Abstract
Interleukin-23 (IL-23) plays an essential role in maintenance of IL-17-producing T helper (Th17) cells that are involved in the pathogenesis of several autoimmune diseases. Regulation of Th17 cells is tightly controlled by multiple factors such as IL-27 and IFN-γ. However, the detailed mechanisms responsible for IFN-γ-mediated Th17 cell inhibition are still largely unknown. In this study, we demonstrate that IFN-γ differentially regulates IL-12 and IL-23 production in both dendritic cells and macrophages. IFN-γ suppresses IL-23 expression by selectively targeting p19 mRNA stability through its 3′untranslated region (3′UTR). Furthermore, IFN-γ enhances LPS-induced tristetraprolin (TTP) mRNA expression and protein production. Overexpression of TTP suppresses IL-23 p19 mRNA expression and p19 3′UTR-dependent luciferase activity. In addition, deletion of TTP completely abolishes IFN-γ-mediated p19 mRNA degradation. We further demonstrate that IFN-γ suppresses LPS-induced p38 phosphorylation and blockade of p38 MAPK signaling pathway with SB203580 inhibits IFN-γ and LPS induced p19 mRNA expression whereas overexpression of p38 increases p19 mRNA expression via reducing TTP binding to the p19 3′UTR. Finally, inhibition of p38 phosphorylation by IFN-γ leads to TTP dephosphorylation that could result in stronger binding of the TTP to the adenosine/uridine-rich elements in the p19 3′UTR and p19 mRNA degradation. In summary, our results reveal a direct link among TTP, IFN-γ and IL-23, indicating that IFN-γ-mediated Th17 cell suppression might act through TTP by increasing p19 mRNA degradation and therefore IL-23 inhibition.
Introduction
IL-23 is an interleukin-12 family cytokine composed of two different subunits, one shared subunit with IL-12, p40, and another unique subunit, p19 (1). IL-23 has drawn much attention in recent years because of its broad effects on the pathogenesis of several diseases (2, 3). Increased levels of IL-23 were observed in inflammatory bowel disease (IBD), multiple sclerosis (MS) and psoriasis patients (4-6). In addition, IL-23 level in synovial fluid (SF) was significantly increased in rheumatoid arthritis (RA) patients with joint destruction compared to those without, and correlated with the concentration of IL-1β, TNF-α and IL-17 either in serum or SF (7), which is believed to be mediated through PI3-kinase/Akt, NF-κB and MAPK-p38 signal transduction pathways (8, 9). Recent studies reveal an association of IL-23 receptor gene haplotypes with severity of several diseases such as IBD, RA, MS and psoriasis, and neutralization of IL-23 by anti-IL-12/23p40 antibody significantly ameliorated the severity of IBD and psoriasis. Furthermore, it has been reported that IL-23 expression was highly expressed in many tumors including breast tumor, prostate cancer, liver and skin cancers and the levels of IL-23 was correlated with tumor progression (10, 11).
IL-23 gene expression is tightly controlled at the transcriptional level. It has been reported that NF-κB c-Rel physically binds to the p19 promoter and induces IL-23 p19 gene expression (12, 13). Conversely, deletion of c-Rel abolished IL-23 p19 expression in both dendritic cells and macrophages (12, 13). In addition, Sma- and Mad-related protein (SMAD)-3, activating transcription factor (ATF)-2, and activating protein-1 (AP-1) also play important roles in control of IL-23 p19 gene expression (14). Silence of SMAD-3 and ATF-2 expression by shRNA reduced p19 promoter activity and protein expression in macrophages infected or treated with Theiler’s murine encephalomyelitis virus or poly (I-C), respectively (15). MAPKs including p38, JNK and ERK are involved in LPS-induced IL-23 p19 gene transcription (14). However, So far, most studies focus on transcriptional regulation of p19 expression, little is known about how IL-23 is regulated at the posttranscriptional level despite the fact that IL-23 p19 mRNA has a long 3′UTR containing multiple putative adenosine/uridine-rich elements (AREs).
Posttranscriptional regulation of many cytokines occurs by modulation of their mRNA stability through AREs in the 3′UTR. One of the best characterized ARE-associated RNA binding decay proteins is tristetraprolin (TTP). TTP (also known as TIS11, ZFP36, and Nup475) is a member of CCCH tandem zinc finger proteins (ZFP) and involved in the regulation of inflammatory responses at the posttranscriptional level (16). TTP binds to AREs within the 3′UTR causing destabilization of mRNAs encoding tumor necrosis factor-alpha (TNF-α) (17), granulocyte-macrophage colony-stimulating factor (GM-CSF) (18), cyclooxygenase 2 (19), interleukin-2 (20), interleukin-10 (21) and the chemokine CXCL1 (22). The mRNAs encoding TNF-α and GM-CSF are stabilized in TTP-deficient mice and in cells derived from these deficient mice (18, 23). Overproduction of these cytokines in TTP knockout mice result in a severe systemic inflammatory response including arthritis, autoimmunity and myeloid hyperplasia (24, 25). Meanwhile, up-regulation of TTP could reduce inflammatory responses in macrophages (26). All evidence to date indicates that TTP is a critical protein involved in the control of inflammation and maintenance of homeostasis. However, the role of TTP in regulation of IL-23 expression is unknown.
IL-12 family cytokines play critical roles in control of adaptive immunity against various pathogens. The first member of the family, IL-12 is essential for differentiation and proliferation of Th1 cells. IL-23 plays an essential role for Th17 cell survival and expansion important for host defense against extracellular infection. Th17 cells are associated with the pathogenesis of certain autoimmune diseases (27-29), mediate neutrophil recruitment, and participate in inflammatory responses (30-35). IL-23 is required in both the draining lymph node of proteolipid protein- or myelin oligodendrocyte glycoprotein-immunized mice, as well as in the CNS, in order to drive pathology in the EAE murine model of MS (36). On the other hand, Th17 cells are negatively controlled by IL-27 and IFN-γ. IL-27 suppresses de novo Th17 cell differentiation in a STAT-1 dependent, IFN-γ-independent pathway (28, 29). Recent studies demonstrate that IFN-γ strongly inhibited Th17 cell development in vitro (29, 37). Consistent with this later observation, IFN-γ deficient mice displayed excessive numbers of Th17 cells compared with wild type control mice (29). The effect of IFN-γ on Th17 cells is at least partially mediated by STAT-1 since the inhibitory effect of IFN-γ on Th17 cells was partially reversed in STAT-1 deficient mice (29). However, the detailed mechanisms of IFN-γ-mediated Th17 cell inhibition are still not fully understood.
In this study, we demonstrate that IFN-γ differentially regulates IL-12 and IL-23 expression by selective suppression of IL-23 production through TTP-mediated p19 mRNA degradation, which could lead to Th17 cell suppression. This novel mechanistic insight helps us better understand the cross-regulation between cytokines produced by different cells and the pathogenic consequence of this regulation.
Materials and Methods
Mice
TTP+/− mouse breeders used in the experiments were at 6~8 week old, housed in cages with filter tops in a laminar flow hood, fed food and water ad libitum at Saint Louis University Animal Facilities in accordance with the principles of Animal Care (NIH publication number 85-23, revised 1985).
Cells
The murine macrophage cell line RAW264.7 (RAW cells) was obtained from American Type Culture Collection, and maintained in RPMI1640 supplemented with 2 mM glutamine, 100 units/ml of penicillin and streptomycin and 10% FBS (Sigma, St. Louis, MO, endotoxin NMT 10.0 EU/ml) (Complete medium). Mouse peritoneal macrophages were obtained by lavage 3 days after injection of sterile 3% thioglycolate broth (1 ml i.p. per mouse) and plated in 24 well tissue culture plates (1 × 106 cells/well) with RPMI1640 complete medium. Mouse bone marrow derived dendritic cells (BMDCs) were generated from bone marrow cells obtained from mouse femurs. 3 × 106 bone marrow cells were cultured with RPMI1640 complete medium containing 40 ng/ml mouse GM-CSF and 10 ng/ml mouse IL-4. GM-CSF and IL-4 containing complete medium was replenished at day 3. After 7-day culture, the fully differentiated BMDCs were used for experiments. Mouse bone marrow-derived macrophages (BMDMs) were generated from bone marrow cells and cultured with complete RPMI1640 medium containing 10 ng/ml of M-CSF for one week.
