Abstract
The archaeal machinery responsible for DNA replication is largely homologous to that of eukaryotes and is clearly distinct from its bacterial counterpart. Moreover, it shows high diversity in the various archaeal lineages, including different sets of components, heterogeneous taxonomic distribution, and a large number of additional copies that are sometimes highly divergent. This has made the evolutionary history of this cellular system particularly challenging to dissect. Here, we have carried out an exhaustive identification of homologs of all major replication components in over 140 complete archaeal genomes. Phylogenomic analysis allowed assigning them to either a conserved and probably essential core of replication components that were mainly vertically inherited, or to a variable and highly divergent shell of extra copies that have likely arisen from integrative elements. This suggests that replication proteins are frequently exchanged between extrachromosomal elements and cellular genomes. Our study allowed clarifying the history that shaped this key cellular process (ancestral components, horizontal gene transfers, and gene losses), providing important evolutionary and functional information. Finally, our precise identification of core components permitted to show that the phylogenetic signal carried by DNA replication is highly consistent with that harbored by two other key informational machineries (translation and transcription), strengthening the existence of a robust organismal tree for the Archaea.
Keywords: Cdc6/Orc1, RPA/SSB, DNA gyrase, primase, phylogeny, nanosized archaea
Introduction
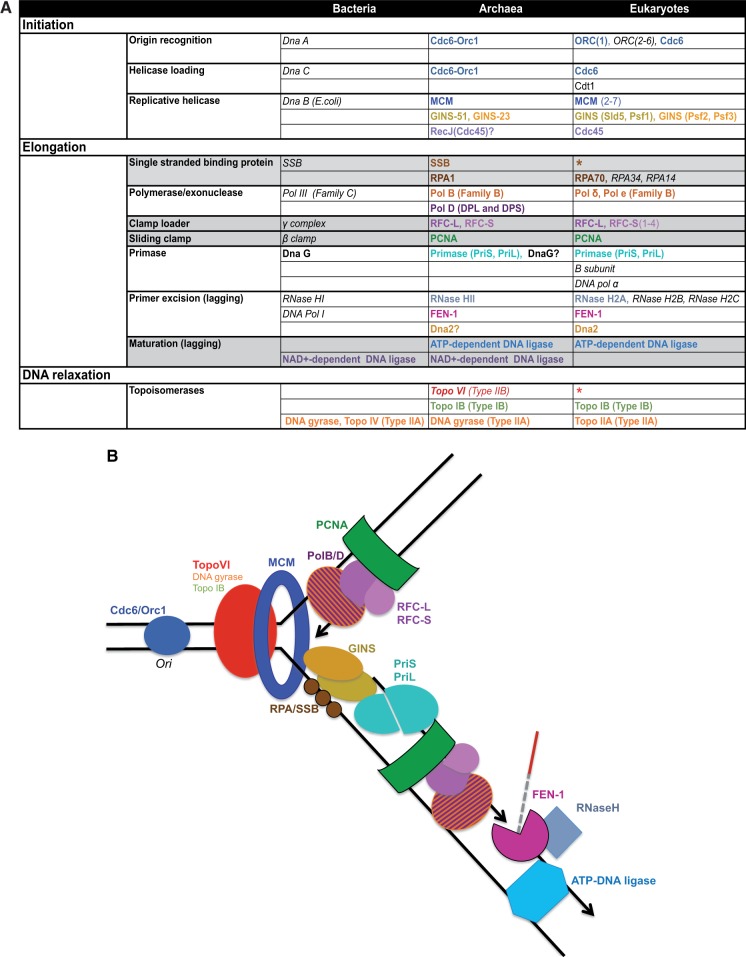
Replication of the genetic material is a crucial step of the cell cycle. All three domains of life replicate their DNA semiconservatively (Meselson and Stahl 1958) and follow basically the same sequence of events (for a recent review see DePamphilis and Bell [2010]): The replication fork is assembled by a specific protein or initiation complex that recognizes the origin of replication on the chromosome and opens up the double-stranded DNA. A helicase is then recruited, producing a replication bubble that is protected by single-stranded DNA-binding proteins. The core replication machinery then assembles at the fork with the help of the sliding clamp, a ring-shaped factor that tethers it to the DNA template. The main replicative polymerase extends DNA replication bidirectionally from short RNA primers made by a primase, with one strand being synthesized continuously (leading strand), and the other discontinuously (lagging strand). The Okazaki fragments produced during synthesis of the lagging strand are joined together by a DNA ligase after excision of the RNA primers. During the whole process, a number of topoisomerases act to resolve topological problems arising from DNA supercoiling in front of the replication fork and chromosome entangling at the end of replication. Despite the overall conservation of these major steps, the machinery used for DNA replication in Archaea and Eukaryotes exhibits striking differences to the bacterial replication machinery, which uses nonhomologous proteins belonging to completely different families (fig. 1) (Grabowski and Kelman 2003; Barry and Bell 2006).
