Abstract
Cartilage has limited repair and regeneration capacity, thus damage of cartilage often results in its dysfunction and even chronic diseases like osteoarthritis (OA). Chondrogenesis induced by tissue-engineering methods is essential to treating cartilage-related diseases. MicroRNAs (miRNAs) are a class of small non-coding single-stranded RNAs which exert their biological effects by binding to the target messenger RNAs (mRNAs), resulting in decay or translation suppression of target mRNAs. There are emerging evidence indicating that miRNAs may play important roles in regulating both prenatal and postnatal chondrogenesis. During embryonic skeletal development, prenatal chondrogenesis is thought to be a precondition for formation of cartilage in developing limbs. Plenty of studies on different types of stem cells have undoubtedly proven their capacity of differentiating into chondrocytes. MiRNAs are found to comprehensively modulate these processes by establishing an interaction network with target genes, transcription factors and cytokines et al. In addition, translational application of miRNA technology has also been explored. In this review, we focus on the up-dated progress on regulatory mechanisms of miRNAs in prenatal and postnatal chondrogenesis. In addition, several miRNA target genes and roles of miRNAs in cartilage-related diseases are also discussed. This will contribute to studies of chondrogenesis mechanisms and development of new treating methods.
Keywords: microRNA, chondrogenesis, stem cell, embryonic development, cartilage-related disease
Introduction
-
MicroRNAs in prenatal chondrogenesis
–MicroRNAs are essential in prenatal chondrogenesis
–Expression patterns of miRNAs in prenatal chondrogenesis
–Functions of miRNAs in prenatal chondrogenesis
MicroRNAs in postnatal chondrogenesis of stem cells in vitro
-
MicroRNAs in chondrogenic differentiation of mesenchymal stem cells
–Expression patterns of miRNAs during MSC chondrogenic differentiation
–Functions of miRNAs during MSC chondrogenic differentiation
MicroRNAs in chondrogenic differentiation of other types of stem cells
Target genes of miRNAs in chondrogenesis
-
MicroRNAs in cartilage-related diseases
–MicroRNAs in OA
–MicroRNAs in RA
–Application of microRNAs in translational medicine
Conclusion and Prospects
Introduction
Providing the characteristics of skeletal systems, cartilage is an avascular tissue without nerves, and it consists of only one cell type—chondrocytes. Thus, due to the simplicity of its formation and construction, the repair and regeneration capacity of cartilage is consequently limited, resulting in its vulnerability to changeable immediate environment. In some cases, damage of cartilage resulted from traumatic injuries or autoimmune reactions leads to its dysfunction, and even severe joint diseases, such as osteoarthritis (OA) and rheumatoid arthritis (RA). In recent years, there has been increasing demand for inducing chondrogenesis and regenerating completely functional cartilage to treat cartilage-related diseases 1–2. A number of tissue-engineering methods including cell therapy, gene therapy and biomaterial have been adopted to induce chondrogenesis and obtain functionally improved cartilage 3. However, it is still a challenge to regenerate completely effective and functional cartilage 4. Many metabolic and regulatory problems related with chondrogenesis and cartilage formation are still unknown. Therefore, the detailed mechanisms of chondrogenic differentiation need to be elucidated.
MicroRNAs (miRNAs) are a crowd of small non-coding single-stranded RNAs whose lengths are about 17–24 nucleotides. They often reside outside genes, or sometimes, within introns of other genes. Usually, miRNAs bind to the 3′-untranslated region (3′-UTR) of the target messenger RNAs (mRNAs) through perfect or imperfect complementary base pairing to induce mRNAs’ decay or translational suppression 5. The synthesis of miRNAs in cells is precisely regulated. Firstly, a miRNA gene is transcribed by RNA polymerase II or III in the nucleus, and the transcriptive product is called pri-miRNA, which contains the sequence of the mature miRNA in a stem-loop structure. Then, a pri-miRNA is regularly transferred to be cleaved by a protein complex comprising RNA endonuclease III (known as Dosha) and DGCR8 (DiGeorge syndrome critical region gene 8). This cleaving produces the precursor miRNA (pre-miRNA), which has a short stem-loop of 70–100 nucleotides in the nucleus 6. The pre-miRNA is exported by exportin-5 to the cytoplasm, in which the short stem-loop is removed by Dicer, another RNA endonuclease III, leaving a miRNA duplex alone 7. The double strands then are incorporated into the RNA-induced silencing complex (RISC), where the duplex binds to proteins of the Argonaute family to degrade one strand—passenger miRNA and release the other strand—mature miRNA. The released mature miRNA with RISC contains the seed sequence which interacts with 3′-UTR, or in unusual cases, 5′-UTR and protein coding regions of the target mRNA through fully or partially complementary base pairing 8. When the base pair match is perfect, RISC degrades the target mRNA; otherwise, RISC inhibits the translation of the target mRNA or deadenylates and/or dacaps the target mRNA 9. Up till now, published data have identified more than 1000 miRNAs in human, and it is estimated that they may target about 60% of total human mRNAs 10. Since miRNAs were first discovered in C. elegans in 1993 11–12, a lot of studies have proven that miRNAs exert their functions in a great variety of biological processes including embryo development, cell proliferation, cell differentiation, signal transduction, immune response and apoptosis 13,14.
