Abstract
Climate change has altered life history events in many plant species; however, little is known about genetic variation underlying seasonal thermal response. In this study, we simulated current and three future warming climates and measured flowering time across a globally diverse set of Arabidopsis thaliana accessions. We found that increased diurnal and seasonal temperature (1°–3°) decreased flowering time in two fall cohorts. The early fall cohort was unique in that both rapid cycling and overwintering life history strategies were revealed; the proportion of rapid cycling plants increased by 3–7% for each 1° temperature increase. We performed genome-wide association studies (GWAS) to identify the underlying genetic basis of thermal sensitivity. GWAS identified five main-effect quantitative trait loci (QTL) controlling flowering time and another five QTL with thermal sensitivity. Candidate genes include known flowering loci; a cochaperone that interacts with heat-shock protein 90; and a flowering hormone, gibberellic acid, a biosynthetic enzyme. The identified genetic architecture allowed accurate prediction of flowering phenotypes (R2 > 0.95) that has application for genomic selection of adaptive genotypes for future environments. This work may serve as a reference for breeding and conservation genetic studies under changing environments.
Keywords: Arabidopsis, genome-wide association studies (GWAS), flowering time, growing season, climate change
CLIMATE change is accelerating, altering growing seasons, and affecting the developmental timings or phenology of plant species. This study investigates the complex genetic basis of Arabidopsis flowering time under seasonal warming. Certain genotypes switch from overwintering to rapid fall flowering in warmer winter seasons. Thermal-sensitive alleles were identified in genes of the heat-shock and hormone response pathways. This genetic model was able to accurately predict flowering time of new genotypes in our future conditions, illustrating an important method for breeding and facilitating adaptation in other species.
The impact of climate change on life history traits has been observed in many species, including earlier leaf and flower bud burst in plants, earlier breeding date in birds and frogs, and first observed flight date for butterflies (Parmesan 2006; Rosenzweig et al. 2008). Flowering time (FT) in plants is an important life history trait underlying reproductive fitness and is sensitive to local growing conditions. FT, like many other adaptive traits, affects survival. For example, over the past 150 years, species with flowering times less responsive to warming temperatures have decreased in local abundance (Willis et al. 2008), suggesting that flowering time sensitivity is important for climate adaptation. Flowering time is influenced by genotype (G), environment (E), and their interaction [genotype by environment (G × E)]. Several studies in plants and animals have looked at how quickly populations can adapt to rapid climate change, given quantitative genetic variation along with reproduction and migration rates (Franks and Hoffmann 2012; Shaw and Etterson 2012), The genetic loci underlying this phenotypic sensitivity can be detected by examining G × E. These alleles would enable a population to adapt to changing environments (Via and Lande 1985). G × E is also important for model prediction of phenotypic variation, species diversity and distribution, and crop yield stability under changing climates (Hoffmann and Willi 2008; Nicotra et al. 2010).
Plants such as Arabidopsis thaliana have evolved different life history strategies to adapt to a wide range of growing regions and seasons through two major life history strategies (Mitchell-Olds and Schmitt 2006). The rapid cycling strategy is to germinate both in spring and early autumn and then flower and set seeds rapidly. Winter annuals germinate in early or late autumn and overwinter as a rosette, while receiving a vernalizing cold period that promotes flowering in spring. Many plants require vernalization to ensure reproductive success.
Several genes have been identified as controlling flowering that are sensitive to the environment. In brassicas, including Arabidopsis, flowering locus C (FLC) is the major gene underlying the vernalization requirement. FLC prevents flowering during winter by repressing the floral integrator genes such as flowering locus T (Simpson and Dean 2002). FLC is activated by a life history switch FRIDIDA (FRI) (Johanson et al. 2000) but repressed by vernalization. Together, functional FRI and FLC alleles promote overwintering while null alleles at either locus allow rapid cycling when conditions allow. In addition to the genetic requirements, the variation in life history depends on the seasonal environment, with different germination times as a result of different seasonal cues that are perceived by plants (Wilczek et al. 2009; Li et al. 2010). For example, accessions with FRI null alleles are rapid cyclers when they germinate in early fall, but overwinter when they germinate in late fall. These alterations indicate the importance of germination timing under seasonal environments as well as genetic modifications (Wilczek et al. 2009). The fall germination time of a population cohort can also affect flowering time under basic greenhouse conditions with only minimal temperature augmentation (Botto and Coluccio 2007).
Knowledge of what genetic loci sense climate cues is important to understand and predict phenotypic responses to future climate change. For example, populations with variation at thermal-sensitive loci would be able to adapt better than those lacking variation. In addition, plant breeders could select for particular alleles for certain environments as conditions change. Li et al. (2010) investigated the natural genetic variation underlying flowering time of A. thaliana under early and late spring simulated climates for both the southern and the northern range. We used a diverse mapping set of accessions typed at approximately 213,000 SNPs to finely map the genetic loci, using genome-wide association studies (GWAS). Here we use the same mapping set to investigate overwintering conditions under current and future warmer seasonal climates. The goal of this study is to investigate the genetic variation underlying FT sensitivity to future seasonal warming within A. thaliana. We hypothesized that genotypes would vary in flowering responses dependent on the extent of future warming but that this may be restricted to particular fall germination times.