Fig. 1.—
(A) General overview of the components of DNA replication in the Archaea compared to the other two domains of life. Same color in a given row indicates homology; gray shading indicates that the bacterial version has only structural similarity with the archaeal/eukaryal component; question marks represent components with unclear implication in archaeal replication, i.e., DnaG, Dna2, and RecJ homologs; asterisks indicate that a eukaryotic homolog exist but is not involved in replication, i.e., SSB and TopoVI. See main text for details. (B) Sketch of the DNA replication machinery in the Archaea. Colors corresponds to those in (A).
The archaeal replication machinery is generally considered to be a simplified version of the eukaryotic apparatus, which usually harbors more components (fig. 1). However, it too has its own peculiar characteristics. Along with a PolB polymerase, most archaea also possess a PolD polymerase whose catalytic subunit has no homologs in Bacteria or Eukaryotes (Cann et al. 1998). Furthermore, to relax positive superturns arising during replication and decatenate the chromosome at the end of replication, two tasks which are performed by Type IIA enzymes in Eukaryotes and Bacteria, most archaea use a topoisomerase of the type IIB family (TopoVI; Forterre et al. 2007). Some archaeal components have homologs in eukaryotes that are not involved in DNA replication (fig. 1). For example, eukaryotic homologs of the catalytic subunit of archaeal TopoVI (Spo11) are involved in the initiation of meiotic recombination (Bergerat et al. 1997). Additionally, homologs of the archaeal single-stranded binding (SSB) proteins were identified in eukaryotes several years ago (Robbins et al. 2005) and are the subject of growing appeal due to their probable yet poorly understood role in genome integrity (Richard et al. 2008; Shi et al. 2012). The role of some homologs of eukaryotic replication components in archaea is not clear and remains to be confirmed by functional studies. For example, Dna2 may be involved in Okazaki fragment maturation, performing the same function as in eukaryotes (Higashibata et al. 2003). Similarly, the role of the archaeal RecJ, a 5′-3′ exonuclease (also found in bacteria and a distant homolog of eukaryotic Cdc45) remains to be verified experimentally, but may fulfill the same function in an archaeal CMG (Cdc45, MCM, GINS) complex (Makarova et al. 2012). Archaea also harbor a few homologs of bacterial replication components such as NAD+-dependent DNA ligase, DNA gyrase, and DnaG (fig. 1). Although ATP-dependent ligases are ubiquitous in Archaea and Eukaryotes (Wilkinson et al. 2001; Martin and MacNeill 2002), bacterial-like NAD+-dependent ligases have been identified in some members of Halobacteriales (Zhao et al. 2006). DNA gyrase, a topoisomerase belonging to the Topo IIA family, is present in a number of euryarchaeal lineages (Forterre et al. 2007). In the case of archaeal homologs of bacterial primase DnaG (Aravind and Koonin 1998), the proposal that they are involved in replication (Bauer et al. 2013) is weakened by strong evidence that suggests a role in RNA metabolism (Hou et al. 2013).
