Dear Sir:
Current evidence indicates that about 5%–10% of pancreatic cancer has a familial component, although the vast majority of pancreatic cancer families remain unexplained. 1 PALB2 is a recently identified breast cancer susceptibility gene whose protein is closely associated with BRCA2, and is essential for BRCA2 anchorage to nuclear structures. This functional relationship made PALB2 a candidate gene for susceptibility to BRCA2-related cancers such as pancreas cancer. Recently, Jones et al2 screened 96 familial pancreatic cancer patients, 16 of whom had 1 first-degree relative with pancreatic cancer and 80 had ≥2 additional relatives with pancreatic cancer, ≥1 of which was first degree.2 Truncating PALB2 mutations were identified in 3 patients (3.1%) and there was no difference in average age of cancer onset between mutation-positive and -negative families. We sought to screen a larger cohort of pancreatic cancer cases, including both familial and sporadic types, to determine the wider contribution of PALB2 gene mutations in pancreatic cancer. We selected a total of 254 individuals with pancreas cancer (148 male, 106 female) at a median age of diagnosis of 61 years. Patients were identified between 1997 and 2007 via clinic-based recruitment in Toronto and Montreal and through either the Familial Gastrointestinal Cancer Registry at Mount Sinai Hospital in Toronto or the population-based Ontario Pancreas Cancer Study. In total, 203 patients were recruited in Toronto and 51 in Montreal. All probands were confirmed to have pancreatic adenocarcinoma by pathology report; 101 pro-bands had a family history of pancreatic cancer, of which 32 had 2 affected first-degree relatives. In these 101 cases, 74 had 1 affected relative, 18 had 2 affected relatives, 7 had 3 affected relatives, and 2 cases had >3 affected relatives (Table 1). Sixty probands had a family history of breast/ovarian cancer, including 21 cases with a family history of pancreatic cancer (included above). Because the cases were ascertained through pancreas cancer studies, the family history of breast cancer or ovarian cancer was not strong, with most families having a single relative affected, and no family having a BRCAPRO score >0.12 (ie, these were not primarily familial breast/ovarian cancer families with 1 or 2 additional cases of pancreas cancer). The median age at diagnosis of pancreatic cancer cases with no family history was 49 years (range, 31–85); 55% of these were <50 years old and 66% were <60 years old at diagnosis. Genomic DNA was obtained from blood, saliva, or buccal cells using standard extraction methods. Before analysis, 20 ng of total genomic DNA was used for whole genome amplification according to the manufacturer’s instructions using the Repli-g Mini Kit (Qiagen, Mississauga, Ontario, Canada). We screened the 13 coding exons of PALB2 by sequencing (n = 83) or high-resolution melt analysis (n = 171), which has similar sensitivity to sequencing.3 Samples with variants were reamplified by polymerase chain reaction (PCR) using the original, non–whole-genome-amplified DNA as template and the PCR product was sequenced in forward and reverse directions for confirmation. We performed multiplex ligation-dependent probe amplification assay (MLPA) for PALB2 on 228 samples where we had sufficient DNA of adequate quality, as previously described by Foulkes et al.4
Table 1.
Pancreas Cancer Cases
| Type of case | No. of cases (total = 254) |
No. of relatives with pancreas cancer |
No. of relatives with breast/ ovarian cancer |
|---|---|---|---|
| Sporadic pancreas | 114 | NA | NA |
| Family history of pancreas cancer | 80 | 1 relative: 74 2 relatives: 18 3 relatives: 7 >3 relatives: 2 |
NA |
| Familial pancreas with breast/ovary cases | 21 | 1 relative: 34 2 relatives: 17 3 relatives: 7 >3 relatives: 2 |
|
| Sporadic pancreas with breast/ovary cases | 39 | NA |
NA, not applicable.