Plasmids
Mouse IL-23 p19 3′UTR plasmid was cloned by inserting p19 3′UTR into the pGL3-control vector (Promega) between Xba I and Fse I sites. Primers used for amplification of p19 3′UTR (655 bp) were ggatgcccaggttcccatggctaccatgataaga (sense) and gccacataaataaatctttattgaaaaaaaaatacat (antisense). Mouse IL-23 p19 promoter was generated by cloning p19 promoter (1348 bp) into pGL2-basic luciferase vector (Promega) between Kpn I and Xho I sites. Primers used for cloning p19 promoter were caggacagccagggatacacagaga for sense strand and ggcacagccaggccctg for antisense strand. MAPK expression plasmids encoding MLK3, MKK3, MKK6, p38, dominant negative mutant MK2 and p38α as well as 3′UTRTNF-α and 3′UTRβ-Actin were kindly provided by Dr. Aihao Ding (Weill Cornell Medical College) with permissions from Drs. John M. Kyriakis (Tufts University School of Medicine, Boston, Massachusetts), Jim Woodgett (Samuel Lunenfeld Research Institute, Toronto, Canada), Jiahuai Han (The Scripps Research Institute, La Jolla, California), and Matthias Gaestel (Medical School Hanover, Hanover, Germany). All plasmid DNA were prepared with QIAGEN Endo-free Maxi-Prep kits.
Reagents
Mouse TTP antibody (N-terminal), Actinomycin D (ACD), 5,6-Dichlorobenzimidazole riboside (DRB), and LPS from Escherichia coli 0217:B8 were purchased from Sigma-Aldrich (St. Louis, MO). Phospho-14-3-3 Binding Motif (4E2) mouse mAb and MAPK antibodies including p38, ERK and JNK antibodies against either phosphorylated or total proteins were purchased from Cell Signaling Technology (Danvers, MA). Other antibodies were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Recombinant mouse IFN-γ was purchased from Genzyme (Boston, MA). IL-4, GM-CSF and M-CSF were purchased from R&D Systems. Lambda protein phosphatase was purchased from New England Biolabs (Ipswich, MA).
Quantitative real-time PCR
Reverse-transcription reactions were carried out as previously described (38), Quantitative real time PCR (qRT-PCR) was performed by a modified protocol. Briefly, cDNA samples converted from 1 μg of total RNA were diluted and studied at several concentrations. Diluted cDNA was mixed with a pair of primers (10 μM) targeting mouse p19, p35, p40, TTP, TNF-α, luciferase, or GAPDH cDNA sequences, with SYBR green PCR master mix (Sigma-Aldrich, St. Louis) in a 15 μl volume. The following primers were used for PCR amplification of the mouse p19 cDNA: sense: tgcaccagcgggacatatgaatct, antisense: tgttgtccttgagtccttgtgggt; mouse p35 cDNA: sense: acctgctgaagaccacagatgaca, antisense: tagccaggcaactctcgttcttgt; mouse p40 cDNA: sense: acctgtgacacgcctgaagaagat, antisense: tcttgtggagcagcagatgtgagt; mouse TTP cDNA: sense: aatccctcggaggactttggaaca, antisense: agttgcagtaggcgaagtaggtga; mouse TNF-α cDNA: sense: agccgatgggttgtaccttgtcta, antisense: tgagatagcaaatcggctgacggt; Luciferase cDNA: sense: atttatcggagttgcagttgcgcc, antisense: acaaac actacggtaggctgcgaa; and mouse GAPDH cDNA: sense: aactttggcattgtggaagg; antisense: acacattgggggtaggaaca.
Enzyme-linked immunosorbent assays (ELISA)
Supernatants from murine peritoneal macrophage and BMDC cultures were harvested at 24 h after IFN-γ and LPS stimulation and stored at −70°C. Mouse IL-12 p70 and TNF-α were detected using BD OptEIA ELISA kits and mouse IL-23 was detected using eBioscience ELISA kit according to the manufacturer’s instructions. Concentrations were calculated by regression analysis of a standard curve.
Transfection assay
Transient transfections were performed as described previously (38). Briefly, for each condition 1 × 107 RAW cell suspension was mixed with 16 μg of total DNA (including reporter, effector, internal control and carrier DNA), and electroporated at 975 microfarade and 300 V in RPMI1640 medium without serum. The transfected cells were resuspended in RPMI1640 complete medium containing 10 μM chloroquine and incubated for 48 h prior to harvesting. To measure luciferase activity, cells were pelleted and resuspended in lysis buffer. Luciferase activity was measured in cell lysates.
Primary transcript
To determine the rate of primary transcription of p19 gene, cDNA were synthesized with random primers using 1 μg of total RNA generated from BMDCs treated with IFN-γ, LPS or LPS plus IFN-γ. The primers used for measuring primary transcript were: p19 intron 1 (sense): tgttgtccttgagtccttgtgggt, p19 exon 2 (antisense): aaaccttcccagtcctccaagtgt.
LightShift Chemiluminescent RNA-EMSA
Single strand RNA sequence of the p19 3′UTR harboring ARE3 site (GGUUAUUUAUUC) was synthesized and labeled with biotin at the 5′ end by Integrated DNA Technology (Coralville, IA). LightShift Chemiluminescent RNA-EMSA was performed according to the manufacture’s protocol (Pierce, Rockford, IL) with optimizations (38).
Co-immunoprecipitation
Whole cell lysates were pre-cleared by adding 20 μl of protein G-agarose and incubating at 4°C for 1 h on a rocker. Pre-cleared lysates were incubated with 2 μg of 14-3-3 antibody in the presence of 20 μl of 50% (v/v) protein G-agarose overnight at 4°C with gentle rocking. After washings three times with PBS, precipitated complexes were solubilized by boiling in SDS buffer, fractionated by 12% SDS-PAGE, and transferred to PVDF membrane. Western blotting was performed using either a TTP or 14-3-3 antibody.
Western Blotting
SDS-PAGE was performed with 100 μg of whole cell lysates. Gels were transferred to PVDF membranes and blocked in 5% nonfat milk in Tris buffer, pH8.0. Primary antibody was added at a concentration of 1 μg/ml in Tris buffer containing 5% milk and left overnight at 4°C. After extensive washing, secondary antibody conjugated to horseradish peroxidase was added at a 1:5000 dilution in 5% nonfat milk in Tris buffer. After extensive washing, blots were subjected to Enhanced Chemiluminescence detection (PerkinElmer Life Sciences Inc, Boston, MA).
Statistical analysis
Student t test was performed wherever applicable. Standard deviation of the mean is shown unless otherwise indicated. *, p<0.05; **, p<0.01; ***, p<0.001.
Results
IFN-γ differentially regulates IL-12 and IL-23 production in dendritic cells
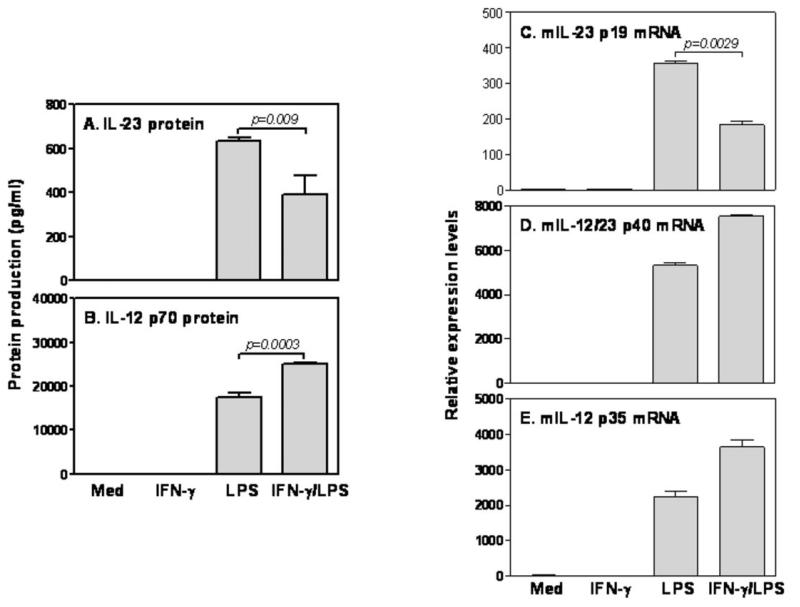
Since dendritic cells (DCs) are the major producer of IL-12 family cytokines and the most potent antigen presentation cells, we first wanted to evaluate the effects of IFN-γ on IL-12 and IL-23 production in DCs stimulated with LPS. As shown in Fig. 1A&B, LPS stimulation induced significant amounts of IL-23 (A) and IL-12 p70 (B) protein production. However, addition of IFN-γ showed differential effects on IL-12 and IL-23 production, inhibiting LPS-induced IL-23 (A) while enhancing IL-12 p70 production (B). Cells stimulated with IFN-γ alone did not produce either IL-23 or IL-12 p70, consistent with our previous studies (39). These data indicate that IFN-γ has reciprocal effects on IL-12 and IL-23 production in DCs. Since p40 is a shared subunit whereas p35 and p19 are unique subunits for IL-12 and IL-23 secretion, respectively, we determined to evaluate the effects of IFN-γ on p35 and p19 mRNA expression in LPS-treated BMDCs. LPS significantly increased p19 (C), p40 (D) and p35 (E) mRNA expression in BMDCs. Addition of IFN-γ suppressed p19 mRNA expression about twofold in LPS-treated cells (C), while significantly increasing p40 (D) and p35 (E) mRNA expression. The differential effects of IFN-γ on IL-12 and IL-23 expression were also seen in mouse peritoneal macrophages (Supple Fig .1). Taken together, these data suggest that IFN-γ differentially regulates IL-12 and IL-23 production by selectively affecting p35 and p19 mRNA expression.