Remarkably, the machinery for DNA replication appears to vary greatly among archaeal lineages, which can harbor various combinations of key components. This variation includes different main replicative polymerases (PolB and PolD), single or multiple replication origins and initiator proteins (Cdc6/Orc1), different SSB proteins (SSB, RPA), and alternative multimeric complexes (PCNA, RFC, and GINS); (Grabowski and Kelman 2003; Barry and Bell 2006; McGeoch and Bell 2008; Bell 2011; Beattie and Bell 2011). There have also been reports of possible replacements of components by nonhomologous proteins, such as the putative initiator protein MJ0774 in Methanococcus jannaschii (Zhang RR and Zhang C-TC 2004) and the putative single-stranding binding protein ThermoDPB in Thermoproteales (Paytubi et al. 2012). Moreover, archaeal genomes can display additional copies of replication components that are often embedded in integrative elements of plasmid and/or viral origin. For example, the integrated element TKV3 of Thermococcus kodakarensis KOD1 encodes a homolog of PCNA (Fukui et al. 2005); Haloferax volcanii harbors three replication origins and nine Cdc6/Orc1 coding genes, with one pair embedded in a 50 kb prophage region (Hartman et al. 2009); Sulfolobales contain three replication origins and three Cdc6/Orc1 copies, one of which is associated with the second origin of replication that was contributed by an integrative element (Samson et al. 2013). Finally, a number of additional divergent MCM homologs originating from integrative elements or plasmids are present in various archaeal taxa (Krupovic, Gribaldo, et al. 2010).
Such extreme diversity has made it particularly challenging to dissect the evolutionary history of archaeal DNA replication. Although some components have been previously analyzed (Chia et al. 2010; Krupovic, Gribaldo, et al. 2010), no attempt has been made to perform a global survey of the complete machinery. Here, we have carried out an in depth phylogenomic analysis of all components of DNA replication in over 140 complete archaeal genomes. We specifically assess the taxonomic distribution of homologs in each of these genomes. In addition, we precisely identify copies arising from integrative elements/decaying paralogs/horizontal gene transfers as opposed to those that constitute a conserved and vertically inherited core replication machinery, providing important information for further evolutionary and functional analysis of these components. Phylogenetic analysis of the core components allowed us to infer the nature of DNA replication in the last archaeal common ancestor (LACA) and the subsequent evolutionary history that shaped this machinery. Finally, our analysis enabled us to investigate, for the first time, the phylogenetic signal carried by DNA replication. It shows remarkable consistency with that harbored by the two other main informational processes (transcription and translation), confirming the existence of a robust phylogenomic core that can be used to reconstruct the tree of the Archaea.
Materials and Methods
Identification of Homologs of DNA Replication Components
Homologs of each archaeal DNA replication component were retrieved from the reference sequence database at the National Center for Biotechnology Information (NCBI) using the BlastP (Altschul et al. 1997) program with different seeds from each archaeal order. The top 100 best hits for each order were then used to create hidden Markov model (HMM) profiles (Johnson et al. 2010; http://www.hmmer.org, last accessed January 16, 2014) that allowed an iterative search of a local database of 142 archaeal genomes including 98 plasmid sequences and a local database of 56 complete archaeal virus genomes downloaded from the Viral Genomes database of NCBI (as of June 20, 2013) (supplementary table S3, Supplementary Material online). The absence of a given homolog in a specific genome was verified by performing additional tBlastN (Altschul et al. 1997) searches. Genomic context was investigated using MaGe (Vallenet et al. 2005), MGV2 (Kerkhoven et al. 2004), and STRING (Szklarczyk et al. 2011).