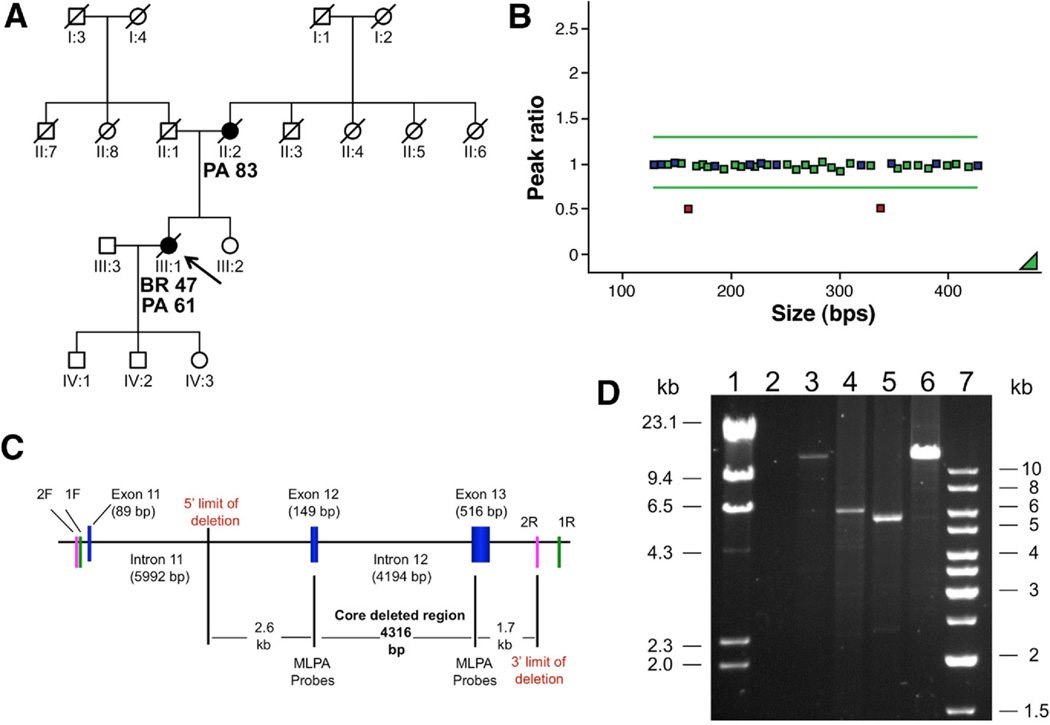
We identified a heterozygous, 6.7-kb germline deletion including exons 12 and 13 of PALB2 in a patient who was affected by breast and then pancreas cancer (ages 47 and 61, respectively) and whose mother died of pancreas cancer at age 83 (Figure 1). This result was confirmed by long range PCR (Takara Bio Inc., Madison, WI) using 2 different primer pairs, which determined the deleted region to span a region from the middle of intron 11 (2.7 kb from the beginning of exon 12), up to 1.8kb after exon 13. This deletion would disrupt the PALB2 WD40 motif, which is required for interaction with the BRCA2 protein. 5 Aside from the exonic deletion, 2 previously unreported missense variants (S285L and T911I) were identified, but neither were predicted to be pathogenic. Both these variants were seen in young-onset pancreas cancer cases (41 years and 48 years) with no family history. A number of previously reported variants were also identified (Table 2).
Figure 1.
(A) Pedigree of the proband (shown by arrow). PA, pancreatic cancer; BR, breast cancer. (B) MLPA showing deletion of exon 12 (161-bp fragment) and exon 3 (338-bp fragment). (C) Diagram of the deleted region showing the obligate core deletion as defined by the MLPA probes and the potential 5= and 3= limits of the deletion estimated from the observed size of the deleted region by long-range PCR (approximately 6.7–7.0 kb). 1F and 1R, forward and reverse primers for long-range fragment 1; 2F and 2R, forward and reverse primers for long-range fragment 2. (D) Long-range PCR, Lane 1, lambda molecular marker; lane 2, no DNA control; lane 3, wt control fragment 1 (expected size 13,141 bp); lane 4, patient fragment 1; lane 5, patient fragment 2; lane 6, wt control fragment 2 (expected size12,633 bp); lane 7, 1 kb molecular marker. Owing to low availability of native genomic DNA, long-range PCR for the patient was performed from whole-genome amplified (WGA) DNA. As a consequence, we expect amplification of large fragments by long-range PCR to be limited by the relative abundance of full-length copies of the region as generated by the WGA reaction. This may explain the absence of the wild-type allele (13 kb) in the patient, who was shown to be heterozygous for the exonic deletion by MLPA, and the correspondingly increased amplification efficiency of the smaller allele carrying the deletion (6 kb).