Figure 1. IFN-γ differentially regulates LPS-induced IL-23 p19 and IL-12 p35 mRNA expression in DCs.
1 × 106 fully differentiated BMDCs were inoculated into each well of 24 well plates with 1 ml complete culture medium and treated with IFN-γ (10 ng/ml), LPS (1 μg/ml) and IFN-γ plus LPS either for 24 hours to measure IL-23 (A) and IL-12 p70 (B) protein levels in the supernatants by ELISA or for 4 hours to detect IL-23 p19 (A), IL-12/23 p40 (B) and IL-12 p35 (C) mRNA expression by qRT-PCR. qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the results from the un-treated group, which was set as 1. Results shown are mean plus SD of three to four independent experiments.
IFN-γ suppresses IL-23 p19 mRNA expression by inhibiting p19 mRNA stability
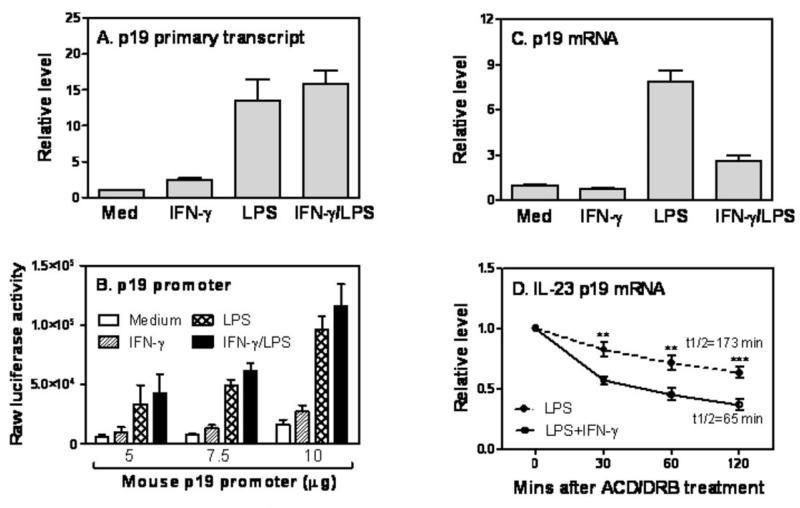
The level of steady-state mRNA is determined by both de novo mRNA synthesis and mRNA stability. To explore the mechanisms of IFN-γ-mediated p19 mRNA inhibition, we measured the nascent p19 primary transcript rate by qRT-PCR using a pair of primers corresponding to intron 1 and exon 2 of mouse p19 gene. As shown in Fig. 2A, LPS strongly induced p19 gene primary transcript and addition of IFN-γ did not cause any inhibitory effect, suggesting that IFN-γ-mediated p19 mRNA inhibition may be regulated at the posttranscriptional level. To confirm this, we cloned the murine p19 promoter into a luciferase expression plasmid and transiently transfected this p19 promoter-luciferase construct into RAW cells by electroporation, and then studied the effects of IFN-γ and LPS treatment on p19 promoter activity. Though LPS activated p19 promoter, addition of IFN-γ to LPS-stimulated cells did not inhibit but slightly increased p19 promoter activity (Fig. 2B) consistent with the primary transcript data. Fig. 2C confirms that RAW cells used in the transfection assay behaved similarly to primary macrophages and DCs, responding to IFN-γ treatment with p19 mRNA inhibition. Since both primary transcript and promoter data suggested that the inhibitory effects of IFN-γ on p19 expression are regulated at the posttranscriptional level, we next measured the half-life of p19 mRNA after blocking de novo RNA synthesis with ACD and DRB. LPS-induced p19 mRNA expression was significantly inhibited by IFN-γ at 0 min after ACD/DRB treatment (Data not shown). More importantly, the half-life of p19 mRNA in LPS-treated cells (t1/2=173 min) was significantly shortened after adding IFN-γ (t1/2=65 min) (Fig. 2D). Taken together, all data indicate that IFN-γ inhibits LPS-induced IL-23 p19 mRNA expression at the posttranscriptional level by promoting p19 mRNA degradation.
Figure 2. Posttranscriptional regulation of IL-23 p19 mRNA by IFN-γ.
(A) Total RNA was extracted from BMDCs treated with IFN-γ, LPS, or IFN-γ plus LPS for 1 h and reverse transcribed into cDNA using random primers. A pair of primers flanking intron1 and exon2 of the p19 gene was used to measure p19 primary transcripts by qRT-PCR. Data represents one of two experiments with similar results. (B) 10 × 106 RAW cells were transiently transfected with various amounts of mouse p19 promoter as indicated. 40 hrs later, the transfected cells were treated with IFN-γ (10 ng/ml), LPS (1 μg/ml) or IFN-γ plus LPS for 7 h, followed by lysis of the cells and measurement of luciferase activity by luminometer. Results shown are mean plus SD of four experiments. (C) Total RNA was extracted from RAW cells treated with IFN-γ (10 ng/ml), LPS (1 μg/ml) and IFN-γ plus LPS for 4 h and p19 mRNA expression measured by qRT-PCR. Data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the results from the un-treated group, which was set as 1. (D) 3 × 106 BMDMs were treated with LPS or IFN-γ plus LPS for 60 min. Then Actinomycin D (10 μg/ml) and DRB (50 μM) were added to all conditions, followed by collection of total RNA at 0, 30, 60, 120 min after ACD and DRB treatment. The remaining p19 mRNA levels were measured by qRT-PCR and compared between LPS and LPS plus IFN-γ treated cells at each time point. Data were normalized relative to GAPDH mRNA expression levels and further normalized to the results collected at 0 min after ACD/DRB treatment in each condition, which was set as 1. p value indicates the difference between two groups treated by LPS and IFN-γ plus LPS at each time point (n=3).