Phylogenetic Analysis
Multiple alignments were performed with MUSCLE v3.8.31 (Edgar 2004) and manually inspected using the ED program from the MUST package (Philippe 1993) to verify that all sequences retrieved at the first step were homologous. Final single protein data sets were trimmed using the software BMGE (Criscuolo and Gribaldo 2010) with default parameters and subjected to phylogenetic analyses by Maximum Likelihood and Bayesian methods. Maximum likelihood analyses were performed with Treefinder (Jobb et al. 2004; version of 2008). For each protein data set, the best-fit parameters and model of amino acid substitution were chosen using the Akaike information criterion with a correction (AICc) for finite sample sizes (Hurvich and Tsai 1989) as implemented in Treefinder (Jobb et al. 2004). Bootstrap supports were calculated based on 100 resamplings of the original alignment. Bayesian analyses were run with MrBayes 3.2 (Ronquist et al. 2012), using the mixed amino acid substitution model and four categories of evolutionary rates. Two independent runs were performed for each data set, and runs were stopped when they reached a standard deviation of split frequency below 0.01 or the log likelihood values reached stationary. The majority rule consensus trees were obtained after discarding first 25% samples as burn-in.
For the analysis of DNA gyrase, alternative tree topologies were statistically evaluated using the following paired-sites tests: expected-likelihood weights, bootstrap probability (BP; Felsenstein 1985), Kishino and Hasegawa (Kishino and Hasegawa 1989), Shimodaira and Hasegawa (SH; Shimodaira and Hasegawa 1999; Goldman et al. 2000), Weighted SH test (Shimodaira and Hasegawa 1999; Buckley et al. 2001), and approximately unbiased (AU) test (Shimodaira 2002) as implemented in Treefinder (Jobb et al. 2004). A total of 500000 RELL (Kishino et al. 1990) replicates were run. Three alternative topologies were tested and it was determined that the data did not reject the topology if the P value was greater than 0.05 for all tests.
Supermatrix Analyses
Fourteen DNA core replication proteins that were present in at least 60% of the archaeal genomes (PriS, MCM, PCNA, Cdc6/Orc1, DPL, DPS, PolB, TopoVI-A, TopoVI-B, RFC-s, RFC-l, RNaseH, DNA ligase, and FEN-1) were retained for supermatrix analysis. To handle species-specific paralogs, we chose one paralog, and the slowest evolving if applicable, to limit possible artifacts due to fast evolutionary rates. In the case of ancient paralogs, we also chose those representing the cluster with larger taxonomic representation and/or showing the slowest evolutionary rates. For example, we chose the Cdc6/Orc1-1 paralog (see Results). Each multiple alignment was independently realigned, trimmed, and concatenated into a character supermatrix comprised of 4,295 amino acid positions and 129 archaeal taxa (after keeping only one representative strain of the same species). PhyloBayes 3.3b (Lartillot et al. 2009) was used to perform Bayesian analysis using the CAT + GTR model and a gamma distribution with four categories of evolutionary rates was used to model the heterogeneity of site evolutionary rates. The concatenated datasets were also recoded using Dayhoff6 and Dayhoff4 recoding schemes as implemented in PhyloBayes 3.3b (Lartillot et al. 2009) and analyzed with the same model parameters. For each data set, two independent chains were run until convergence (max diff < 0.01). The first 25% of trees were discarded as burn in and the posterior consensus was computed by selecting one tree out of every two to compute the 50% majority consensus tree. Maximum likelihood analysis was carried out by PhyML (Guindon et al. 2010), the LG model and a gamma correction with four categories of evolutionary rates. Bootstrap support was calculated based on 100 resamplings of the original alignment.