Table 2.
Other Variants Detected in PALB2
| Sequence variant | Protein change | No. of times observed | Additional information |
|---|---|---|---|
| c.1 – 47 G>A | 5′ UTR | 13 | rs8053188a |
| c.212 –58 A>C | Intron 3 | 16 | non-codinga |
| c.1684 +42 ins.TGA | Intron 4 | 1 | non-codingc |
| c.854 C>T | S285L | 1 | possibly damaging,c,d toleratede |
| c.1010 T>C | L337S | 4 | rs45494092a |
| c.1572 A>G | S524S | 1 | rs45472400a |
| c.1194 G>A | V398V | 1 | Silenta |
| c.1676 A>G | Q559R | 40 | rs152451a |
| c.1743 A>G | L581L | 1 | Silentc |
| c.2014 G>C | E672Q | 18 | rs45532440a |
| c.2586 +58 C>T | Intron 6 | 57 | rs249954b |
| c.2590 C>T | P864S | 1 | rs45568339a |
| c.2732 C>T | T911I | 1 | possibly damaging,c,d toleratede |
| c.2794 G>A | V932M | 4 | rs45624036a |
| c.2816 T>G | L939W | 2 | rs45478192a |
| c.2993 G>A | G998E | 11 | rs45551636a |
| c.3300 T>G | T1100T | 16 | rs45516100a |
The main purpose of the study by Jones et al2 was to illustrate that exomic sequencing could be used to identify genes conferring a substantial risk of cancer, in the absence of linkage or other localizing information. After identification of a PALB2 germ-line mutation in a pancreas cancer case with a family history of pancreatic cancer, truncating mutations were found in 3 other individuals, all of whom had a personal and family history of pancreas cancer. Here, we studied a more varied set of pancreas cancer cases, with fewer probands with very strong family histories of pancreas cancer, but more cases with family histories of other BRCA2-related cancers, and 13 cases diagnosed under the age of 40. We found only 1 deleterious mutation, a 2-exon deletion, which is only the second described case of a PALB2 exonic deletion, the other being reported in a case of Fanconi anemia.6 To date, PALB2 exonic deletions have not been reported in breast cancer families.7
Taking the 2 sets of data together, we conclude that germ-line mutations in PALB2 are a rare cause of pancreas cancer, whether hereditary, familial, or sporadic. Of note, 4 of the 5 PALB2-related pancreas families identified to date include ≥1 case of breast cancer and in 2 families mutations were seen in individuals with both breast and pancreatic cancer (a total of 9 cases in our study had both breast and pancreas cancer). It may be the case that investigating PALB2 will be worthwhile in individuals with very strong family histories of pancreas cancer, or where there is a concomitant history of breast cancer, but based on the findings presented herein, routine screening of PALB2 in individuals with, or at risk of pancreas cancer is difficult to justify. If PALB2 mutation screening is undertaken, it should include MLPA for exonic deletions. As is seen in breast cancer,8 the penetrance for these very rare disease-causing variants seems to be high.
Acknowledgment
Funding
Work carried out in Montreal was supported by grants from the Jewish General Hospital Weekend to End Breast Cancer, Susan G. Komen Foundation for the Cure and the Turner Family Cancer Research Fund. Work carried out in Toronto was supported by grants from the National Institutes of Health (R01 CA97075, as part of the PACGENE consortium), The Lustgarten Foundation for Pancreatic Cancer Research and the Ontario Cancer Research Network. We acknowledge the Pancreatic Cancer Canada Foundation (www.pancreaticcancercanada.ca) for their continued support of research into the early detection of pancreatic cancer, and the Pancreas Cancer Screening Study at Mount Sinai Hospital and Princess Margaret Hospital in Toronto.