IL-23 p19 3′UTR controls p19 mRNA stability
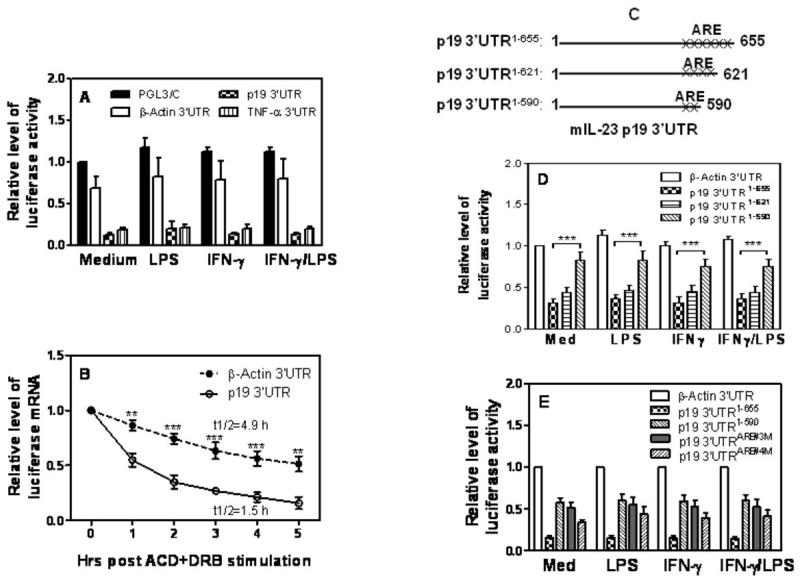
It is well known that the 3′UTR in the mRNA controls mRNA stability. Since IL-23 p19 mRNA contains a long 3′UTR (Supple Fig. 2), we decided to evaluate the role of p19 3′UTR in control of p19 mRNA degradation. We cloned the p19 3′UTR immediately downstream of the luciferase gene and transfected this construct into RAW cells by electroporation, followed by IFN-γ and LPS treatment. Since TNF-α mRNA stability is known to be controlled by its 3′UTR, we used TNF-α 3′UTR cloned into the same luciferase plasmid as a positive control. The same construct carrying β-Actin 3′UTR served as a negative control since its mRNA expression is quite stable. As shown in Fig. 3A, cells transfected with β-actin 3′UTR displayed a robust luciferase activity at a level similar to the activity observed in cells transfected with empty PGL3-control vector, indicating that β-actin 3′UTR does not affect luciferase gene stability. In contrast, luciferase activity was significantly reduced in cells transfected with the TNF-α 3′UTR, consistent with the notion that TNF-α 3′UTR enhances mRNA degradation. More importantly, the lucifease activity was reduced to an even lower level in cells transfected with p19 3′UTR compared to cells transfected with TNF-α 3′UTR (Fig. 3A). Since Luciferase activity detected in the reporter assay depends on translational efficiency as well as transcript stability. The reduced luciferase activity only indicates a possibility of decreased transcript stability. To directly test transcript stability, we measured actual decay rates of the reporter mRNA by measuring the half-life of luciferase mRNA. As shown in Fig. 3B, the half-life of luciferase mRNA was significantly shortened in cells transfected with the p19 3′UTR compared with β-actin-transfeced cells, confirming that p19 3′UTR promotes mRNA decay.
Figure 3. Degradation of p19 mRNA is mediated through AREs in the 3′UTR.
(A) 5 μg of luciferase constructs carrying the 3′UTR of genes encoding β-actin (β-Actin 3′UTR), TNF-α (TNF-α 3′UTR), or p19 (p19 3′UTR) or empty vector (PGL3/C) were transfected into RAW cells by electroporation. The transfected cells were treated with IFN-γ (10 ng/ml), LPS (1 μg/ml), or IFN-γ and LPS for 7 hours, followed by detection of luciferase activity in cell lysates. Luciferase activities were normalized to the activities obtained in PGL3/C-transfected cells without treatment (Medium), which was set as 1. (B) 5 μg of β-Actin 3′UTR or p19 3′UTR plasmids were respectively transfected into RAW cells by electroporation. 24 hrs later the transfected cells were treated with ACD (10 μg/ml) and DRB (50 μM), followed by collection of total RNA at different time points as indicated for measurement of luciferase mRNA expression by qRT-PCR. Data were normalized to the results at 0 min after ACD and DRB treatment. p value indicates the difference between two groups at each time point. (C) Schematics of the p19 3′UTR deletion constructs containing different AREs. The numbers of “x” represent the numbers of putative AREs in p19 3′UTR adjacent to the 3′ end. (D) A series of p19 3′UTR deletion constructs were transfected into RAW cells by electroporation. The transfected cells were treated with IFN-γ (10 ng/ml), LPS (1 μg/ml), or IFN-γ and LPS for 7 hours, followed by measurement of luciferase activity in cell lysates. (E) The full-length p19 3′UTR construct (p19 3′UTR1-655), deletion construct (p19 3′UTR1-590), and two ARE mutants (p19 3′UTRARE#3M and p19 3′UTRARE#4M) were transiently transfected into RAW cells by electroporation as described above. The transfected cells were treated with IFN-γ (10 ng/ml), LPS (1 μg/ml), or IFN-γ plus LPS for 7 hours, followed by measurement of luciferase activities in cell lysates. Luciferase activities were normalized to the activities obtained in the β-actin 3′UTR transfected cells without treatment which was set as 1. All result shown represents mean plus SD of three to four separate experiments.
IL-23 p19 3′UTR contains six adenine-uridine rich elements (AUUUA) close to its 3′end (Fig. 3C and supple Fig. 2). To determine which ARE mediates p19 mRNA degradation, we generated two deletion constructs, p19 3′UTR1-621 and p19 3′UTR1-590, by deleting each two AREs from the 3′end as elucidated in Fig. 3C. These two deletion constructs and the full-length plasmid (p19 3′UTR1-655) were respectively transfected into RAW cells followed by IFN-γ and LPS treatment and measurement of luciferase activity. As shown in Fig. 3D, cells transfected with the p19 3′UTR1-655 and p19 3′UTR1-621 (two AREs being deleted from the 3′ end) both displayed reduced luciferase activity compared to cells transfected with the β-Actin 3′UTR. However, cells transfected with the p19 3′UTR1-590 plasmid (four AREs being deleted from the 3′end) showed much higher luciferase activity than cells transfected with the plasmid deleting only two p19 3′UTR, suggesting that the two AREs (ARE#3 & #4) localized between 590 and 621 sites in the 3′UTR are critical for p19 mRNA degradation. To further confirm the importance of ARE#3 & #4 in control of p19 mRNA degradation, we individually mutated ARE#3 and ARE#4 in the p19 3′UTR and transfected these two mutants into RAW cells with the full-length p19 3′UTR as a control. Cells transfected with the ARE#3 & #4 mutants displayed higher luciferase activity similar to the level of cells transfected with p19 3′UTR1-590, with a stronger recovery in ARE#3 mutant than ARE#4 mutant (Fig. 3E), suggesting that the ARE#3 site in the p19 3′UTR is more important than ARE#4 site in control of p19 mRNA degradation.
Tristetraprolin enhances IL-23 p19 mRNA degradation
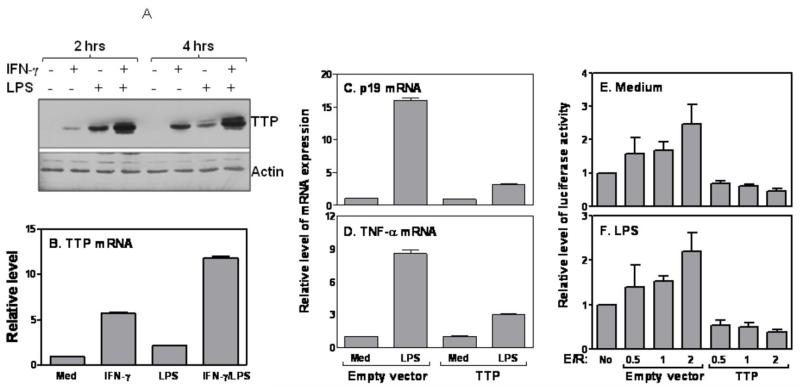
Since Tristetraproline (TTP) is known to mediate mRNA degradation of many cytokines, we wanted to test whether the TTP might be also responsible for IFN-γ-mediated p19 mRNA degradation. We stimulated macrophages with IFN-γ or LPS for either 2 or 4 hrs, followed by extraction of whole cell lysates for measurement of TTP protein by Western blot. Indeed, as reported previously (26) IFN-γ significantly increased LPS-induced TTP protein production at both time points (Fig. 4A). Interestingly, LPS appeared to induce TTP expression earlier than IFN-γ; TTP was hardly induced by IFN-γ at 2 hrs after stimulation and reached an equivalent level as LPS-treated cells 4 hrs after treatment (Fig. 4A). This phenomenon was also seen in BMDMs and RAW cells (Data not shown). TTP mRNA expression was also further enhanced by addition of IFN-γ to LPS-stimulated cells (Fig. 4B). To determine if TTP could suppress p19 mRNA expression, we transfected TTP and control plasmids into RAW cells followed by measurement of p19 mRNA expression after LPS stimulation. TNF-α, a major cytokine known to be regulated by TTP (25), was used as a positive control. Overexpression of TTP markedly inhibited LPS-induced p19 (Fig. 4C) and TNF-α (Fig. 4D) mRNA expression, indicating that TTP may regulate p19 and TNF-α mRNA expression similarly. To further determine the effect of TTP on p19 3′UTR-mediated mRNA degradation, we co-transfected TTP with p19 3′UTR-linked luciferase vector into RAW cells followed by LPS treatment. Overexpression of TTP suppressed p19 3′UTR dependent luciferase activity in both untreated (Fig. 4E) and LPS-treated conditions (Fig. 4F), supporting that TTP promotes p19 3′UTR-mediated luciferase gene degradation. Consistent with previous studies (17), TTP destabilized TNF-α 3′UTR-dependent luciferase activity in both basal and LPS-treated conditions (Data not shown).