For many years, the urgent demand of setting up interdisciplinary tissue-engineering strategies to treat cartilage-related diseases has always been addressed by both patients and doctors. Much evidence has proven that miRNAs play important roles in regulating chondrogenesis and pathogenesis of cartilage-related diseases 16. MiRNAs may be developed as new candidates for diagnostic markers and therapeutic targets in treating cartilage-related diseases. So in this review, we focus on miRNA functions in regulating prenatal chondrogenesis of embryonic development and postnatal chondrogenesis of different types of stem cells in vitro. In addition, we also discussed several miRNA target genes and roles of miRNAs in cartilage-related diseases.
MicroRNAs in prenatal chondrogenesis
MicroRNAs are essential in prenatal chondrogenesis
Chondrogenesis is believed to be the onset of embryonic skeletal development and a premise for formation of cartilage in developing limbs. During this process, mesenchymal cells are recruited and undergo a succession of alterations, including proliferation, progenitor condensation, differentiation, hypertrophy, mineralization, apoptosis and displacement by bone, resulting in cartilage formation and endochondral ossification 17–18. Many factors participate in the regulatory mechanisms of prenatal chondrogenesis, such as transcription factors, growth factors, extracellular matrix (ECM) and miRNAs. The depletion of RNA endonuclease Dicer severely injured proliferating chondrocytes and accelerated post-mitotic chondrocytic hypertrophy, leading to skeletal growth defects, and even deaths of mice 19. This result indicated that miRNAs were essential to chondrocyte survival and inhibition of premature hypertrophic differentiation of chondrocytes in skeletal development.
Expression patterns of miRNAs in prenatal chondrogenesis
The expression patterns of miRNAs in prenatal chondrogenesis are both species-specific and tissue-specific. In axolotls, two different microarray platforms identified overexpression of miR-21 in mid-bud blastemas compared with stump tissue 20. During chick development, miR-455-3p was expressed in developing long bones 21. In terms of tissue-specific expression patterns of miRNAs, a miRNA expression profiling study of medaka embryos found specific expression of miR-140 and miR-199a in developing cartilage compared with other tissues 19. In the developing zebrafish embryos, 10 miRNAs, including miR-23a, miR-27a, miR-27b, miR-140, miR-140*, miR-145, miR-146, miR-199a, miR-199a* and miR-214, were mainly found in pharyngeal arches, indicating that these miRNAs were highly specific in cartilaginous tissues 22. Moreover, the Solexa sequencing identified both increased and decreased miRNAs only in developing articular cartilage of rats 23. The aforementioned results displayed the species-specific and tissue-specific characteristics of miRNA expression and may provide research targets in the developmental biology.
Functions of miRNAs in prenatal chondrogenesis
MicroRNAs are critical regulators in prenatal chondrogenesis. In prenatal chondrogenesis, the morphological transition of mesenchymal cells from a fibroblastoid shape to a round one is a persuasive indicator of differentiation 24. In the chick embryonic development, the inhibition of miR-34a led to the rearrangement of actin cytoskeleton by modulating cross-talk between RhoA and Rac1. At the same time, miR-34a overexpression could cause intensified stress fibres 25. In addition, miR-34a could strongly constrain chondroblasts from migrating by targeting EphA5 and assist in Interleukin-1β-induced damage of chondrocytes. These findings suggested multiple functions of miR-34a in chick prenatal chondrogenesis 26. Position-dependent chondrogenesis is a common phenomenon in limb development 27. Kim et al. showed that by enhancing Adam9 transcription, miR-142-3p inhibited chondrocyte migration and pre-cartilage condensation and increased cell death. These alterations contributed to the limb type-specific outcome of transforming growth factor-β3 (TGF-β3) upon chick leg versus wing mesenchymal cells 28. Similarly, miR-375 was also found to negatively regulate chick chondrocyte migration and pre-cartilage condensation by down-regulating cadherin-729. Moreover, Kim et al. also confirmed that blockade of miR-221 obviously promoted proliferation of chick chondrogenic progenitors and repression of target Mdm2 by inhibiting ubiquitylation and proteosomal degradation of Slug protein 30. In terms of control of miRNAs, miR-140 was modulated by transcription factor Sox9 in the developing zebrafish, however, its function was independent of Sox9b 31. These research findings described above persuasively show that miRNAs play multifunctional and comprehensive roles in prenatal chondrogenesis of mammalian animals.
MicroRNAs in postnatal chondrogenesis of stem cells in vitro
The postnatal chondrogenic differentiation of different types of stem cells in vitro has been comprehensively studied and it is under a complex regulatory regime. The important roles of miRNAs in various types of stem cells, such as mesenchymal stem cells (MSCs), adipose-derived stem cells (ADSCs) and unrestricted somatic stem cells (USSCs), have been recognized and discussed. Up to now, a number of significant findings have presented individuals with a clearer picture of miRNA integral part of regulatory mechanisms in chondrogenic differentiation of stem cells.
MicroRNAs in chondrogenic differentiation of MSCs
Mesenchymal stem cells, also referred to as mesenchymal stromal cells, have the capacity of self-renew and differentiation into multiple cell lineages including osteocytes, chondrocytes, adipocytes, whereas the differentiation capacity into other cell types remains to be controversial 32,33. Many studies have identified miRNA expression profiling during chondrogenic differentiation of MSCs and these findings provide new insights into the detailed mechanisms of MSC chondrogenic differentiation.