Materials and Methods
Experimental design
Seeds of a genetic and geographically diverse core set of accessions were obtained from a previous study (Li et al. 2010) and were grown in the same conditions in 3 × 3 × 3-inch pots. Specifically, the seeds were stratified for 6 days in the dark at 4° in 5 mg/liter gibberellic acid (GA) water to promote germination (GA3; Sigma-Aldrich). They were then transferred to soil under 24-hr light at 23° for 10 days to synchronize the germination (most seeds germinated within 3 days). Plants were thinned to one in each pot and moved into each of four simulated fall seasonal climate conditions that represent current and future years (2010, 2025, 2040, and 2055) with an increased diurnal and seasonal temperature. Flowering time was recorded daily as days to flower (DTF) bud after germination.
A typical Northern Hemisphere continental climate, where populations have been well studied, was selected for our study. We simulated this climate using SolarCalc 2.0 (Spokas and Forcella 2006) with latitude 41.84, longitude −87.68, and elevation 182 m (Chicago), using the 30-year average air temperature within the program for the current prediction. SolarCalc uses the local maximum and minimum daily air temperatures derived using Global TempSIM (Legates and Willmott 1990, 2006; Willmott and Matsuura 1995; New et al. 1999). Predicted warming values from the Intergovernmental Panel on Climate Change fourth assessment report (medium A1B scenario) were used in the generation of the predicted air temperatures. Two walk-in growth chambers (AR-916; Percival Scientific, Perry, IA) were programmed to cycle the simulated climates with adjustments made every 5 min to light spectrum, light intensity, temperature, and relative humidity throughout the day and the season. For each planting, one chamber ran simulated 2010 conditions while the other chamber ran simulated 2040 conditions. Within chambers, the top shelf matched the target temperature while the bottom shelf was recorded as 1° warmer and thus simulated additional future conditions of 2025 and 2055. Together, these conditions reflect current (2010) and predicted future warmer growing seasons for 2025, 2040, and 2055 (Supporting Information, Figure S1, A and B). The maximum humidity was 75% and the minimum temperature was 5°, due to the chamber limitations. However, 5° is an effective temperature for vernalization (Wilczek et al. 2009). To reduce the number of winter days with minimal growth, a shortened winter season was simulated by running the simulation only every second day in November and March and only every third day in December, January, and February, compressing the treatment down from 5 months to 2 months.
Germination cohorts in the wild are triggered by rainfall and/or soil tilling and can occur at various times during fall. In this study, we simulated fall climate conditions by performing synchronized plantings in September and October to bracket the growth season window. Synchronized germination is necessary to measure relative flowering time variation among genotypes and their sensitivity to temperature in early and late fall growing seasons. The final September data set contained 1479 plants that survived and ultimately flowered. This represented 417 distinct accessions, with 288 flowering in all four conditions, 85 in three conditions, 28 in two conditions, and 16 in a single condition. All data in this study are available in File S1, File S2, File S3, File S4 and at http://borevitzlab.anu.edu.au/resources/association-studies.
Association mapping analysis
We considered the following model for our data analysis,
| (1) |
where yij is the phenotypic value of the jth accession in the ith environment (i.e., one of the four climate conditions simulated for years 2010, 2025, 2040, and 2055, respectively), xij represents intercept and covariates (if any) with effects β, is the effect of the ith environment, is a coding variable (with a value 0 or 1) of two genotypes at the scanning locus and a∗ is the effect of the putative quantitative trait locus (QTL), is the interactive effect of the ith environment with the putative QTL, uij represents polygenic variation, and εij denotes the residual effect. While uij and εij are random, the rest are fixed effects. We assumed that , , and with K = (Kij) being the kinship matrix, and The kinship matrix K was estimated from genotypic data by using the software EMMA (Kang et al. 2008).
Under model (1) without the putative QTL effects, the environmental effect was statistically significant at 0.05. Its estimate, , was approximately linear in the temperature across the 2010, 2025, 2040, and 2055 thermal seasonal environments (R2 = 0.9976). Considering temperature is a major gauge for the environmental conditions in our simulations, it was reasonable to replace the environmental effect with temperature or rather the estimated environmental effect because of the approximate linearity. Therefore, we used the following model for genome-wide association and model selection,
| (2) |
where β∗ was a parameter corresponding to the environmental-effect ’s when their scores were used. The advantages of using ’s rather than ’s in model (2) included the following: (1) we were interested in the genetic basis of FT and its sensitivity across temperature environments (G × E) rather than to each specific temperature environment; and (2) the number of parameters was reduced, which was potentially beneficial for the statistical power to detect G × E effects with a reduced number of total tests.
In the analysis of the DTF data, ≥10,000 SNPs with a minor allele frequency <5% were filtered out, and the genome-wide scan for FT QTL was performed separately for the September and October planting data, using the R package QTLRel (Cheng et al. 2011). Forward model selection was then performed to determine the number and locations of putative QTL among SNPs with LOD scores larger than the 0.05 empirical genome-wide significance threshold, which was estimated from 5000 permutations of the genotype data (Cheng and Palmer 2013). The entry value to include a new QTL in the model selection procedure was the 0.05 empirical genome-wide significance threshold. Next, the genome-wide scan was performed to identify G × E QTL [or thermal sensitivity (THERM) QTL] by using model (2) with the identified FT QTL being included as covariates, and the model selection procedure was performed to determine the number and locations of THERM QTL in the same manner as above with the FT QTL being added to THERM QTL candidates.