Figure 4. Tristetraprolin controls IL-23 p19 mRNA degradation.
(A) 5 × 10 RAW cells were stimulated with IFN-γ (10 ng/ml), LPS (1 μg/ml) or IFN-γ plus LPS for different times as indicated, followed by collection of whole cell lysates for Western blot with anti-TTP antibody. The same blot was stripped and reanalyzed with anti-Actin antibody. (B) 5 × 106 RAW cells were treated with IFN-γ (10 ng/ml), LPS (1 μg/ml) or IFN-γ plus LPS for 4 h followed by extraction of total RNA to measure TTP mRNA expression by qRT-PCR. 5 μg of TTP expression or empty vectors were transiently transfected into RAW cells and the transfected cells were stimulated with or without LPS (1 μg/ml) for 4 h, followed by measurement of p19 (C) and TNF-α (D) mRNA expression by qRT-PCR. 5 μg of p19 3′UTR1-655 luciferase construct was co-transfected with different amounts of TTP expression or empty vectors (effector vs. reporter= 0.5, 1 & 2) into RAW cells and the transfected cells were either left untreated (E) or treated with LPS (F) for 7 h. The luciferase activities were measured in cell lysates and normalized to the activities obtained in cells transfected with empty vector with no treatment. All qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the results from the un-treated group, which was set as 1. The results shown are mean plus SD of three to four independent experiments.
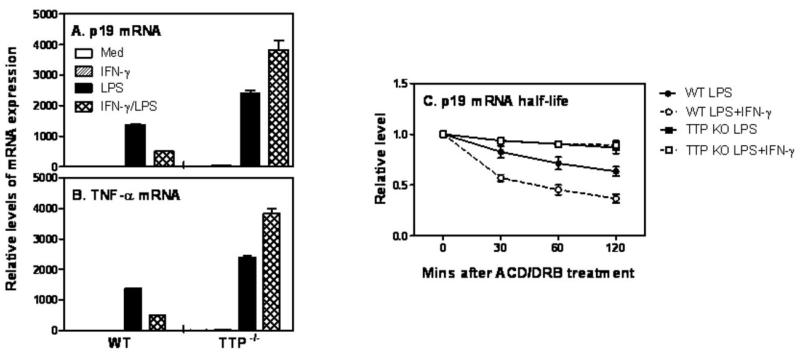
IFN-γ-mediated p19 mRNA degradation is rescued in TTP deficient mice
To further confirm the destabilization effect of TTP on p19 mRNA expression, we isolated peritoneal macrophages from TTP deficient mice and their littermates, and stimulated these cells with IFN-γ and LPS followed by measurement of p19 mRNA expression. Consistent with the previous data in BMDCs (Fig. 1) and macrophages (Supple Fig. 1), IFN-γ inhibited LPS-induced p19 mRNA expression in WT cells (Fig. 5A). More importantly, this inhibitory effect was completely reversed in TTP deficient cells (Fig. 5A), demonstrates that TTP is indeed required for IFN-γ-mediated inhibition of p19 mRNA expression. Interestingly, p19 mRNA expression induced by LPS was also significantly increased in TTP deficient cells compared to WT cells (Fig. 5A), Consistent with previous reports (17, 25), TNF-α expression was significantly increased in TTP−/− cells stimulated with either LPS or IFN-γ plus LPS compared with WT cells (Fig. 5B). To further confirm the effect of TTP on p19 mRNA decay, we measured the half-life of p19 mRNA in WT and TTP deficient cells stimulated with LPS or IFN-γ plus LPS. IFN-γ promoted p19 mRNA decay in WT cells treated with LPS (Fig. 5C). More importantly, the destabilized p19 mRNA induced by IFN-γ was stabilized in TTP deficient cells and even LPS-induced p19 mRNA was stabilized in TTP knockout cells (Fig. 5C), demonstrate that TTP is essential for IFN-γ-mediated p19 mRNA degradation.
Figure 5. IFN-γ-mediated p19 mRNA degradation is reversed in TTP deficient macrophages.
(A) 1 × 106 mouse peritoneal macrophages isolated from WT and TTP−/− mice were inoculated into each well of 24 well-plates with 1 ml RPMI1640 complete medium. The cells were stimulated with IFN-γ (10 ng/ml), LPS (1 μg/ml) or IFN-γ plus LPS for 4 hours, followed by extraction of total RNA for measurement of p19 (A) and TNF-α (B) mRNA expression by qRT-PCR. qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the results from the un-treated group. (C) 3×106 BMDMs differentiated from bone marrow cells of WT and TTP knockout mice were treated with LPS (1 μg/ml) or IFN-γ (10 ng/ml) plus LPS for 60 min. Then Actinomycin D (10 μg/ml) and DRB (50 μM) were added to all conditions, followed by collection of total RNA at 0, 30, 60, 120 min after ACD and DRB treatment. The remaining p19 mRNAs were measured by qRT-PCR and normalized relative to GAPDH mRNA expression levels and further normalized to the results obtained at 0 min after ACD/DRB treatment in each condition, which was set as 1. Data shown are mean plus SD from three experiments.
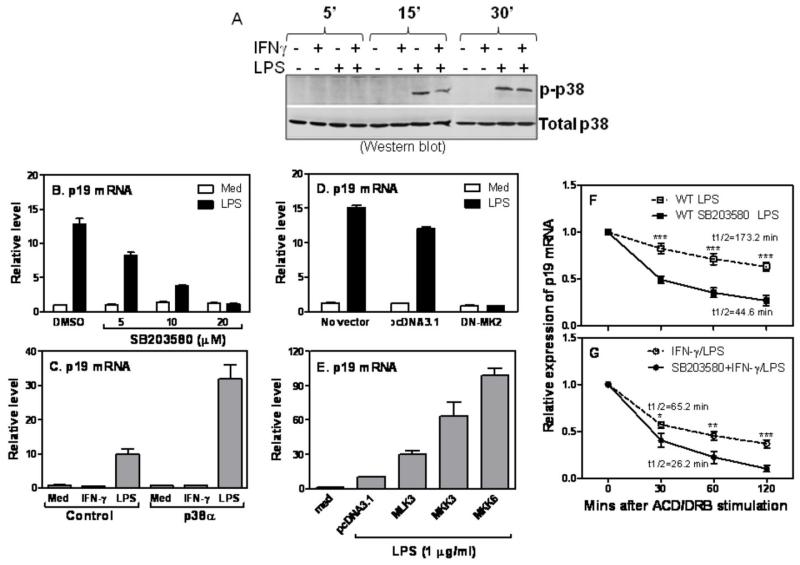
p38 MAPK is involved in stabilizing p19 mRNA expression
It has been shown that p38 MAPK is involved in the control of mRNA stability of many cytokines (40, 41). To determine whether p38 MAPK was involved in IFN-γ-mediated p19 mRNA degradation, we first measured phosphorylated p38 upon LPS and IFN-γ stimulation. The level of total p38 was used as loading control. As shown in Fig. 6A, LPS induced p38 phosphorylation in a time-dependent manner starting as early as 15 min and increasing up to 30 min after stimulation. Addition of IFN-γ inhibited LPS-induced p38 phosphorylation with the most dramatic effects at 15 min after stimulation (Fig. 6A). We next studied the involvement of p38 MAPK in p19 mRNA expression by blocking p38 signaling with SB203583. Blockade of p38 pathway with 20 μM SB203580 completely abrogated LPS-induced p19 mRNA expression (Fig. 6B). On the other hand, overexpression of p38-α significantly enhanced LPS-induced p19 mRNA expression compared with empty vector-transfected cells (Fig. 6C). To dissect which component in p38 pathway is involved in p19 mRNA induction, we first transfected a dominant negative mutant of MK2 (DN-MK2) or empty vector into RAW cells, followed by LPS treatment. Blocking MK2 by DN-MK2 completely abolished p19 mRNA expression induced by LPS (Fig. 6D), suggesting that p38 acts through the downstream molecule MK2 to induce p19 expression. To determine which upstream molecule of p38 pathway involves in p19 expression, we individually transfected MLK3, MKK3 and MKK6 or empty vector into RAW cells followed by LPS treatment. As shown in Fig. 6E, all three upstream molecules enhanced LPS-induced p19 mRNA expression, with MKK6 the strongest. To further confirm the role of p38 signaling in p19 mRNA degradation, we measured the half-life of p19 mRNA after blocking p38 pathway in LPS- or IFN-γ plus LPS-treated cells. Blockade of p38 by SB203580 promoted p19 mRNA decay in LPS-treated cells (Fig. 6F) and further shortened the half-life of p19 mRNA in cells treated with IFN-γ plus LPS (Fig. 6G), indicating that p38 signaling pathway plays a critical role in stabilizing p19 mRNA expression.