Expression patterns of miRNAs during MSC chondrogenic differentiation
The aberrant expression patterns of miRNAs are identified during MSC chondrogenic differentiation. By using miRNA microarrays, five miRNAs (miR-26b, miR-28, miR-130b, miR-152, miR-193b) were significantly overexpressed in chondrogenic differentiated human MSCs compared with undifferentiated MSCs in two samples 35. Similarly, miRNA microarray analysis identified 10 up-regulated miRNAs (miR-15b, miR-16, miR-23b, miR-27b, miR-140, miR-148, miR-197, miR-222, miR-328 and miR-505) through comparison between human MSCs and human articular chondrocytes 36. Furthermore, in isolated human CD146+ MSCs, 12 miRNAs were up-regulated and 24 miRNAs were down-regulated during chondrogenic differentiation 37. In terms of mouse MSCs, eight miRNAs were strongly overexpressed and five miRNAs were under-expressed during TGF-β3-induced chondrogenic differentiation 38. In addition, the expression levels of seven miRNAs altered more than fivefold during chondrogenic differentiation of mouse MSCs 39. It should be noted that different studies found more or less different changes of miRNA expression patterns during chondrogenic differentiation of human MSCs, and so was the case in mouse MSCs. This inconsistency in different findings may be attributed to the various sources of MSCs 40–41, diverse culturing methods and different micro-environments of MSCs.
Functions of miRNAs during MSC chondrogenic differentiation
The special expression patterns of miRNAs during MSC chondrogenic differentiation are essential to miRNA function. Laine et al. found that during chondrogenic differentiation of human MSCs, miR-124 down-regulated ACAN expression, whereas miR-199a up-regulated ACAN expression 42. ACAN is a marker gene of chondrogenic differentiation. Besides, miR-455-3p acted on TGF-β signalling by targeting ACVR2B, SMAD2 and CHRDL1 21. In addition, miR-449a inhibited Lymphoid Enhancer Binding Factor-1(LEF-1) expression in a dose-dependent and sequence-specific manner during chondrogenesis of human MSCs. And inhibition of LEF-1 by miR-449a repressed expression of SOX9, leading to delayed progression of chondrogenesis 43. Moreover, a small molecule H-89 could induce miR-23b expression, and miR-23b was a positive regulator which therefore promoted chondrogenic differentiation of human MSCs by inhibiting protein kinase A (PKA) signalling 44. During chondrogenic differentiation of mouse MSCs, miR-145 repressed Sox9 expression and down-regulated mRNA levels such as Col2a1, Comp, Acan, Col9a2, which were all chondrogenic marker genes 45. In addition, through modulating transcription factor Smad1, miR-199a* decreased expression of mouse early chondrogenic marker genes 46. These findings clearly showed that by targeting certain genes and transcription factors, miRNAs contributed a lot to the regulation and modulation of chondrogenic differentiation of MSCs.
MicroRNAs in chondrogenic differentiation of other types of stem cells
Although MSCs show a variety of favourable cellular capacities in tissue engineering, there are other types of stem cells which have their own advantages and could also be good candidates. One example comes from the unrestricted somatic stem cell (hUSSC). USSCs can also differentiate into many cell lineages. Compared with MSCs, USSCs are less mature and have a longer life span and extended telomere length 47–48. By using miRNA microarray and RT-qPCR, Bakhshandeh et al. identified many up-regulated and down-regulated miRNAs and their families during USSC chondrogenic differentiation, such as miR-376, miR-624, miR-630 and miR-1268. Particularly, expression of miR-16 decreased about threefold in chondrocytes compared to hUSSCs. However, expression of miR-16 was increased in primary chondrocytes compared with hMSCs 36. The conflicting results for miR-16 may be due to the different characteristic of hUSSCs and hMSCs. Human MSC populations are quite heterogenous. Another reason may be that miR-16 is modulated by some other factors except for chondrogenesis 49. Another example is human adipose-derived stem cell (hADSC). ADSCs are capable of self-renewing and differentiating into multiple cell lineages. Besides, ADSCs are easily available since adipose tissue is abundant 50–51. MiRNA microarray analysis confirmed 12 differentially expressed miRNAs during chondrogenic differentiation of ADSCs, including miR-193b, miR-199a-3p/miR-199b-3p, miR-455-3p, miR-210, miR-381, miR-92a, miR-320c, miR-136, miR-490-5p, miR-4287, miR-BART8* and miR-US25-1* (Table 1). The predicted target genes of these microRNAs were also analysed, which exerted huge effects on chondrogenic differentiation of ADSCs 52. In addition, miR-194 inhibited chondrogenic differentiation of ADSCs by targeting SOX5 53. The functions of major miRNAs in chondrogenesis are listed in Table 2. These findings are significant and instructive, but there are still many unknown details which remain to be elucidated in the mechanisms of miRNA function.
Table 1.