Footnotes
Conflicts of interest
The authors disclose no conflicts.
Contributor Information
Marc D. Tischkowitz, Program in Cancer Genetics, Departments of Oncology and Human Genetics, McGill University, and Segal Cancer Centre, Jewish General Hospital, Montreal, Quebec, Canada
Nelly Sabbaghian, Program in Cancer Genetics, Departments of Oncology and Human Genetics, McGill University, and Segal Cancer Centre, Jewish General Hospital, Montreal, Quebec, Canada.
Nancy Hamel, Program in Cancer Genetics, Departments of Oncology and Human Genetics, McGill University, and The Research Institute, McGill University Health Centre, Montreal, Quebec, Canada.
Ayelet Borgida, Dr Zane Cohen Digestive Diseases Clinical, Research Centre, Mount Sinai Hospital, Toronto, Ontario, Canada.
Chaim Rosner, Program in Cancer Genetics, Departments of Oncology and Human Genetics, McGill University, and Segal Cancer Centre, Jewish General Hospital, Montreal, Quebec, Canada.
Nassim Taherian, Program in Cancer Genetics, Departments of Oncology and Human Genetics, McGill University, and Segal Cancer Centre, Jewish General Hospital, and The Research Institute, McGill University Health Centre, Montreal, Quebec, Canada.
Archana Srivastava, Program in Cancer Genetics Departments of Oncology and Human Genetics, McGill University, and Segal Cancer Centre, Jewish General Hospital, Montreal, Quebec, Canada.
Spring Holter, Dr Zane Cohen Digestive Diseases Clinical, Research Centre, Mount Sinai Hospital, Toronto, Ontario, Canada.
Heidi Rothenmund, Dr Zane Cohen Digestive Diseases Clinical, Research Centre, Mount Sinai Hospital, Toronto, Ontario, Canada.
Parviz Ghadirian, Epidemiology Research Unit, Research Centre, Centre Hospitalier Université de Montréal, (CRCHUM), Hôtel-Dieu, Montréal, Quebec, Canada.
William D. Foulkes, Program in Cancer Genetics, Departments of Oncology and Human Genetics, McGill University, and Segal Cancer Centre, Jewish General Hospital, and The Research Institute, McGill University Health Centre, Montreal, Quebec, Canada
Steven Gallinger, Dr Zane Cohen Digestive Diseases Clinical, Research Centre, Mount Sinai Hospital, and Department of Surgery, University Health Network, University of Toronto Toronto, Ontario, Canada.
References
- 1.Shi C, Hruban RH, Klein AP. Familial pancreatic cancer. Arch Pathol Lab Med. 2009;133:365–374. doi: 10.5858/133.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Hum Mutat. 2009;30:857–859. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- 4.Foulkes WD, Ghadirian P, Akbari MR, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 Cellular and Clinical Functions by a Nuclear Partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 6.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 7.Ameziane N, van den Ouweland AM, Adank MA, et al. Lack of large genomic deletions in BRIP1, PALB2, and FANCD2 genes in BRCA1/2 negative familial breast cancer. Breast Cancer Res Treat. 2009 Jun 6; doi: 10.1007/s10549-009-0428-8. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 8.Erkko H, Dowty JG, Nikkila J, et al. Penetrance analysis of the PALB2 c.1592delT founder mutation. Clin Cancer Res. 2008;14:4667–4671. doi: 10.1158/1078-0432.CCR-08-0210. [DOI] [PubMed] [Google Scholar]
- 9.Garcia MJ, Fernandez V, Osorio A, et al. Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative Spanish breast cancer families. Breast Cancer Res Treat. 2009;113:545–551. doi: 10.1007/s10549-008-9945-0. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Liang J, Wang Z, et al. Association of common PALB2 polymorphisms with breast cancer risk: a case-control study. Clin Cancer Res. 2008;14:5931–5937. doi: 10.1158/1078-0432.CCR-08-0429. [DOI] [PubMed] [Google Scholar]