Results
Archaeal DNA Replication: The Core Component and the Variable Shell
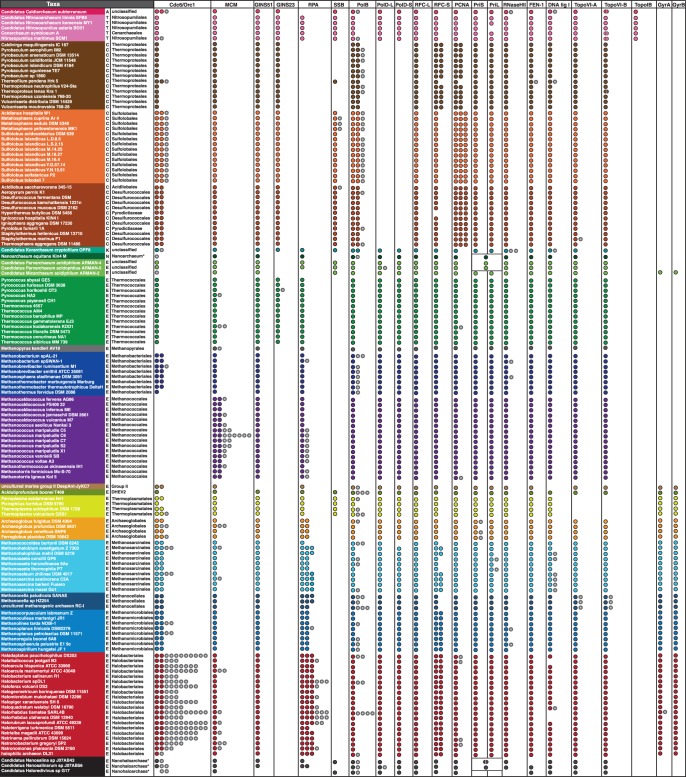
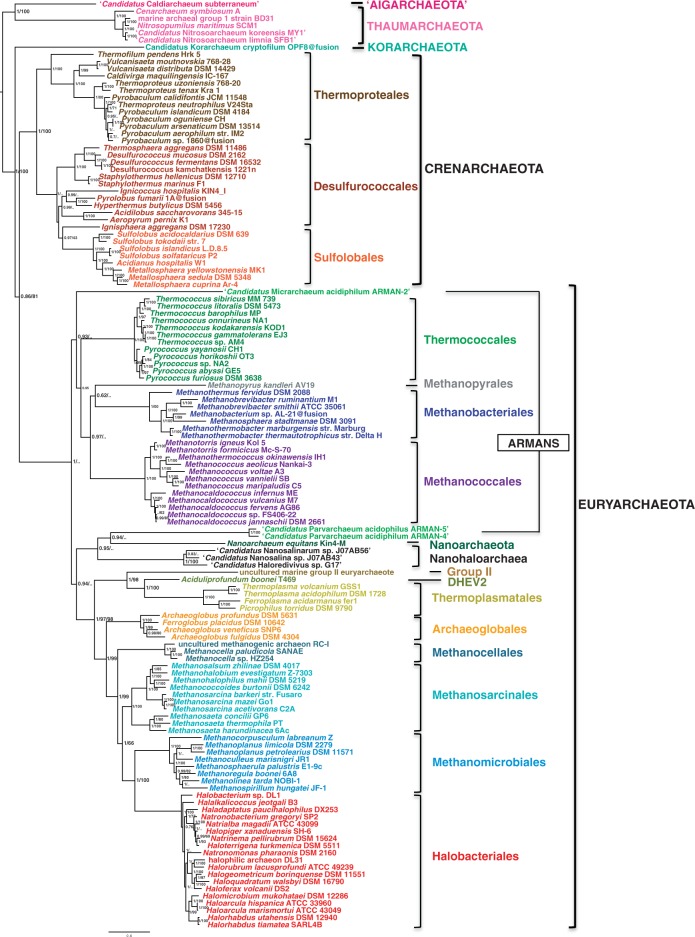
We performed an exhaustive search for homologs of the 16 major components of the DNA replication machinery (22 proteins considering subunits) in 142 complete archaeal genomes (fig. 2; supplementary table S1, Supplementary Material online). The taxonomic distribution of these proteins shows a highly dynamic pattern along the different archaeal lineages. Some components are present essentially in a single copy and in the majority of genomes (e.g., GINS 51, TopoVI A and B, RFC-L, DNA ligase 1, Fen1, RNase HII, PriS, and PriL), whereas others are missing altogether from a number of archaeal lineages (e.g., Cdc6/Orc1 in Methanococcales and Methanopyrales, TopoVI in Thermoplasmatales, PolD and RPA in Crenarchaeota, SSB in most Euryarchaeota and Thermoproteales). Incomplete assembly of some genomes, such as the Nanohaloarchaea, uncultured marine group II, Candidatus Caldiarchaeum subterraneum (Aigarchaeota), and the ARMANS (Archaeal Richmond Mine Acidophilic Nanoorganisms) suggests that some absences in these taxa must be taken with caution. Finally, a few components display a large number of extra copies in some taxa (e.g., Cdc6/Orc1 in Halobacteriales, MCM in Methanococcales, RPA in many Euryarchaeota, PolB in many Euryarchaeota and Crenarchaeota, PCNA in Crenarchaeota).