Finally, the identified FT QTL and THERM QTL were included in model (2) to estimate QTL effects as well as other parameters and to make best linear unbiased prediction (BLUP) of the phenotype. The BLUPs were used to predict phenotypes compared to observed phenotypes in particular environments. If E, G, G × E, and polygenic effects were effectively modeled, the prediction should be reasonably accurate and provide useful information about the performance of a plant in a future environment.
Results
Impact of future seasonal warming on flowering time at the population level
We simulated changing diurnal light intensity, quality, and day lengths for early and late fall planting times for a typical location where Arabidopsis grows naturally. Diurnal and seasonal temperature fluctuations were set to current (2010) and predicted future climates (2025, 2040, and 2055) increased by ∼1° for each 15 years into the future (Figure S1, A and B). This simulation extends the growing season by days to weeks, due to shorter winters (Figure S2A). Under these conditions, the average flowering date of our global population advanced as a consequence of the warmer growing seasons for both September and October planting cohorts (Figure S1, C and D, and Figure S2B).
The phenotypic variation in FT was largely hidden in the late fall germination cohort as essentially all accessions overwintered. In contrast, the September germination cohort displayed both life history strategies (Figure 1 and Figure S1, Figure S2, Figure S3, Figure S4, and Figure S5). Rapid cyclers flowered in fall while overwintering annuals flowered in spring. A third maladapted minority class flowered in winter. The proportion of rapid cycling plants (flowering in fall) was incrementally higher for each 1° temperature increase (Figure S2B).
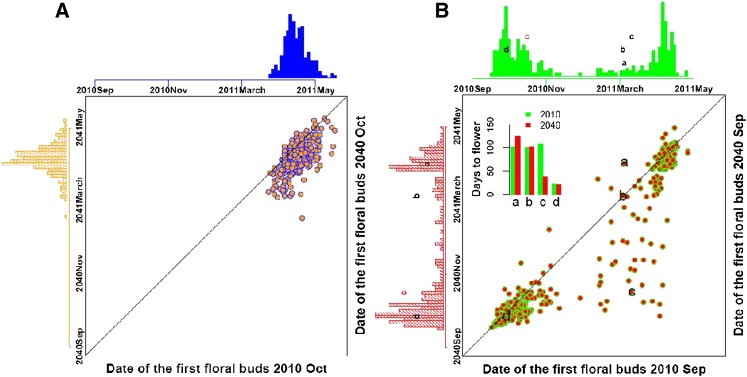
Figure 1.
(A and B) Natural variation in flowering time sensitivity to future warming among A. thaliana accessions in (A) September and (B) October germination cohorts. Each circle represents the flowering time of one accession in two environments. The accessions labeled a, b, c, and d in B are Lu-1, Ts-5, N7, and Col-0, respectively.
Natural variation in flowering responses to future warming among individuals
To evaluate the variation in flowering response to warmer climates among individual genotypes, we compared FT of each accession in current vs. future environments (e.g., 2010 vs. 2040). Substantial genetic variation in thermal sensitivity of FT was observed in the September cohort (Figure 1 and Figure S5). Some accessions showed strong FT responses to warming. For example, the accession labeled c flowered in spring (108 days after germination) in the 2010 climate, but flowered in fall (37 days after germination) in the simulated 2040 climate, displaying altered life history in the warmer future climate. The accessions that switched their life history strategy contribute to the increased frequency of rapid cycling plants under warmer climates. However, other accessions displayed unaltered life history in the 2040 climate (accessions a, b, and d).
In contrast, the October germination cohort, composed of the same global set of accessions, showed little variation in the FT sensitivity to future warming. Here, all accessions overwintered, revealing the dramatic plasticity of potentially rapid cycling accessions. The October cohort did respond quantitatively to future seasonal climates with advanced flowering time under warmer climates (Figure 1A and Figure S1, Figure S3, and Figure S5). Taken together, the flowering time response was influenced by germination date, genotype, and climate (diurnal and seasonal temperature).