Figure 6. p38 MAPK is involved in stabilizing p19 mRNA expression.
(A) 5 × 106 RAW cells were stimulated with IFN-γ (10 ng/ml), LPS (1 μg/ml), or IFN-γ plus LPS for 5, 15 and 30 min, followed by collection of whole cell lysates to measure phosphorylated p38 expression by Western blot (upper panel). The same blot was stripped and reanalyzed with anti-total p38 antibody (lower panel). (B) 3 × 106 BMDMs were pre-treated with 5, 10 and 20 μM of SB203580 for 60 min prior to LPS stimulation. The same amount of dissolvent DMSO was used as a negative control. 4 h after LPS stimulation, total RNA was extracted and p19 mRNA expression was measured by qRT-PCR. (C) 5 μg of p38 expression or empty plasmids were transiently transfected into RAW cells followed by IFN-γ (10 ng/ml) or LPS (1 μg/ml) treatment for 4 h to detect p19 mRNA expression by qRT-PCR. (D) 5 μg of DN-MK2 expression or empty plasmids were transiently transfected into RAW cells followed by LPS (1 μg/ml) treatment for 4 h to detect p19 mRNA expression by qRT-PCR. (E) 5 μg of MLK3, MKK3 and MKK6 expression or empty plasmids were transiently transfected into RAW cells. The transfected cells were treated by LPS for 4 h followed by detection of p19 mRNA expression by qRT-PCR. All qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the results from the un-treated group. 3×106 BMDMs were pre-treated with SB203580 (10 μM) for 60 min, followed by treatment with LPS (F) or IFN-γ plus LPS (G) for additional 60 min. Then Actinomycin D (10 μg/ml) and DRB (50 μM) were added to all conditions, followed by collection of total RNA at 0, 30, 60, 120 min after ACD and DRB treatment. The remaining p19 mRNAs were measured by qRT-PCR and normalized relative to GAPDH mRNA expression levels and further normalized to the results obtained at 0 min after ACD/DRB treatment in each condition, which was set as 1. Data shown are mean plus SD from three experiments. p value indicates the difference between two groups at each time point.
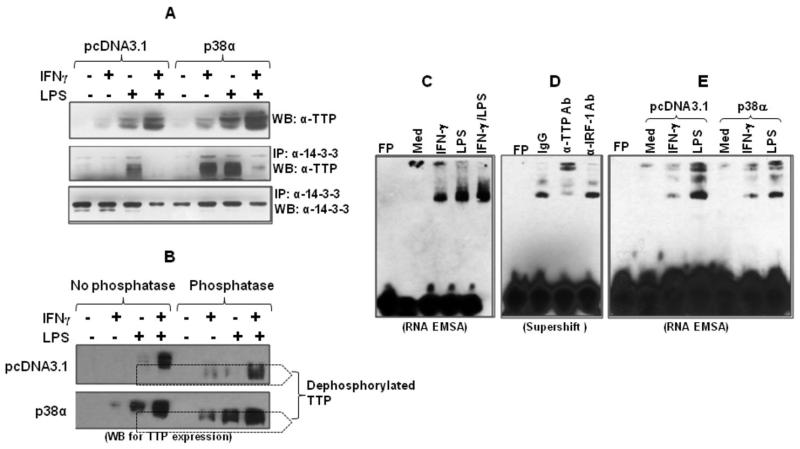
p38 inhibits TTP binding to the p19 3′UTR
The binding of TTP to AREs is controlled by protein phosphorylation. Once phosphorylated, TTP dissociates from ARE leading to stabilization of targeted mRNA. On the other hand, dephosphorylation of TTP leads to stronger TTP binding to the ARE causing enhanced mRNA degradation and less cytokine production (42). To determine the effects of IFN-γ and p38 on TTP dephosphorylation, we transfected p38α into RAW cells and treated the cells with IFN-γ and LPS, followed by extraction of whole cell lysates. The phosphorylated TTP was immunoprecipitated first with phospho-14-3-3 antibody followed by detection of TTP protein by western blot. As shown in Fig. 7A, IFN-γ enhanced LPS-induced TTP protein expression and overexpression of p38 further increased TTP protein expression (upper panel). Interestingly, overexpression of p38α significantly increased TTP phosphorylation induced by IFN-γ or LPS while suppressing TTP phosphorylation in cells co-stimulated with IFN-γ plus LPS (middle panel). The expression of total phosphorylated protein was similar among all groups (lower panel). In addition, as shown in figure 7B, digestion of whole cell lysates by lambda phosphatase further confirmed that IFN-γ increased TTP dephosphorylation in LPS-treated cells (upper panel) and overexpression of p38 enhanced TTP phosphorylation (lower panel). More importantly, IFN-γ treatment increased the expression of dephosphorylated TTP in p38-overexpressed cells (lower panel in Fig. 7B), suggesting that IFN-γ suppresses p38-mediated TTP phosphorylation resulting in increased TTP dephosphorylation.
Fig. 7. Overexpression of p38 MAPK inhibits TTP binding to the p19 3′UTR.
(A) 5 μg of p38 expression or empty vectors were transiently transfected into RAW cells. 24 h later the cells were stimulated with IFN-γ (10 ng/ml), LPS (1 μg/ml), or IFN-γ plus LPS for 4 h, followed by extraction of whole cell lysates to detect TTP protein by Western blot (upper panel). 100 μg of cytoplasmic protein was immunoprecipitated with 2 μg of anti phospho-14-4-3 antibody and the IPed products were used to detect TTP expression with anti-TTP antibody (middle panel). The same blot was stripped and reanalyzed with anti-phospho-14-3-3 antibody (lower panel). (B) RAW cells were transiently transfected with p38-α or empty vector pcDNA3.1 as described above and stimulated by IFN-γ (10 ng/ml), LPS (1 μg/ml) or LPS plus IFN-γ for 2 h, followed by extraction of whole cell lysates. 50 μg of lysates were digested with 50 U lambda protein phosphates for 2 h at 30°C. The digested product and equal amount of undigested lysates were used for detection of TTP expression by western blot. (C) RNA-LightShift Chemiluminescent EMSA was performed by incubating biotin-labeled RNA probe with 10 μg cytoplasmic protein. Cytoplasmic proteins were isolated from RAW cells treated with IFN-γ (10 ng/ml), LPS (1 μg/ml) and IFN-γ plus LPS for 4 hrs as we previously described. Med: medium condition. (D) 10 μg of cytoplasmic protein was incubated with 1 ug of anti-TTP antibody and unrelated anti-IRF-1 antibody for 20 min, followed by performing RNA-EMSA assay. (E) 5 μg of p38 expression or empty vectors were transiently transfected into RAW cells. 24 h later the cells were stimulated with IFN-γ (10 ng/ml) or LPS (1 μg/ml) for 4 h, followed by extraction of cytoplasmic protein to detect TTP binding by RNA-EMSA. 10 μg of protein was used for each sample. Data shown are one of two-three experiments with similar results.
To identify if TTP physically bound to the p19 3′UTR, we performed RNA-EMSA using a RNA sequence encompassing the ARE #3 site in the p19 3′UTR as a probe and incubated the biotin-labeled probe with cytoplasmic protein extracted from the cells treated with IFN-γ or IFNγ plus LPS. As shown in Fig. 7C, there was a unique RNA-cytoplasmic protein binding complex formed with the p19 3′UTR after IFN-γ or LPS treatment. More importantly, the binding intensity was increased by addition of IFN-γ to LPS-treated cells (Fig. 7C). To confirm this unique binding complex contained TTP, we added anti-TTP antibody to the cytoplasmic protein collected from the cells stimulated with IFN-γ plus LPS, followed by supershift RNA-EMSA. The binding intensity was significantly reduced and supershifted by adding anti-TTP antibody but not anti-IRF-1 antibody, confirming that the IFN-γ-enhanced binding complex indeed contains TTP. We next wanted to determine whether p38 actually affected TTP binding to the p19 3′UTR. LPS induced a strong TTP binding in empty vector-transfected cells whereas overexpression of p38α reduced TTP binding to p19 3′UTR (Fig. 7E). Taken together, these data indicate that IFN-γ inhibits p38-mediated TTP phosphorylation in cells stimulated with LPS, resulting in TTP dephosphorylation and stronger binding of TTP to the p19 3′UTR and p19 mRNA degradation.