Changes of microRNA expression during chondrogenic differentiation of different stem cells
| Name | Cell types | References | |
|---|---|---|---|
| Up-regulated microRNAs | miR-26b | hMSCs | 35 |
| miR-28 | hMSCs | 35 | |
| miR-130b | hMSCs | 35 | |
| miR-152 | hMSCs | 35 | |
| miR-193b | hMSCs | 35 | |
| miR-15b | hMSCs | 36 | |
| miR-16 | hMSCs | 36 | |
| miR-23b | hMSCs | 36 | |
| miR-27b | hMSCs | 36 | |
| miR-140 | hMSCs | 36 | |
| miR-148 | hMSCs | 36 | |
| miR-197 | hMSCs | 36 | |
| miR-222 | hMSCs | 36 | |
| miR-328 | hMSCs | 36 | |
| miR-505 | hMSCs | 36 | |
| miR-127 | Mouse MSCs | 38 | |
| miR-140 | Mouse MSCs | 38 | |
| miR-125b* | Mouse MSCs | 38 | |
| miR-99a | Mouse MSCs | 38 | |
| miR-140* | Mouse MSCs | 38 | |
| miR-181a-1 | Mouse MSCs | 38 | |
| let-7f | Mouse MSCs | 38 | |
| miR-30a | Mouse MSCs | 38 | |
| miR-24 | Mouse MSCs | 39 | |
| miR-101 | Mouse MSCs | 39 | |
| miR-124a | Mouse MSCs | 39 | |
| miR-199a | Mouse MSCs | 39 | |
| miR-199b | Mouse MSCs | 39 | |
| miR-1207-5p | hUSSCs | 49 | |
| miR-1225-5p | hUSSCs | 49 | |
| miR-1246 | hUSSCs | 49 | |
| miR-1268 | hUSSCs | 49 | |
| miR-1275 | hUSSCs | 49 | |
| miR-1287 | hUSSCs | 49 | |
| miR-1290 | hUSSCs | 49 | |
| miR-146a | hUSSCs | 49 | |
| miR-181a-1 | hUSSCs | 49 | |
| miR-188-5p | hUSSCs | 49 | |
| miR-1915 | hUSSCs | 49 | |
| miR-210 | hUSSCs | 49 | |
| miR-630 | hUSSCs | 49 | |
| miR-638 | hUSSCs | 49 | |
| miR-193b | hADSCs | 52 | |
| miR-199a-3p/miR-199b-3p | hADSCs | 52 | |
| miR-455-3p | hADSCs | 52 | |
| miR-210 | hADSCs | 52 | |
| miR-381 | hADSCs | 52 | |
| miR-92a | hADSCs | 52 | |
| miR-320c | hADSCs | 52 | |
| miR-136 | hADSCs | 52 | |
| Down-regulated microRNAs | miR-145 | Mouse MSCs | 38 |
| miR-212 | Mouse MSCs | 38 | |
| miR-132 | Mouse MSCs | 38 | |
| miR-143 | Mouse MSCs | 38 | |
| miR-125b-3p | Mouse MSCs | 38 | |
| miR-18 | Mouse MSCs | 38 | |
| miR-96 | Mouse MSCs | 38 | |
| miR-100 | hUSSCs | 49 | |
| miR-10b | hUSSCs | 49 | |
| miR-127-3p | hUSSCs | 49 | |
| miR-130a | hUSSCs | 49 | |
| miR-143 | hUSSCs | 49 | |
| miR-145 | hUSSCs | 49 | |
| miR-16 | hUSSCs | 49 | |
| miR-221 | hUSSCs | 49 | |
| miR-376a | hUSSCs | 49 | |
| miR-376c | hUSSCs | 49 | |
| miR-377 | hUSSCs | 49 | |
| miR-379 | hUSSCs | 49 | |
| miR-490-5p | hUSSCs | 49 | |
| miR-490-5p | hADSCs | 52 | |
| miR-4287 | hADSCs | 52 | |
| miR-BART8* | hADSCs | 52 | |
| miR-US25-1* | hADSCs | 52 |
Table 2.
Functions of major microRNA in chondrogenesis
| Name | Types of chondrogenesis | Targets | Function | Reference |
|---|---|---|---|---|
| miR-34a | Prenatal | RhoA/Rac1 | Rearrange actin cytoskeleton | 25 |
| EphA5 | Inhibit chondroblast migration | 26 | ||
| miR-142-3p | Prenatal | Adam9 | Limb type-specific chondrogenesis | 28 |
| miR-375 | Prenatal | Cadherin-7 | Inhibit pre-cartilage condensation | 29 |
| miR-221 | Prenatal | Mdm2 | Promote progenitor proliferation | 30 |
| miR-455-3p | Prenatal | ACVR2B, SMAD2, CHRDL1 | Affect TGFβ Signalling | 21 |
| miR-449a | Postnatal | LEF-1 | Delayed Chondrogenesis | 43 |
| miR-23b | Postnatal | PKA signalling | Promote chondrogenic differentiation | 44 |
| miR-145 | Postnatal | Sox9 | Affect chondrogenic differentiation | 45 |
| miR-199a* | Postnatal | Smad1 | Decrease chondrogenic marker genes | 46 |
| miR-194 | Postnatal | Sox5 | Inhibit chondrogenic differentiation | 53 |
Target genes of miRNAs in chondrogenesis
Regulating certain target genes is one of the most important ways for miRNAs to exert their multiple functions in chondrogenesis. One miRNA may have many target genes, and one gene may be regulated by many miRNAs. The genes targeted by miRNAs often play vital roles in the overall progression of chondrogenesis.
One of the target genes is C/EBPβ. C/EBPβ was regulated by several miRNAs during chondrogenic differentiation of hADSCs and its expression level was up-regulated after chondrogenesis 52. In addition, C/EBPβ was confirmed to facilitate transition of chondrocytes from proliferative state to hypertrophic state and down-regulation of C/EBPβ inhibited progression of OA 54. Moreover, C/EBPβ could degrade type II collagen and aggrecan and induce expression of pro-inflammatory cytokines and chemokines 55–56. Besides, by usage of alternative translation, the multi-isoforms of C/EBPβ could be obtained from a single mRNA 55,57.