Genetic loci for fall flowering time and thermal sensitivity in early fall planting
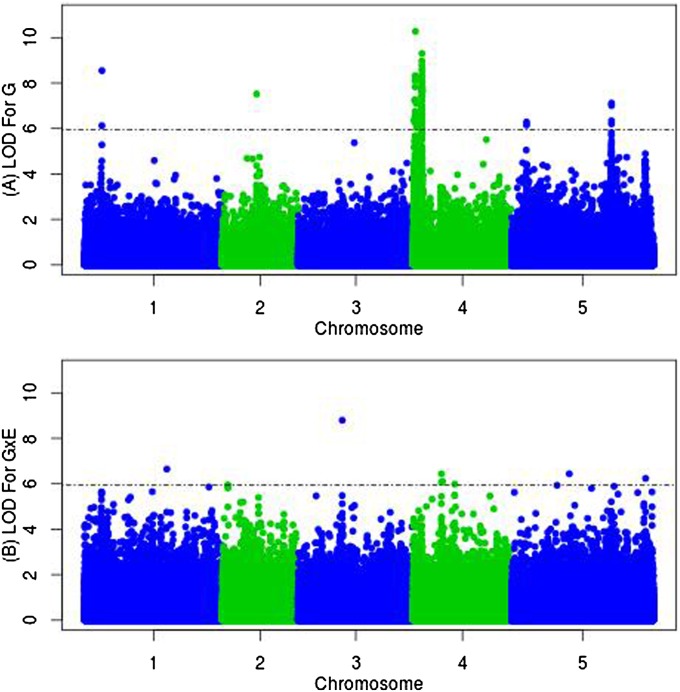
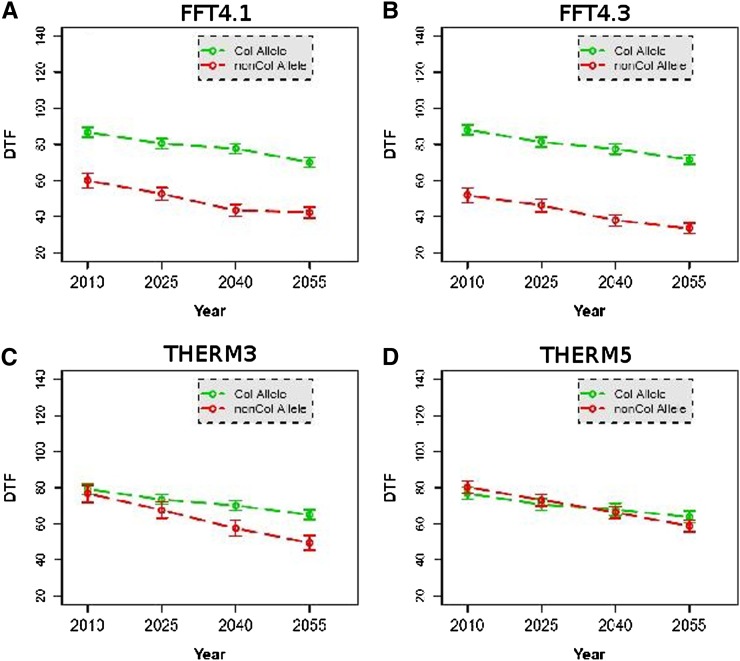
Genetic variation was hidden in the October cohort but revealed in the September cohort. Therefore, we chose to focus on the discovery of the genetic basis for fall flowering time (FFT) and THERM as G × E, in the September planting. Figure 2, A and B, displays Manhattan plots of a single SNP and a temperature–environment interaction scan, respectively, showing several potential FFT QTL and THERM QTL. QQ plots are shown in Figure S6 and Figure S7. The model selection process, described in Materials and Methods, resulted in five FFT QTL: snp1_3978064, snp2_8272902, snp4_383752, snp4_493905, and snp4_1330749. Three of these five FFT QTL (snp1_3978064, snp4_383752, and snp4_1330749) were also identified as spring FT QTL when the same accessions were phenotyped in our previous climate study (Li et al. 2010) (Figure 3, A and B, and Table 1). Another, FFT4.2b (snp4_493905), was tightly linked to the candidate gene Light Responsive Bric-a-Brac3, At4g01160, with family members shown to interact with phytochrome B and D diurnal and seasonal signaling pathways (Christians et al. 2012).
Figure 2.
(A and B) Genome-wide association of flowering time (days to flower) for main FT QTL (A) and THERM QTL (B) across four environments (2010, 2025, 2040, and 2055). The dashed horizontal line represents the 5% empirical genome-wide significance threshold.
Figure 3.
(A and B) The effects of major fall flowering time QTL across simulated climates; (C and D) the differential effects of THERM QTL.
Table 1. Identified QTL and their estimated effects.