Discussion
IFN-γ has differential effects on Th1 and Th17 cell development (promoting Th1 and suppressing Th17 cells), but the underlining mechanisms are not fully understood. Our results indicate that the differential effects of IFN-γ on Th1 and Th17 cells may act through differential regulation of IL-12 and IL-23 production. Since the shared p40 subunit between IL-12 and IL-23 is always produced in large excess compared with the p19 and p35 subunits, we focused our study on the regulation of limiting factor p19 subunit. Consistent with previous reports that IL-23 p19 gene transcription depends on MyD88 (14) and NF-κB (12, 13), our data confirm that LPS-induced p19 mRNA expression was completely abolished in macrophages isolated from MyD88−/− (Supple. Fig. 3A) and NF-κB c-Rel−/− (Supple. Fig. 3B) mice, demonstrate the importance of transcriptional control of p19 gene expression. To determine whether IFN-γ-mediated p19 mRNA inhibition depended on new protein synthesis, we used cyclohexamide (CHX) to block de novo protein synthesis prior to IFN-γ treatment, followed by measurement of p19 mRNA expression. The inhibition of p19 mRNA expression by IFN-γ was intact in CHX-treated cells (Supple. Fig. 4A) whereas the induction of p40 expression by IFN-γ and LPS was abolished after CHX pre-treatment (Supple. Fig. 4B), suggesting that IFN-γ-mediated p19 mRNA inhibition is a primary response and does not need new protein synthesis whereas IFN-γ-mediated p40 induction needs new protein synthesis through a secondary response.
Our data indicated that IFN-γ-mediated p19 mRNA inhibition is regulated at the posttranscriptional level since IFN-γ did not suppress LPS-induced p19 primary transcript (Fig. 2A) or promoter activity (Fig. 2B), instead enhanced p19 mRNA degradation (Fig. 2D). In fact, data from other labs also indicated that IFN-γ did not inhibit LPS-induced p19 promoter activity (13, 14). These results are different from a recent report that IFN-γ inhibited IL-23 expression at the promoter level through reduction of NF-κB binding to the p19 promoter (43). We reason that this difference could be due to the alteration of NF-κB binding and activity affected by TTP. This is not surprising because IFN-γ enhanced LPS-induced TTP expression (Fig. 4A&B) that could inhibit NF-κB activation through either blocking NF-κB p65 nuclear translocation (44) or directly binding to the p65 subunit (45). Therefore, it is possible that the increased TTP induced by IFN-γ suppresses NF-κB activation and/or nuclear translocation that results in reduced p19 transcription and IL-23 production.
Posttranscriptional regulation of cytokine production is mediated by multiple steps, including nuclear export, cytoplasmic localization, translation initiation and mRNA decay (46). These regulatory pathways are coordinated to control the expression of pro-inflammatory and anti-inflammatory cytokines to turn on or off inflammatory responses (46). One of mRNA decay mechanisms is ARE-binding proteins binding to the 3′UTR of target genes causing mRNA degradation. In deed, IL-23 p19 mRNA has a long 3′UTR containing at least six AREs close to its 3′ end (Supple Fig. 2). A series of deletions of these AREs in the p19 3′UTR further indicated that the ARE3 and ARE4 are the major sites that mediate p19 mRNA degradation. However, other AREs in the p19 3′UTR may also participate in regulating mRNA stability since deletion of four AREs adjacent to the 3′end did not result in a complete recovery of p19 3′UTR dependent luciferase activity (Fig. 3D&E). Thought the luciferase reporter assays are useful for mapping a negative regulatory element in the p19 3′UTR, we notice that none of the reporters appear to show responses to LPS stimulation. This lack of response of the p19 3′UTR-luciferase reporters to LPS stimulation could be due to a lack of LPS response element in the p19 3′UTR. It has been reported that interferons could limit inflammatory responses through induction of TTP that promotes degradation of several ARE-containing mRNAs. IFN-γ-mediated TTP induction is dependent on transcription factor STAT1 and the co-stimulatory stress signal requires p38 signaling pathway, suggesting an immunomodulatory role for IFN-γ during inflammation (26). We found that overexpression of TTP reduced p19 3′UTR dependent luciferase activity (Fig. 4E&F), indicating that TTP inhibits p19 mRNA stability through p19 3′UTR. In line with this finding, IL-23 p19 mRNA expression was significantly enhanced (Fig. 5A) and stabilized (Fig. 5C) in macrophages isolated from the TTP deficient mice. Interestingly, LPS-induced p19 mRNA expression was also stabilized in TTP knockout cells (Fig. 5C), indicating that the expression of p19 mRNA in response to LPS stimulation is a result of dynamic balance between transcription and decay. It has been reported that TTP−/− mice appeared normal at birth, but soon developed patchy alopecia, dermatitis, erosive arthritis, cachexia, conjunctivitis, and other autoimmunity manifestations as well as a severe loss of body weight (25). Similar to these previous findings, we observed swollen ankles and reduced body weight in TTP−/− mice (Supple Fig. 5A&B).
Activation of p38 MAPK by pro-inflammatory stimuli such as LPS causes p38 phosphorylation that affects p19 gene expression through transcription factor AP-1 (14). Our results demonstrate that IFN-γ inhibited p38 phosphorylation as early as 15 min after LPS stimulation (Fig. 6A) and p38 significantly increased LPS-induced p19 mRNA expression (Fig. 6 B&C). Our data further indicate that MKK3/6 and MK2 are involved in p38 signal transduction (Fig. 6D&E). p38 can induce TTP phosphorylation, resulting in disassociation of TTP from its targeted 3′UTR and stabilized p18 mRNA. Conversely, dephosphorylation of TTP leads to firm binding of TTP to 3′UTR and causes mRNA degradation. Consistent with this notion, LPS induced TTP phosphorylation (Fig. 7A), led to disassociation of TTP from p19 3′UTR. In contrast, IFN-γ inhibited LPS-induced TTP phosphorylation and caused TTP dephosphorylation (Fig. 7A&B), which led to stronger binding of TTP to the p19 3′UTR and p19 mRNA degradation. Interestingly, overexpresson of p38 increased TTP phosphorylation induced by either IFN-γ or LPS alone but had no effect on IFN-γ plus LPS treated cells (Fig. 7A), further indicating that IFN-γ inhibits LPS-induced TTP phosphorylation. Moreover, overexpression of p38 reduced TTP binding to the p19 3′UTR (Fig. 7E). It is worth noting that despite overexpression of p38 enhanced IFN-γ-induced TTP phosphorylation (Fig. 7A&B), there was no p19 mRNA expression and IL-23 protein production in the absence of TLR signaling (Fig. 1 & Supple Fig. 1), indicating the importance of TLR signaling pathway in induction of IL-23 expression.
In summary, our study for the first time demonstrates that IFN-γ inhibits LPS-induced IL-23 production in DCs and macrophages through promoting p19 mRNA degradation. IFN-γ-induced p19 mRNA degradation is mediated via p38 signaling pathway through induction of TTP dephosphorylation resulting in stronger binding of TTP to the p19 3′UTR. The differential effects of IFN-γ on IL-12 and IL-23 expression help us understand the differential roles of IFN-γ on Th1 and Th17 cell development.
Supplementary Material
Acknowledgments
This project was supported, in whole or in part, by Grant Number AR055353 from the National Institutes of Health (to J.L)
Footnotes
Disclosures
The authors declare no competing financial interests.