Sox gene family includes over 30 members and plays a master role in chondrogenesis. Smits et al. displayed the importance of Sox5 in chondrogenesis by use of genetically manipulated mice with knockout of Sox5 59. Sox5 and Sox6 could be found in differentiating cartilage and they were essential to the control of ECM by enhancing expression of Col2a1 and Acan 60–61. Furthermore, Sox9 is a strong regulator of chondrogenesis. It induced Col2a1 expression by activating a 48-bp enhancer residing in the first intron 62 and promoted differentiation of MSCs into chondrocytes.
In terms of target gene Adam9, up-regulation of Adam9 could lead to apoptosis of chondrogenic progenitors and inhibition of mesenchymal cell migration 28. Besides, Adam9 could bind to myeloma cells with the help of ανβ5 integrin 63 and promote fibroblast adhesion by interacting with α6β1 integrin 64. Moreover, by targeting several integrins, ADAM9 enhanced invasion of cancer cells 65. The regulation mechanisms of prenatal and postnatal chondrogenesis by miRNAs through targeting genes are presented in Figure 1.
Figure 1.
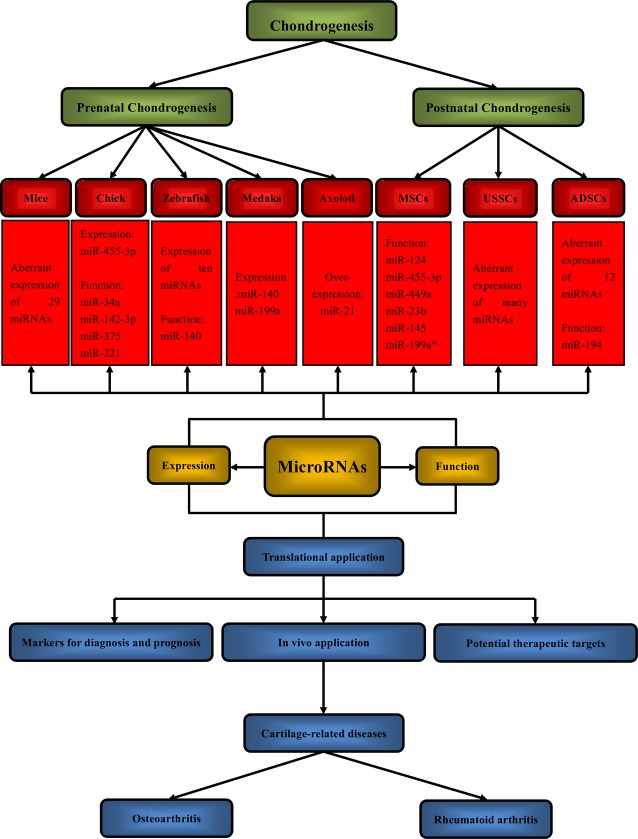
Roles of miRNAs in prenatal chondrogenesis, postnatal chondrogenesis and cartilage-related diseases. Expression and function are two major aspects of miRNAs. During embryonic skeletal development, prenatal chondrogenesis is thought to be a precondition for formation of cartilage in developing limbs. Plenty of studies on different types of adult stem cells have undoubtedly proven their capacity of differentiating into chondrocytes. MiRNAs are found to comprehensively modulate these processes by establishing an interaction network with target genes. In terms of translational application, miRNA technology has great potential to be applied to treat cartilage-related diseases, such as osteoarthritis and rheumatoid arthritis.
MicroRNAs in cartilage-related diseases
MicroRNAs are highly involved in cartilage-related diseases. These diseases, such as OA, RA and psoriatic arthritis, often lead to severe joint pain and even disability of elderly persons 66,67. A lot of studies have been done to elucidate the miRNA roles in the pathogenesis and progression of these diseases.
MicroRNAs in OA
Osteoarthritis is a commonly seen chronic joint disease. The pathogenesis of OA is closely related to the disrupted homeostatic balance between synthesis and degradation of ECM, resulting in cartilage dysfunction and repair failure 69,70.Up till now, effective treatments in OA therapies are limited and the molecular mechanisms underlying the onset and development of OA need to be elucidated urgently. There have been evidences that miRNAs are highly involved in the pathogenesis and progression of OA. Microarray analysis found nine up-regulated miRNAs (miR-16, miR-22, miR-23b, miR-30b, miR-103, miR-223, miR-377, miR-483 and miR-509) and seven down-regulated miRNAs (miR-25, miR-26a, miR-29a, miR-140, miR-210, miR-337 and miR-373) in OA cartilage compared with normal cartilage 72. Inflammatory mediators often participate in OA pathogenesis. Miyaki et al. found that the expression of miR-140 was inhibited when chondrocytes were treated with IL-1β, indicating the role of miR-140 in OA pathogenesis 36. Moreover, by generating a mouse line with miR-140 deletion, this group proved that miR-140−/− mice showed age-related OA-like changes. Meanwhile, transgenic mice with overexpressed miR140 showed resistance to joint damage from OA. This study clearly revealed the critical roles of miR-140 in OA 73.