| Candidatea | QTL | Chr | Positionb | Freqc (%) | Effect | SE | % variation |
|---|---|---|---|---|---|---|---|
| Intercept | 55.8 | 38.1 | |||||
| Environment | 5.1 | 0.8*** | 0.15 | ||||
| SFT1 | FFT1 | 1 | 3,978,064 | 19 | −25.0 | 4.9*** | 12.03 |
| FFT2 | 2 | 8,272,902 | 17 | 21.9 | 4.3*** | ||
| SFT4.2 | FFT4.2 | 4 | 383,752 | 31 | 19.4 | 3.8*** | |
| LRB3 | FFT4.2b | 4 | 493,905 | 44 | −18.0 | 3.5*** | |
| SFT4.3 | FFT4.3 | 4 | 1,330,749 | 24 | 24.0 | 4.6*** | |
| CRP | 4 | 219,919 | 15 | 9.4 | 5.4 | 1.40 | |
| FRller | 4 | 268,809 | 91 | 8.9 | 5.9 | ||
| FRlcol | 4 | 269,962 | 6 | −7.3 | 6.1 | ||
| FLC(−) | 5 | 3,188,327 | 22 | −17.7 | 4.6*** | ||
| GA2ox7 | THERM1 | 1 | 18,897,058 | 17 | 16.8 | 4.5*** | 0.15 |
| ROF1 | THERM3 | 3 | 9,185,447 | 25 | 8.5 | 4.3** | |
| ARCK1 | THERM4.1 | 4 | 7,148,335 | 6 | −14.3 | 7.8* | |
| THERM4.2 | 4 | 7,626,840 | 48 | 8.0 | 3.6** | ||
| PLC1 | THERM5 | 5 | 23,690,898 | 51 | −6.4 | 3.7* | |
| GA2ox7 | THERM1.e | 1 | 18,897,058 | 17 | −1.5 | 0.3*** | 7.04 |
| ROF1 | THERM3.e | 3 | 9,185,447 | 25 | −1.2 | 0.2*** | |
| ARCK1 | THERM4.1.e | 4 | 7,148,335 | 6 | −2.3 | 0.4*** | |
| THERM4.2.e | 4 | 7,626,840 | 48 | 0.9 | 0.2*** | ||
| PLC1 | THERM5.e | 5 | 23,690,898 | 51 | −0.9 | 0.2*** | |
| Polygenic | All | All | >5 | 76.59 |
***P = 0.01; **P = 0.05; *observed effect of SNP allele is opposite to that of candidate allele based on t test.
Candidate gene or QTL identified by Li et al. (2010).
Physical position (in base pairs) according to TAIR9.
Reference Col-0 allele frequency.
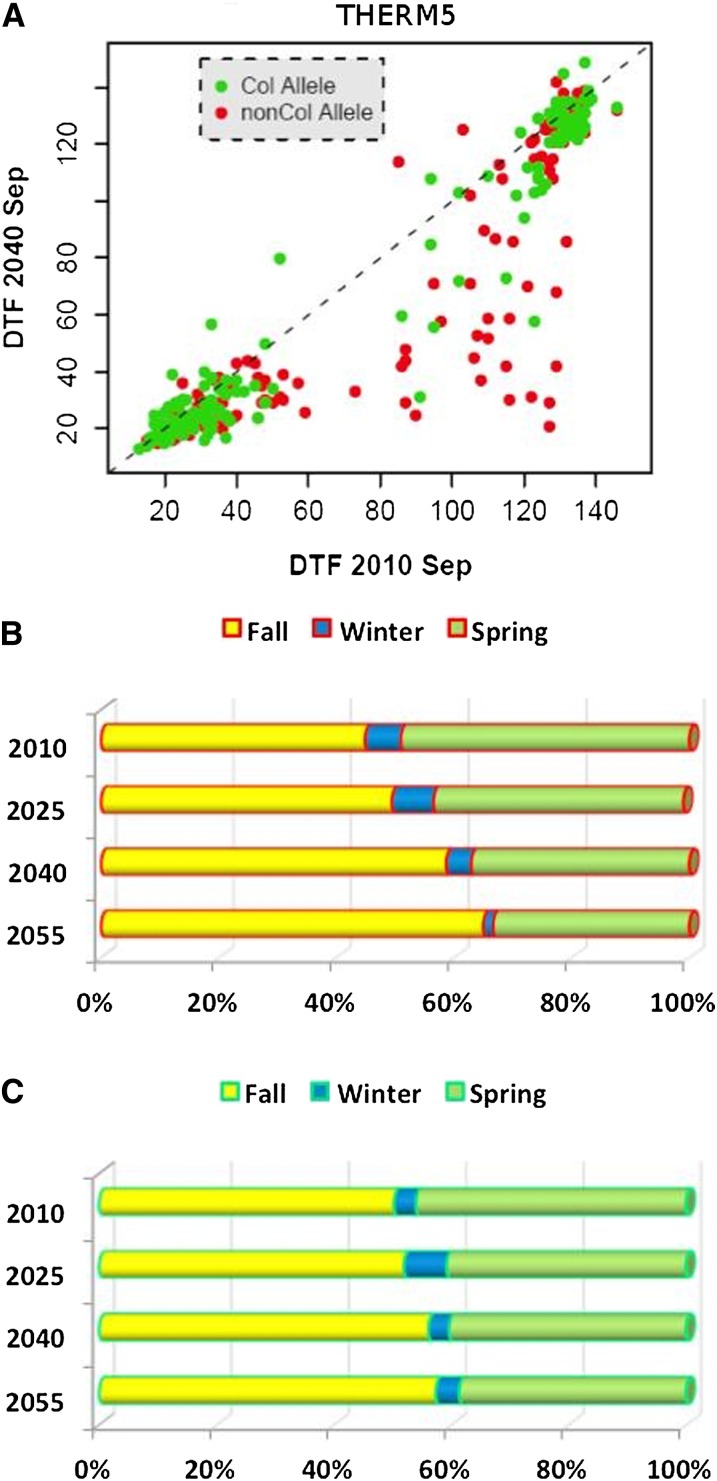
Two additional SNPs were identified as QTL at a significance level of 5% in a genome-wide scan but they were not retained by the selection procedure. They are located within 10 kb of a priori candidate genes (FLC and CRP) so we included them subsequently in the model. We also included the two known major loss-of-function alleles at FRI (Li et al. 2010). These four SNPs were included with the initial five FFT QTL as covariates in a subsequent G × E scan. In this run, the model selection process identified a further five THERM QTL: snp1_18900758, snp3_9185447, snp4_7148335, snp4_7626840, and snp5_23690898. For four of five THERM QTL, the main effect on FT (Table 1) was not significant while the interaction with temperature was considerable. The THERM3 QTL SNP3_9185447 is located within 10 kb of the Rotamase FKBP 1 gene, ROF1 (AtFKBP62, AT3G25230; Figure 3C). THERM4.2 and THERM5 QTL displayed a large deviation in allele frequencies between the thermal-sensitive and -insensitive groups. Accessions that contain the thermal-sensitive (nonreference) allele at this SNP showed a substantial increase in the frequency of the rapid cyclers under future warming, compared to accessions with the Col allele (Figure 4). THERM5 (SNP5_23690898) is located within 10 kb of a phospholipase C1 gene (PLC1, AT5G58670). Another strong candidate gene was identified for THERM QTL at SNP1_18900758, which is within 10 kb of gibberellin 2-oxidase (GA2OX7, AT1G50960), a flowering-time hormone biosynthetic gene.