References
- 1.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu M, Gorman D, Wagner J, Zurawski S, Liu Y, Abrams JS, Moore KW, Rennick D, de Waal-Malefyt R, Hannum C, Bazan JF, Kastelein RA. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 2.Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu Rev Immunol. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 3.Cocco C, Canale S, Frasson C, Di Carlo E, Ognio E, Ribatti D, Prigione I, Basso G, Airoldi I. Interleukin-23 acts as antitumor agent on childhood B-acute lymphoblastic leukemia cells. Blood. 2010;116:3887–3898. doi: 10.1182/blood-2009-10-248245. [DOI] [PubMed] [Google Scholar]
- 4.Lee E, Trepicchio WL, Oestreicher JL, Pittman D, Wang F, Chamian F, Dhodapkar M, Krueger JG. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J Exp Med. 2004;199:125–130. doi: 10.1084/jem.20030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaknin-Dembinsky A, Balashov K, Weiner HL. IL-23 is increased in dendritic cells in multiple sclerosis and down-regulation of IL-23 by antisense oligos increases dendritic cell IL-10 production. J Immunol. 2006;176:7768–7774. doi: 10.4049/jimmunol.176.12.7768. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt C, Giese T, Ludwig B, Mueller-Molaian I, Marth T, Zeuzem S, Meuer SC, Stallmach A. Expression of interleukin-12-related cytokine transcripts in inflammatory bowel disease: elevated interleukin-23p19 and interleukin-27p28 in Crohn’s disease but not in ulcerative colitis. Inflamm Bowel Dis. 2005;11:16–23. doi: 10.1097/00054725-200501000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Kim HR, Kim HS, Park MK, Cho ML, Lee SH, Kim HY. The clinical role of IL-23p19 in patients with rheumatoid arthritis. Scand J Rheumatol. 2007;36:259–264. doi: 10.1080/03009740701286813. [DOI] [PubMed] [Google Scholar]
- 8.Kim HR, Cho ML, Kim KW, Juhn JY, Hwang SY, Yoon CH, Park SH, Lee SH, Kim HY. Up-regulation of IL-23p19 expression in rheumatoid arthritis synovial fibroblasts by IL-17 through PI3-kinase-, NF-kappaB- and p38 MAPK-dependent signalling pathways. Rheumatology (Oxford) 2007;46:57–64. doi: 10.1093/rheumatology/kel159. [DOI] [PubMed] [Google Scholar]
- 9.Liu FL, Chen CH, Chu SJ, Chen JH, Lai JH, Sytwu HK, Chang DM. Interleukin (IL)-23 p19 expression induced by IL-1beta in human fibroblast-like synoviocytes with rheumatoid arthritis via active nuclear factor-kappaB and AP-1 dependent pathway. Rheumatology (Oxford) 2007;46:1266–1273. doi: 10.1093/rheumatology/kem055. [DOI] [PubMed] [Google Scholar]
- 10.Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, Basham B, McClanahan T, Kastelein RA, Oft M. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Orozco N, Dong C. The IL-17/IL-23 axis of inflammation in cancer: friend or foe? Curr Opin Investig Drugs. 2009;10:543–549. [PubMed] [Google Scholar]
- 12.Carmody RJ, Ruan Q, Liou HC, Chen YH. Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J Immunol. 2007;178:186–191. doi: 10.4049/jimmunol.178.1.186. [DOI] [PubMed] [Google Scholar]
- 13.Mise-Omata S, Kuroda E, Niikura J, Yamashita U, Obata Y, Doi TS. A proximal kappaB site in the IL-23 p19 promoter is responsible for RelA- and c-Rel-dependent transcription. J Immunol. 2007;179:6596–6603. doi: 10.4049/jimmunol.179.10.6596. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Ouyang X, Yang J, Liu J, Li Q, Gu Y, Fukata M, Lin T, He JC, Abreu M, Unkeless JC, Mayer L, Xiong H. AP-1 activated by toll-like receptors regulates expression of IL-23 p19. J Biol Chem. 2009;284:24006–24016. doi: 10.1074/jbc.M109.025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Salleeh F, Petro TM. Promoter analysis reveals critical roles for SMAD-3 and ATF-2 in expression of IL-23 p19 in macrophages. J Immunol. 2008;181:4523–4533. doi: 10.4049/jimmunol.181.7.4523. [DOI] [PubMed] [Google Scholar]
- 16.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 17.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 19.Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetraprolin binds to the 3′-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J Biol Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 20.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 21.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta S, Biswas R, Novotny M, Pavicic PG, Jr., Herjan T, Mandal P, Hamilton TA. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J Immunol. 2008;180:2545–2552. doi: 10.4049/jimmunol.180.4.2545. [DOI] [PubMed] [Google Scholar]
- 23.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 24.Phillips K, Kedersha N, Shen L, Blackshear PJ, Anderson P. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc Natl Acad Sci U S A. 2004;101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 26.Sauer I, Schaljo B, Vogl C, Gattermeier I, Kolbe T, Muller M, Blackshear PJ, Kovarik P. Interferons limit inflammatory responses by induction of tristetraprolin. Blood. 2006;107:4790–4797. doi: 10.1182/blood-2005-07-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakae S, Nambu A, Sudo K, Iwakura Y. Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J Immunol. 2003;171:6173–6177. doi: 10.4049/jimmunol.171.11.6173. [DOI] [PubMed] [Google Scholar]
- 28.Stumhofer JS, Laurence A, Wilson EH, Huang E, Tato CM, Johnson LM, Villarino AV, Huang Q, Yoshimura A, Sehy D, Saris CJ, O’Shea JJ, Hennighausen L, Ernst M, Hunter CA. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat Immunol. 2006;7:937–945. doi: 10.1038/ni1376. [DOI] [PubMed] [Google Scholar]
- 29.Batten M, Li J, Yi S, Kljavin NM, Danilenko DM, Lucas S, Lee J, de Sauvage FJ, Ghilardi N. Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nat Immunol. 2006;7:929–936. doi: 10.1038/ni1375. [DOI] [PubMed] [Google Scholar]
- 30.Aggarwal S, Gurney AL. IL-17: prototype member of an emerging cytokine family. J Leukoc Biol. 2002;71:1–8. [PubMed] [Google Scholar]
- 31.Fossiez F, Djossou O, Chomarat P, Flores-Romo L, Ait-Yahia S, Maat C, Pin JJ, Garrone P, Garcia E, Saeland S, Blanchard D, Gaillard C, Das Mahapatra B, Rouvier E, Golstein P, Banchereau J, Lebecque S. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. J Exp Med. 1996;183:2593–2603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones CE, Chan K. Interleukin-17 stimulates the expression of interleukin-8, growth-related oncogene-alpha, and granulocyte-colony-stimulating factor by human airway epithelial cells. Am J Respir Cell Mol Biol. 2002;26:748–753. doi: 10.1165/ajrcmb.26.6.4757. [DOI] [PubMed] [Google Scholar]
- 33.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolls JK, Kanaly ST, Ramsay AJ. Interleukin-17: an emerging role in lung inflammation. Am J Respir Cell Mol Biol. 2003;28:9–11. doi: 10.1165/rcmb.2002-0255PS. [DOI] [PubMed] [Google Scholar]
- 35.Moseley TA, Haudenschild DR, Rose L, Reddi AH. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 2003;14:155–174. doi: 10.1016/s1359-6101(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 36.Tato CM, Cua DJ. Reconciling id, ego, and superego within interleukin-23. Immunol Rev. 2008;226:103–111. doi: 10.1111/j.1600-065X.2008.00715.x. [DOI] [PubMed] [Google Scholar]
- 37.Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Qian X, Ning H, Yang J, Xiong H, Liu J. Activation of IL-27 p28 gene transcription by interferon regulatory factor 8 in cooperation with interferon regulatory factor 1. J Biol Chem. 2010;285:21269–21281. doi: 10.1074/jbc.M110.100818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Cao S, Herman LM, Ma X. Differential regulation of interleukin (IL)-12 p35 and p40 gene expression and interferon (IFN)-gamma-primed IL-12 production by IFN regulatory factor 1. J Exp Med. 2003;198:1265–1276. doi: 10.1084/jem.20030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JC, Young PR. Role of CSB/p38/RK stress response kinase in LPS and cytokine signaling mechanisms. J Leukoc Biol. 1996;59:152–157. doi: 10.1002/jlb.59.2.152. [DOI] [PubMed] [Google Scholar]
- 42.Anderson P, Phillips K, Stoecklin G, Kedersha N. Post-transcriptional regulation of proinflammatory proteins. J Leukoc Biol. 2004;76:42–47. doi: 10.1189/jlb.1103536. [DOI] [PubMed] [Google Scholar]
- 43.Sheikh SZ, Matsuoka K, Kobayashi T, Li F, Rubinas T, Plevy SE. Cutting edge: IFN-gamma is a negative regulator of IL-23 in murine macrophages and experimental colitis. J Immunol. 2010;184:4069–4073. doi: 10.4049/jimmunol.0903600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schichl YM, Resch U, Hofer-Warbinek R, de Martin R. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. J Biol Chem. 2009;284:29571–29581. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang J, Lei T, Song Y, Yanes N, Qi Y, Fu M. RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. J Biol Chem. 2009;284:29383–29390. doi: 10.1074/jbc.M109.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.