neubert M. G., van den Driessche P., 2002. Invasion theory and biological control. Ecol. Lett. 5: 148–157. [Google Scholar]
- Fisher R., 1937. The wave of advance of advantageous gene. Ann. Eugen. 7: 353–369. [Google Scholar]
- Fox R., Zhong X., Krone S. M., Top E. M., 2008. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2: 1024–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich P., Waclaw B., Allen R. J., 2012. Mutational pathway determines whether drug gradients accelerate evolution of drug-resistant cells. Phys. Rev. Lett. 109: 088101. [DOI] [PubMed] [Google Scholar]
- Hallatschek O., 2009. Fisher waves in the strong noise limit. Phys. Rev. Lett. 103: 108103. [DOI] [PubMed] [Google Scholar]
- Hallatschek O., 2011. The noisy edge of traveling waves. Proc. Natl. Acad. Sci. USA 108: 1783–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O., Nelson D. R., 2008. Gene surfing in expanding populations. Theor. Popul. Biol. 73: 158–170. [DOI] [PubMed] [Google Scholar]
- Hallatschek O., Nelson D. R., 2009. Life at the front of an expanding population. Evolution 64: 193–206. [DOI] [PubMed] [Google Scholar]
- Hallatschek O., Hersen P., Ramanathan S., Nelson D. R., 2007. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl. Acad. Sci. USA 104: 19926–19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hethcote H. W., 2000. The mathematics of infectious diseases. SIAM Rev. 42: 599–653. [Google Scholar]
- Holt R. D., Gomulkiewicz R., 1997. The influence of immigration on local adaptation: a re-examination of a familiar paradigm. Am. Nat. 149: 563–572. [Google Scholar]
- Holzer, M., 2012 Anomalous spreading in a system of coupled Fisher-KPP equations. Physica D (in press). [Google Scholar]
- Holzer, M., and A. Scheel, 2012 Accelerated fronts in a two stage invasion process. SIAM J. Math. Anal. (in press). [Google Scholar]
- Kirkpatrick M., Barton N. H., 1997. Evolution of a species’ range. Am. Nat. 150: 1–23. [DOI] [PubMed] [Google Scholar]
- Klopfstein S., Currat M., Excoffier L., 2006. The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23: 482–490. [DOI] [PubMed] [Google Scholar]
- Kolmogorov A., Petrovsky I., Piskunov N., 1937. Study of the diffusion equation with growth of the quantity of matter and its application to a biological problem. Bull. Univ. Moscow Ser. Int. Sec. A 1: 1–25. [Google Scholar]
- Korolev K. S., Avlund M., Hallatschek O., Nelson D. R., 2010. Genetic demixing and evolution in linear stepping stone models. Rev. Mod. Phys. 82: 1691–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone S. M., Lu R., Fox R., Suzuki H., Top E. M., 2007. Modeling the spatial dynamics of plasmid transfer and persistence. Microbiology 153: 2803–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D. A., 1978. Transient bounds and time-asymptotic behaviour of solutions of nonlinear equations of Fisher type. SIAM J. Appl. Math. 34: 93–103. [Google Scholar]
- Lipsitch M., Nowak M., Ebert D., May R., 1995. The population dynamics of vertically and horizontally transmitted parasites. Proc. R. Soc. Lond. B. 260: 321–327. [DOI] [PubMed] [Google Scholar]
- Majumdar S. N., Krapivsky P. L., 2002. Extreme value statistics and traveling fronts: application to computer science. Phys. Rev. E. 65: 036127. [DOI] [PubMed] [Google Scholar]
- Majumdar S. N., Krapivsky P. L., 2003. Extreme value statistics and traveling fronts: various applications. Physica A 318: 161–170. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McKean H. P., 1975. Application of Brownian motion to the equation of Kolmogorov-Petrovskii-Piskunov. Commun. Pure Appl. Math. 28: 323–331. [Google Scholar]
- Merkey B. V., Lardon L. A., Seoane J. M., Kreft J. U., Smets B. F., 2011. Growth dependence of conjugation explains limited plasmid invasion in biofilms: an individual-based modeling study. Environ. Microbiol. 13: 2435–2452. [DOI] [PubMed] [Google Scholar]