Figure 4.
THERM5 shows a common G × E effect. (A) Scatterplot of FT for accessions in 2010 vs. 2040 color coded by genotype at THERM5 QTL. (B and C) Life history stages of the different genotypes across each of the four simulated climates.
We investigated epistasis among all the identified QTL; however, none of the interactions exceeded the 0.05 empirical significance threshold. Table 1 shows the results of a fit with the full model with all five FFTs, four a priori loci, and five THERM QTL. The proportion of variation explained by factor groups is also shown. Note that together the major FFT QTL explain only 12% while the THERM QTL explain a similar 7% of the variation. The dominant factor (76%) is the polygenic background, estimated as a random effect, using the pairwise kinship matrix.
Phenotype prediction
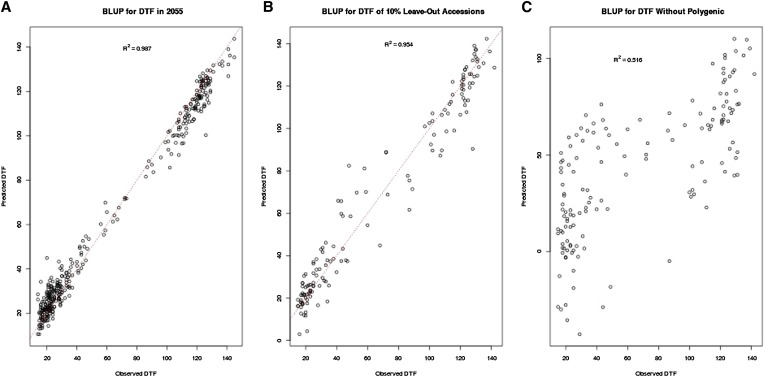
To assess the potential to forecast phenotypes from genotypes under future climates, we calculated the BLUPs of QTL effects in two different scenarios. First, we used the phenotype and genotype data from three simulated climates (2010, 2025, and 2040) to fit model (2), including all identified QTL, and then treated the estimates of the model parameters as their true values to predict flowering time in the final simulated climate (Figure 5A). Second, we randomly selected a subset of 90% of the accessions across all environments to fit model (2) in the same way and then predicted flowering time for the remaining 10% of accessions. Figure 5B displays the BLUP results. The prediction was quite accurate in both scenarios because model (2) could account for ∼95% of the total phenotypic variation (Table 1). However, if the polygenic term is not included, the predictive power is largely reduced (Figure 5C). This highlights the importance of background genetic variation on phenotype, with positive implications for genomic selection even when major QTL are not known.
Figure 5.
(A) Flowering time in 2055 predicted from data in the first three environments 2010, 2025, and 2040; (B) flowering time of a random subset of 10% of accessions predicted from data of the remaining 90% of accessions. (C) Flowering times that would be obtained for the QTL alone without polygenic terms vs. the actual flowering times.
Discussion
A general trend of advanced flowering time has been seen in many species due to climate warming (Parmesan 2006), but little was known about the natural variation within a species or the genetic architecture underlying these responses. The genetic factors allow predictions on population persistence and/or species range shifts under environmental change (Chevin et al. 2010; Duputié et al. 2012). In the field, germination is highly variable and may change in future climates. In this study we selected an early and a late fall time point for germination to bracket the natural range to study seasonal flowering time response. The genetic loci sensing climate variation (G × E loci) can reveal alleles involved in local adaptation (Fournier-Level et al. 2011; Hancock et al. 2011; Horton et al. 2011; Méndez-Vigo et al. 2011) and provide an opportunity to breed crops for future climate change (Nicotra et al. 2010).
In this study, we identify candidate genes and pathways underlying the thermal variation in seasonal flowering time (THERM QTL, G × E). ROF1 modulates thermotolerance by interacting with heat-shock protein 90 (HSP90) and affects the accumulation of small HSPs (Meiri and Breiman 2009). HSPs are well known for their importance in thermotolerance, but, more interestingly, they are chaperones of many regulatory proteins and buffer genetic variation. Recent studies in A. thaliana have shown that HSP90 contributes to phenotypic variation and plays a role in developmental plasticity by buffering or releasing variation under shifting environments (Queitsch et al. 2002; Sangster et al. 2007, 2008). PLC1 is induced by various environmental stresses such as dehydration, salinity, and low temperature. Finally, variation in QTL near two gibberellin oxidases suggests GA as the important pathway underlying seasonal thermal sensitivity in flowering time, previously known to be important only in constant temperature sensing (Blazquez et al. 2003).