MicroRNAs in RA
Rheumatoid arthritis is an autoimmune disease which is characterized by chronically inflammatory synovial tissue and irreversible joint damage 67. Recent studies have suggested that miRNAs play crucial roles in RA pathogenesis. Overexpression of miR-16, miR-132, miR-146a and miR-155 was detected in peripheral blood mononuclear cells (PBMCs) from RA patients 74. In addition, the increased expression level of miR-124 was identified in RA synoviocytes 75. MiR-124 was found to play a role in cell proliferation. In in vivo experiments, Nakasa et al. administered double-stranded miR-146a to prevent joint destruction in arthritic mice 76. This finding indicated the therapeutic potential of miR-146a in treating RA.
Application of microRNAs in translational medicine
It is crucial to develop methods for in vivo application of miRNA technology to regenerate damaged cartilage tissue in clinics. Up till now, intra-articular injection and intravenous injection have showed their great potential. Intra-articular injection of double-stranded miRNA-15a induced cell apoptosis in the synovium of mice with autoantibody-mediated arthritis 77. In addition, administration of miR-210 by intra-articular injection effectively promoted the healing of partly damaged anterior cruciate ligaments (ACLs) 78. Moreover, intravenous injection of miR-146a could protect joint from inflammatory damage in arthritic mice 76. Individuals may learn a lot from these trials and develop new methods for in vivo application of miRNA technology. Roles of miRNAs in prenatal chondrogenesis, postnatal chondrogenesis and cartilage-related diseases are presented in Figure 1.
Conclusion and prospects
Chondrogenesis is critical in cartilage repair and regeneration. A great amount of studies have proven that miRNAs are important multifunctional regulators in chondrogenesis of embryos and stem cells. MiRNAs exert their functions through interactions with their targets, which could be genes, transcription factors, signalling pathways and intracellular functional molecules. Despite the numerous findings about miRNAs, there are still many questions on regulatory mechanisms of miRNAs which remain to be illustrated. The expression levels of miRNAs during chondrogenesis are not static, so information about the continuous variation of miRNA expression levels would help know more about miRNA roles in chondrogenesis. In addition, it should be noted that miRNAs themselves are modulated in various manners. Thus in most cases, miRNAs and their targets often interact with each other to form a whole regulatory network to affect multiple cellular activities. Much more attention should be paid on this network. Besides, several studies found that miRNAs can be packed into microvesicles and secreted outside as exosomes from one cell to another 79–80. This finding indicated extracellular regulation of miRNAs. In terms of disorders, many cartilage-related diseases are accompanied by various pathological processes such as hypoxia, inflammation and stress. So the exact roles of miRNAs in these pathological conditions need further description and elucidation. To sum up, miRNAs are highly involved in the prenatal and postnatal chondrogenesis, and also play important roles in cartilage-related diseases. We are sincerely hopeful that more breakthroughs in regulatory mechanisms of miRNAs will be made, and effective and applicable therapeutic methods can be developed to treat cartilage-related diseases to benefit countless patients.
Acknowledgments
This work was supported by a grant from the Natural Science Foundation of China (No’s. 81271982 and 81071498), the key project of Chinese Ministry of Health (No. 201002018). Yue Zhou and Huan Liu designed main topics of this article. Jin Shang collected needed articles and wrote this article.
Conflicts of interest
The authors confirm that there are no conflicts of interest.
References
- Freyria AM, Mallein-Gerin F. Chondrocytes or adult stem cells for cartilage repair: the indisputable role of growth factors. Injury. 2012;43:259–65. doi: 10.1016/j.injury.2011.05.035. [DOI] [PubMed] [Google Scholar]
- Reddi AH. Role of morphogenetic proteins in skeletal tissue engineering and regeneration. Nat Biotechnol. 1998;16:247–52. doi: 10.1038/nbt0398-247. [DOI] [PubMed] [Google Scholar]
- Zhai LJ, Zhao KQ, Wang ZQ, et al. Mesenchymal stem cells display different gene expression profiles compared to hyaline and elastic chondrocytes. Int J Clin Exp Med. 2011;4:81–90. [PMC free article] [PubMed] [Google Scholar]
- Hollander AP, Dickinson SC, Kafienah W. Stem cells and cartilage development: complexities of a simple tissue. Stem Cells. 2010;28:1992–6. doi: 10.1002/stem.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ferguson J, Chang JT, et al. Inter- and intra-combinatorial regulation by transcription factors and microRNAs. BMC Genomics. 2007;8:396. doi: 10.1186/1471-2164-8-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguda BD, Kim Y, Piper-Hunter MG, et al. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci USA. 2008;105:19678–83. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Shalgi R, Liran A, et al. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring MB, Marcu KB. Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med. 2012;18:109–18. doi: 10.1016/j.molmed.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res C Embryo Today. 2005;75:200–12. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA. 2008;105:1949–54. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman EC, Campbell LJ, Hines J, et al. Microarray analysis of microRNA expression during axolotl limb regeneration. PLoS ONE. 2012;7:e41804. doi: 10.1371/journal.pone.0041804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingler TE, Wheeler G, Carmont V, et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64:1909–19. doi: 10.1002/art.34314. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhong N, Li Q, et al. MicroRNAs of rat articular cartilage at different developmental stages identified by Solexa sequencing. Osteoarthritis Cartilage. 2011;19:1237–45. doi: 10.1016/j.joca.2011.07.002. [DOI] [PubMed] [Google Scholar]