- Merkin J. H., Needham D. J., 1989. Propagating reaction-diffusion waves in a simple isothermal quadratic autocatalytic chemical system. J. Eng. Math. 23: 343–356. [Google Scholar]
- Merkin J. H., Needham D. J., Scott S. K., 1993. Coupled reaction-diffusion waves in an isothermal autocatalytic chemical system. IMA J. Appl. Math. 50: 43–76. [Google Scholar]
- Molin S., Tolker-Nielsen T., 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14: 255–261. [DOI] [PubMed] [Google Scholar]
- Mollinson D., 1991. Dependence of epidemic and population velocities on basic parameters. Math. Biosci. 107: 255–287. [DOI] [PubMed] [Google Scholar]
- Murray J. D., 2004. Mathematical Biology, Vol. I Springer-Verlag, New York. [Google Scholar]
- Prenter J., MacNeil C., Dick J. T. A., Dunn A. M., 2004. Roles of parasites in animal invasions. Trends Ecol. Evol. 19: 385–390. [DOI] [PubMed] [Google Scholar]
- Riordan J., Doering C. R., and D. ben-Avraham, 1995. Fluctuations and stability of Fisher waves. Phys. Rev. Lett. 75: 565–568. [DOI] [PubMed] [Google Scholar]
- Rouzine I. M., Brunet E., Wilke C. O., 2008. The traveling-wave approach to asexual evolution: Muller’s ratchet and speed of adaptation. Theor. Popul. Biol. 73: 24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Datta M., Korolev K. S., Cvijovic I., Dudley C., Gore J., 2013. Range expansion promotes cooperation in an experimental microbial metapopulation Proc. Natl. Acad. Sci. USA 110: 7354–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt J. A., 1998a On the transition from initial data to travelling waves in the F-KPP equation. Dyn. Stab. Syst. 13: 167–174. [Google Scholar]
- Sherratt J. A., 1998b Invading wave fronts and their oscillatory wakes are linked by a modulated travelling phase resetting wave. Physica D 117: 145–166. [Google Scholar]
- Sherratt J. A., Smith M. J., Radenacher D. M., 2009. Locating the transition from periodic oscillations to spatiotemporal chaos in the wake of invasion. Proc. Natl. Acad. Sci. USA 106: 10890–10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L., Gordon D. M., Stewart F. M., Levin B. R., 1990. Estimating the rate of plasmid transfer: an end-point method. J. Gen. Microbiol. 136: 2319–2325. [DOI] [PubMed] [Google Scholar]
- Skellam J. G., 1951. Random dispersals in theoretical populations. Biometrika 38: 196–218. [PubMed] [Google Scholar]
- Summers D. K., 2009. The Biology of Plasmids. Blackwell Scientific, Oxford. [Google Scholar]
- van den Bosch F., Hengeveld R., Metz J. A. J., 1992. Analyzing the velocity of animal range expansion. J. Biogeogr. 19: 135–150. [Google Scholar]
- van Saarloos W., 2003. Front propagation into unstable states. Phys. Rep. 386: 29–222. [Google Scholar]
- Waclaw B., Allen R. J., Evans M. R., 2010. A dynamical phase transition in a model for evolution with migration. Phys. Rev. Lett. 105: 268101. [DOI] [PubMed] [Google Scholar]
- Weinberger H. F., Lewis M. A., Li B., 2007. Anomalous spreading speeds of cooperative recursion systems. J. Math. Biol. 55: 207–222. [DOI] [PubMed] [Google Scholar]
- Wilson P. D. G., Brocklehurst T. F., Arino S., Thuault D., Jakobsen M., et al. , 2002. Modelling microbial growth in structured foods: towards a unified approach. Int. J. Food Microbiol. 73: 275–289. [DOI] [PubMed] [Google Scholar]
- Wimpenny J. W. T., Leistner L., Thomas L. V., Mitchell A. J., Katsaras K., et al. , 1995. Submerged bacterial colonies within food and model systems: their growth, distribution and interactions. Int. J. Food Microbiol. 28: 299–315. [DOI] [PubMed] [Google Scholar]
- Young D. A., Bettinger R. L., 1995. Simulating the global human expansion in the Late Pleistocene. J. Archaeol. Sci. 22: 89–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.