Subtle, but consistent, changes in environmental cues perceived by A. thaliana are amplified through THERM QTL alleles, affecting downstream signaling cascades that result in a qualitative shift from overwintering to rapid cycling, which would have dramatic effects on reproduction. Alleles at unlinked THERM QTL can be combined to provide varying degrees of overall thermal sensitivity, depending on the environment and standing genetic variation. The candidate genes reveal the molecular signaling underlying the altered life history outcomes and include crosstalk among previously characterized photoperiod, gibberellin, vernalization, and autonomous pathways of FT regulation (Henderson and Dean 2004). Together, FFT and THERM QTL, environmental change, and the polygenic background cause a highly predictable switch to rapid cycling.
Some accessions and many common alleles in this mapping population were originally collected in the United States and represent genotypes that occur as local populations in the Chicago area where climates were simulated. Among these, both thermal-sensitive and -nonsensitive genotypes have been found. The common alleles at THERM QTL discovered in this study represent the natural variation in specific local populations (Platt et al. 2010). If FFT and THERM QTL played an important role in the distribution of Arabidopsis across its range, we would expect to find a strong association with latitude of origin; however, this was not the case for the FFT and THERM QTL identified in this study. Future studies could use multiple local populations and seasonal environmental gradients to more carefully assess their role in local adaptation.
In summary, this study focuses on the natural genetic variation in thermal sensitivity of the life history trait, flowering time, under simulated current and future fall growing seasons, in a large diverse mapping set of A. thaliana. The average FT of the population advanced with increasing temperature. Qualitative, genotype-level variation in flowering responses to future warming was observed. Select genotypes of an early fall germination cohort switched from overwintering to rapid cycling in future warmer climates. These observations suggest interactions between light and thermosensory pathways controlling FT. Here, we report several major QTL for natural variation in the FT and thermal sensitivity for this life history transition. By simulating seasonal temperature increases, in climate chambers, we identified a genetic model that accurately predicts future flowering phenotypes (Figure 5). Further, we identify candidate genes responding to temperature that are part of the gibberellin and heat-shock pathways. The QTL identify allelic variation, which may provide A. thaliana with the potential to adapt to warming climates. Genotypes with a switched life history strategy (from overwintering to rapid cycling) can set seeds rapidly before winter and germinate again in spring, which would enable them to reproduce multiple times per year, a clear fitness advantage in many conditions. Indeed, the thermal sensitivity loci identified may provide the plasticity in flowering time to help A. thaliana continue to adapt to a wider range of locations and future growing seasons. Further experiments, such as the one presented here, on standing genetic variation in crop plants and foundation species can reveal the source populations and alleles that should be utilized and conserved to keep pace with future climate change.
Early warnings about the limits of genetic adaptation under rapid climate change (Davis and Shaw 2001) stated that the limited connectivity and large range shifts required leave populations and species vulnerable to extinction. Specialists are particularly at risk, while generalists showing a higher degree of environmental plasticity may fare better (Anderson et al. 2012). Variation in climate sensitivity also exists within species and between populations (Angert et al. 2011), complicating migration under climate change. An understanding of the genetic basis of climate sensitivity can aid breeder and land managers in finding a match between genotype and environment. Here we identify quantitative trait loci differing in seasonal thermal sensitivity. This allows predictions about the resilience of a given genotype or population to a particular environmental shift and can guide collection and restoration efforts in foundation species through managed relocation (Schwartz et al. 2012). Our study suggests a pathway to facilitate adaptation into new and variable climates.
Supplementary Material
Acknowledgments
We thank John Zdenek and Sandra A. M. Suwanski at the University of Chicago greenhouse for help in preparing soils and watering plants, Eleni Boikoglou for collaborating on preparing the simulated weather files, and Nina Noah and Yi Ren for their help in scanning barcodes. This work was funded by National Institutes of Health (NIH) grant R01GM073822 (to J.O.B. and Y.L.) and NIH grants R01DA021336, R01MH079103, and R21DA024845 (to A.A.P. and R.C.) and Australian National University startup funds (to J.O.B. and R.C.).