Fig. 2.—
Distribution of homologs of 22 main replication components in 142 archaeal genomes. Filled circles represent homologs that we assigned to the core replication machinery, whereas gray circles represent homologs assigned to the shell component (see text for details). Split genes are indicated by half circles, and the fused primases by a box (see text for details). Letters in first column indicate the phylum (A, Aigarchaeota; T, Thaumarchaeota; C, Crenarchaeota; K, Korarchaeota; N, Nanoarchaeota; E, Euryarchaeota). Asterisks indicate classes instead of orders. Full accession numbers are given in supplementary table S1, Supplementary Material online.
Inspection of multiple alignments, phylogenies, and genome synteny allowed us to highlight two categories of homologs: 1) slow-evolving homologs lying within chromosomal regions that are syntenic among closely related taxa and whose phylogeny is overall consistent with the archaeal phylogeny, as opposed to 2) highly divergent copies that lie within nonconserved genetic contexts and/or display more restricted taxonomic sampling and inconsistent phylogenetic affiliations. We reasoned that the first category represents components that were primarily vertically inherited during archaeal diversification and form what we called the conserved core replication components (fig. 2, filled circles; for full accession numbers see supplementary table S1, Supplementary Material online), whereas the second category represents horizontally transferred genes, decaying paralogs, or homologs arising from integration of extrachromosomal elements that form a variable pool of proteins that we called the shell replication components (fig. 2, open circles; for full accession numbers see supplementary table S1, Supplementary Material online).
An example of our approach is provided by the analysis of Cdc6/Orc1. Except for the previously mentioned absence in Methanococcales and Methanopyrales, all archaeal genomes contain at least one homolog of the initiation protein Cdc6/Orc1. Most lineages harbor at least two copies, and a very large number of homologs are present in Halobacteriales (fig. 2). We found that in each genome only one or two Cdc6/Orc1 homologs are slow evolving and show conserved synteny among closely related taxa. Additional copies, when present, are very divergent and display nonconserved genomic contexts. When a phylogenetic tree was built from all homologs (not shown) the first category formed two clearly distinct clusters representing a large taxonomic coverage, which, albeit not completely resolved, is globally consistent with archaeal phylogeny. In contrast, the second category fell into an unresolved group showing very long branches, restricted taxonomic coverage and highly inconsistent phylogenetic relationships. The first category was therefore assigned to the core replication machinery (fig. 2, filled circles; supplementary table S1, Supplementary Material online), and the second to the shell (fig. 2, open circles; supplementary table S1, Supplementary Material online). For validation, among the three Cdc6/Orc1 copies present in Sulfolobales, we correctly assigned the copy corresponding to the origin of replication embedded in an integrative element as a shell component (Robinson and Bell 2007). Similarly, among the large number of Cdc6/Orc1 copies present in Halobacteriales, only two were identified as part of the core replication, whereas all others fell into the shell component (fig. 2; supplementary table S1, Supplementary Material online).