- von der Mark K, von der Mark H. The role of three genetically distinct collagen types in endochondral ossification and calcification of cartilage. J Bone Joint Surg Br. 1977;59-B:458–64. doi: 10.1302/0301-620X.59B4.72756. [DOI] [PubMed] [Google Scholar]
- Kim D, Song J, Kim S, et al. MicroRNA-34a modulates cytoskeletal dynamics through regulating RhoA/Rac1 cross-talk in chondroblasts. J Biol Chem. 2012;287:12501–9. doi: 10.1074/jbc.M111.264382. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim D, Song J, Kim S, et al. MicroRNA-34a regulates migration of chondroblast and IL-1beta-induced degeneration of chondrocytes by targeting EphA5. Biochem Biophys Res Commun. 2011;415:551–7. doi: 10.1016/j.bbrc.2011.10.087. [DOI] [PubMed] [Google Scholar]
- Bell E, Saunders JW, Jr, Zwilling E. Limb development in the absence of ectodermal ridge. Nature. 1959;184(Suppl. 22):1736–7. doi: 10.1038/1841736a0. [DOI] [PubMed] [Google Scholar]
- Kim D, Song J, Kim S, et al. MicroRNA-142-3p regulates TGF-beta3-mediated region-dependent chondrogenesis by regulating ADAM9. Biochem Biophys Res Commun. 2011;414:653–9. doi: 10.1016/j.bbrc.2011.09.104. [DOI] [PubMed] [Google Scholar]
- Song J, Kim D, Chun CH, et al. MicroRNA-375, a new regulator of cadherin-7, suppresses the migration of chondrogenic progenitors. Cell Signal. 2012;25:698–706. doi: 10.1016/j.cellsig.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Kim D, Song J, Jin EJ. MicroRNA-221 regulates chondrogenic differentiation through promoting proteosomal degradation of slug by targeting Mdm2. J Biol Chem. 2010;285:26900–7. doi: 10.1074/jbc.M110.115105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nakamura Y, He X, Kato H, et al. Sox9 is upstream of microRNA-140 in cartilage. Appl Biochem Biotechnol. 2012;166:64–71. doi: 10.1007/s12010-011-9404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–4. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- Caplan AI. Why are MSCs therapeutic? New data: new insight. J Pathol. 2009;217:318–24. doi: 10.1002/path.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Zhao RC, Wu Y. The role of microRNAs in self-renewal and differentiation of mesenchymal stem cells. Exp Hematol. 2011;39:608–16. doi: 10.1016/j.exphem.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Han J, Yang T, Gao J, et al. Specific microRNA expression during chondrogenesis of human mesenchymal stem cells. Int J Mol Med. 2010;25:377–84. doi: 10.3892/ijmm_00000355. [DOI] [PubMed] [Google Scholar]
- Miyaki S, Nakasa T, Otsuki S, et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723–30. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino A, Ferracin M, Castelli G, et al. Isolation and characterization of CD146+ multipotent mesenchymal stromal cells. Exp Hematol. 2008;36:1035–46. doi: 10.1016/j.exphem.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Yang B, Guo H, Zhang Y, et al. The microRNA expression profiles of mouse mesenchymal stem cell during chondrogenic differentiation. BMB Rep. 2011;44:28–33. doi: 10.5483/BMBRep.2011.44.1.28. [DOI] [PubMed] [Google Scholar]
- Suomi S, Taipaleenmaki H, Seppanen A, et al. MicroRNAs regulate osteogenesis and chondrogenesis of mouse bone marrow stromal cells. Gene Regul Syst Bio. 2008;2:177–91. doi: 10.4137/grsb.s662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porada CD, Zanjani ED, Almeida-Porad G. Adult mesenchymal stem cells: a pluripotent population with multiple applications. Curr Stem Cell Res Ther. 2006;1:365–9. doi: 10.2174/157488806778226821. [DOI] [PubMed] [Google Scholar]
- da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–13. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- Laine SK, Alm JJ, Virtanen SP, et al. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2012;113:2687–95. doi: 10.1002/jcb.24144. [DOI] [PubMed] [Google Scholar]
- Paik S, Jung HS, Lee S, et al. miR-449a regulates the chondrogenesis of human mesenchymal stem cells through direct targeting of lymphoid enhancer-binding factor-1. Stem Cells Dev. 2012;21:3298–308. doi: 10.1089/scd.2011.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham O, Song BW, Lee SY, et al. The role of microRNA-23b in the differentiation of MSC into chondrocyte by targeting protein kinase A signaling. Biomaterials. 2012;33:4500–7. doi: 10.1016/j.biomaterials.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Yang B, Guo H, Zhang Y, et al. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE. 2011;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EA, Kong L, Bai XH, et al. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284:11326–35. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogler G, Sensken S, Airey JA, et al. A new human somatic stem cell from placental cord blood with intrinsic pluripotent differentiation potential. J Exp Med. 2004;200:123–35. doi: 10.1084/jem.20040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshandeh B, Soleimani M, Ghaemi N, et al. Effective combination of aligned nanocomposite nanofibers and human unrestricted somatic stem cells for bone tissue engineering. Acta Pharmacol Sin. 2011;32:626–36. doi: 10.1038/aps.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshandeh B, Soleimani M, Paylakhi SH, et al. A microRNA signature associated with chondrogenic lineage commitment. J Genet. 2012;91:171–82. doi: 10.1007/s12041-012-0168-0. [DOI] [PubMed] [Google Scholar]