Footnotes
Communicating editor: K. M. Nichols
Literature Cited
- Anderson J. T., Panetta A. M., Mitchell-Olds T., 2012. Evolutionary and ecological responses to anthropogenic climate change. Plant Physiol. 160: 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angert A. L., Sheth S. N., Paul J. R., 2011. Incorporating population-level variation in thermal performance into predictions of geographic range shifts. Integr. Comp. Biol. 51: 733–750. [DOI] [PubMed] [Google Scholar]
- Blazquez M. A., Ahn J. H., Weigel D., 2003. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33: 168–171. [DOI] [PubMed] [Google Scholar]
- Botto J., Coluccio M., 2007. Seasonal and plant-density dependency for quantitative trait loci affecting flowering time in multiple populations of Arabidopsis thaliana. Plant Cell Environ. 30: 1465–1479. [DOI] [PubMed] [Google Scholar]
- Cheng R., Palmer A. A., 2013. A simulation study of permutation, bootstrap, and gene dropping for assessing statistical significance in the case of unequal relatedness. Genetics 193: 1015–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Abney M., Palmer A. A., Skol A. D., 2011. QTLRel: an r package for genome-wide association studies in which relatedness is a concern. BMC Genet. 12: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin L.-M., R. Lande, and G. M. Mace, 2010. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 8: e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians M. J., Gingerich D. J., Hua Z., Lauer T. D., Vierstra R. D., 2012. The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol. 160: 118–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. B., Shaw R. G., 2001. Range shifts and adaptive responses to quaternary climate change. Science 292: 673–679. [DOI] [PubMed] [Google Scholar]
- Duputié A., Massol F., Chuine I., Kirkpatrick M., Ronce O., 2012. How do genetic correlations affect species range shifts in a changing environment? Ecol. Lett. 15: 251–259. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A., Korte A., Cooper M. D., Nordborg M., Schmitt J., et al. , 2011. A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89. [DOI] [PubMed] [Google Scholar]
- Franks S., Hoffmann A., 2012. Genetics of climate change adaptation. Annu. Rev. Genet. 46: 185–208. [DOI] [PubMed] [Google Scholar]
- Hancock A. M., Brachi B., Faure N., Horton M. W., Jarymowycz L. B., et al. , 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86. [DOI] [PubMed] [Google Scholar]
- Henderson I. R., Dean C., 2004. Control of Arabidopsis flowering: the chill before the bloom. Development 131: 3829–3838. [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Willi Y., 2008. Detecting genetic responses to environmental change. Nat. Rev. Genet. 9: 421–432. [DOI] [PubMed] [Google Scholar]
- Horton M. W., Hancock A. M., Huang Y. S., Toomajian C., Atwell S., et al. , 2011. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the regmap panel. Nat. Genet. 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Michaels C. L. S., Amasino R., Dean C., 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347. [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legates D., Willmott C., 1990. Mean seasonal and spatial variability in global surface air temperature. Theor. Appl. Climatol. 41: 11–21. [Google Scholar]
- Legates D., Willmott C., 2006. Mean seasonal and spatial variability in gauge-corrected, global precipitation. Int. J. Climatol. 10: 111–127. [Google Scholar]
- Li Y., Huang Y., Bergelson J., Nordborg M., Borevitz J., 2010. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21199–21204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri D., Breiman A., 2009. Arabidopsis rof1 (fkbp62) modulates thermotolerance by interacting with hsp90. 1 and affecting the accumulation of hsfa2-regulated shsps. Plant J. 59: 387–399. [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B., Picó F., Ramiro M., Martínez-Zapater J., Alonso-Blanco C., 2011. Altitudinal and climatic adaptation is mediated by flowering traits and fri, flc, and phyc genes in Arabidopsis. Plant Physiol. 157: 1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T., Schmitt J., 2006. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441: 947–952. [DOI] [PubMed] [Google Scholar]
- New M., Hulme M., Jones P., 1999. Representing twentieth-century space-time climate variability. Part i: Development of a 1961–90 mean monthly terrestrial climatology. J. Clim. 12: 829–856. [Google Scholar]
- Nicotra A., Atkin O., Bonser S., Davidson A., Finnegan E., et al. , 2010. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15: 684. [DOI] [PubMed] [Google Scholar]
- Parmesan C., 2006. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37: 637–669. [Google Scholar]
- Platt A., Horton M., Huang Y., Li Y., Anastasio A., et al. , 2010. The scale of population structure in Arabidopsis thaliana. PLoS Genet. 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C., Sangster T., Lindquist S., 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624. [DOI] [PubMed] [Google Scholar]
- Rosenzweig C., Karoly D., Vicarelli M., Neofotis P., Wu Q., et al. , 2008. Attributing physical and biological impacts to anthropogenic climate change. Nature 453: 353–357. [DOI] [PubMed] [Google Scholar]
- Sangster T., Bahrami A., Wilczek A., Watanabe E., Schellenberg K., et al. , 2007. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced hsp90 levels. PLoS ONE 2: e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster T., Salathia N., Lee H., Watanabe E., Schellenberg K., et al. , 2008. Hsp90-buffered genetic variation is common in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. W., Hellmann J. J., Mclachlan J. M., Sax D. F., Borevitz J. O., et al. , 2012. Managed relocation: integrating the scientific, regulatory, and ethical challenges. Bioscience 62: 732–743. [Google Scholar]
- Shaw R., Etterson J., 2012. Rapid climate change and the rate of adaptation: insight from experimental quantitative genetics. New Phytol. 195: 752–765. [DOI] [PubMed] [Google Scholar]
- Simpson G., Dean C., 2002. Arabidopsis, the rosetta stone of flowering time? Science 296: 285–289. [DOI] [PubMed] [Google Scholar]
- Spokas K., Forcella F., 2006. Estimating hourly incoming solar radiation from limited meteorological data. Weed Sci. 54: 182–189. [Google Scholar]
- Via S., Lande R., 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39: 505–522. [DOI] [PubMed] [Google Scholar]
- Wilczek A., Roe J., Knapp M., Cooper M., Lopez-Gallego C., et al. , 2009. Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934. [DOI] [PubMed] [Google Scholar]
- Willis C., Ruhfel B., Primack R., Miller-Rushing A., Davis C., 2008. Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. Proc. Natl. Acad. Sci. USA 105: 17029–17033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmott C., Matsuura K., 1995. Smart interpolation of annually averaged air temperature in the United States. J. Appl. Meteorol. 34: 2577–2586. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.