- Guilak F, Lott KE, Awad HA, et al. Clonal analysis of the differentiation potential of human adipose-derived adult stem cells. J Cell Physiol. 2006;206:229–37. doi: 10.1002/jcp.20463. [DOI] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Mizuno H, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–28. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kang Y, Zhang H, et al. Expression of microRNAs during chondrogenesis of human adipose-derived stem cells. Osteoarthritis Cartilage. 2012;20:1638–46. doi: 10.1016/j.joca.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Xu J, Kang Y, Liao WM, et al. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS ONE. 2012;7:e31861. doi: 10.1371/journal.pone.0031861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata M, Kugimiya F, Fukai A, et al. C/EBPbeta Promotes transition from proliferation to hypertrophic differentiation of chondrocytes through transactivation of p57. PLoS ONE. 2009;4:e4543. doi: 10.1371/journal.pone.0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Bryan JL, DeLassus E, et al. CCAAT/enhancer-binding protein beta and NF-kappaB mediate high level expression of chemokine genes CCL3 and CCL4 by human chondrocytes in response to IL-1beta. J Biol Chem. 2010;285:33092–103. doi: 10.1074/jbc.M110.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xing X, Hensley G, et al. Resistin induces expression of proinflammatory cytokines and chemokines in human articular chondrocytes via transcription and messenger RNA stabilization. Arthritis Rheum. 2010;62:1993–2003. doi: 10.1002/art.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–82. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- Imamura T, Imamura C, Iwamoto Y, et al. Transcriptional Co-activators CREB-binding protein/p300 increase chondrocyte Cd-rap gene expression by multiple mechanisms including sequestration of the repressor CCAAT/enhancer-binding protein. J Biol Chem. 2005;280:16625–34. doi: 10.1074/jbc.M411469200. [DOI] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–90. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier MC, Martin JF, et al. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–28. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimal-Monroy J, Rodriguez-Leon J, Montero JA, et al. Analysis of the molecular cascade responsible for mesodermal limb chondrogenesis: Sox genes and BMP signaling. Dev Biol. 2003;257:292–301. doi: 10.1016/s0012-1606(03)00066-6. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–33. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Graham R, Russell G, et al. MDC-9 (ADAM-9/Meltrin gamma) functions as an adhesion molecule by binding the alpha(v)beta(5) integrin. Biochem Biophys Res Commun. 2001;280:574–80. doi: 10.1006/bbrc.2000.4155. [DOI] [PubMed] [Google Scholar]
- Nath D, Slocombe PM, Webster A, et al. Meltrin gamma(ADAM-9) mediates cellular adhesion through alpha(6)beta(1)integrin, leading to a marked induction of fibroblast cell motility. J Cell Sci. 2000;113(Pt 12):2319–28. doi: 10.1242/jcs.113.12.2319. [DOI] [PubMed] [Google Scholar]
- Josson S, Anderson CS, Sung SY, et al. Inhibition of ADAM9 expression induces epithelial phenotypic alterations and sensitizes human prostate cancer cells to radiation and chemotherapy. Prostate. 2011;71:232–40. doi: 10.1002/pros.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier JP, Martel-Pelletier J, Mehraban F, et al. Immunological analysis of proteoglycan structural changes in the early stage of experimental osteoarthritic canine cartilage lesions. J Orthop Res. 1992;10:511–23. doi: 10.1002/jor.1100100406. [DOI] [PubMed] [Google Scholar]
- Klareskog L, Catrina AI, Paget S. Rheumatoid arthritis. Lancet. 2009;373:659–72. doi: 10.1016/S0140-6736(09)60008-8. [DOI] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Lotz MK, Kraus VB. New developments in osteoarthritis. Posttraumatic osteoarthritis: pathogenesis and pharmacological treatment options. Arthritis Res Ther. 2010;12:211. doi: 10.1186/ar3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Nakasa T, Hikata T, et al. Molecular network of cartilage homeostasis and osteoarthritis. Med Res Rev. 2008;28:464–81. doi: 10.1002/med.20113. [DOI] [PubMed] [Google Scholar]
- Goldring MB, Marcu KB. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res Ther. 2009;11:224. doi: 10.1186/ar2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Malizos KN, Oikonomou P, et al. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE. 2008;3:e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki S, Sato T, Inoue A, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–85. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley KM, Satoh M, Chan AL, et al. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamachi Y, Kawano S, Takenokuchi M, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60:1294–304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- Nakasa T, Shibuya H, Nagata Y, et al. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011;63:1582–90. doi: 10.1002/art.30321. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Nakasa T, Mochizuki Y, et al. Induction of apoptosis in the synovium of mice with autoantibody-mediated arthritis by the intraarticular injection of double-stranded MicroRNA-15a. Arthritis Rheum. 2009;60:2677–83. doi: 10.1002/art.24762. [DOI] [PubMed] [Google Scholar]
- Shoji T, Nakasa T, Yamasaki K, et al. The effect of intra-articular injection of microRNA-210 on ligament healing in a rat model. Am J Sports Med. 2012;40:2470–8. doi: 10.1177/0363546512458894. [DOI] [PubMed] [Google Scholar]
- Bruno S, Grange C, Deregibus MC, et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol. 2009;20:1053–67. doi: 10.1681/ASN.2008070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collino F, Deregibus MC, Bruno S, et al. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS ONE. 2010;5:e11803. doi: 10.1371/journal.pone.0011803. [DOI] [PMC free article] [PubMed] [Google Scholar]