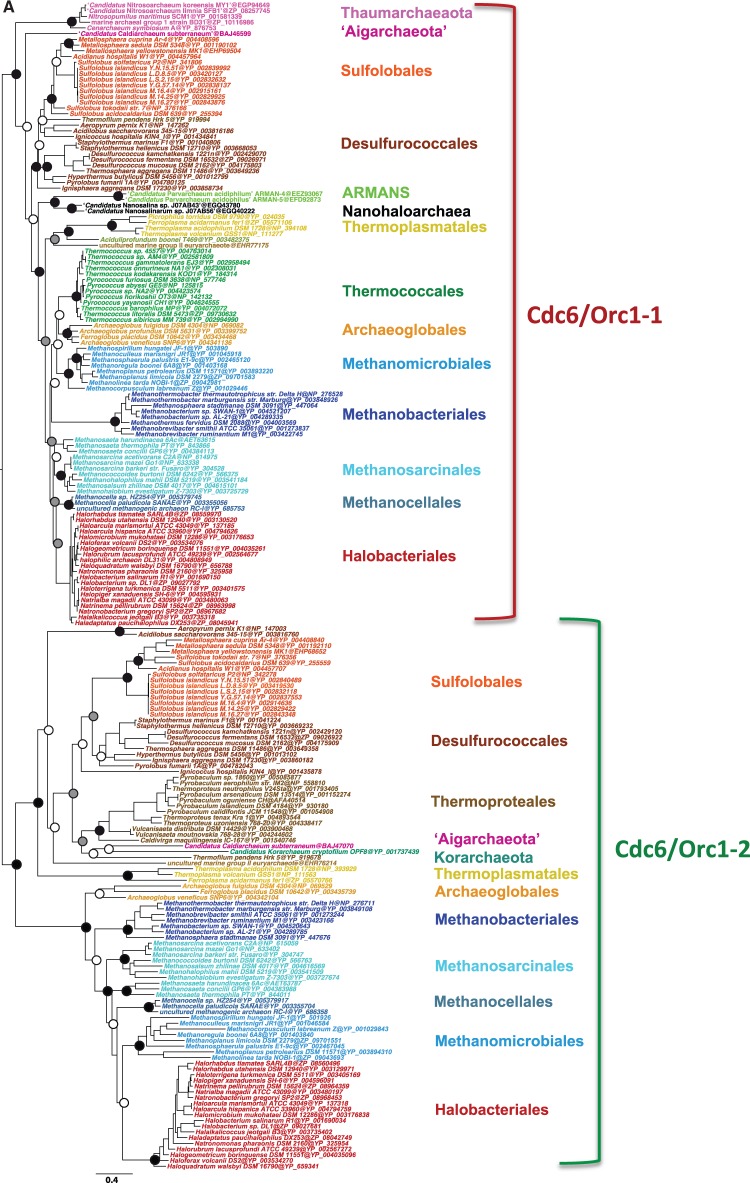
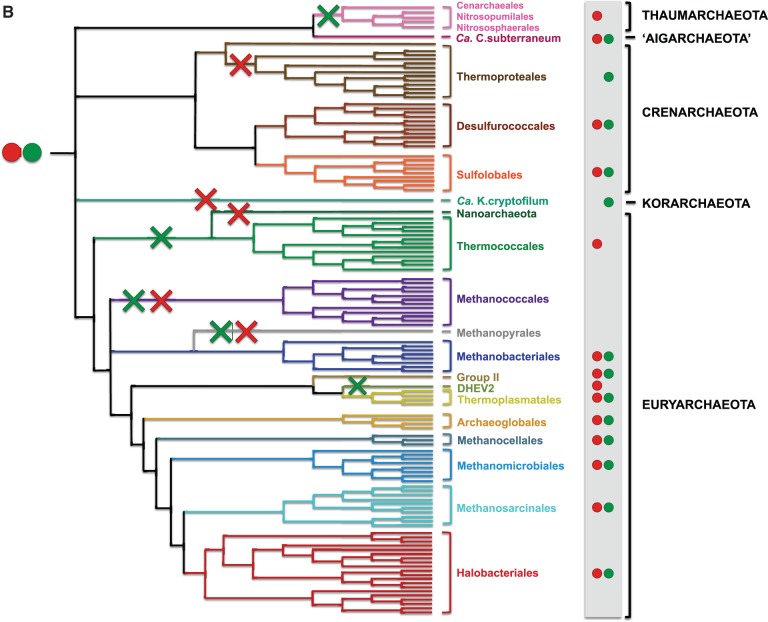
The identification of the fast-evolving shell components allowed for a finer analysis of the precise evolutionary history of core Cdc6/Orc1 proteins (fig. 3). Although the tree was not completely resolved due to the limited number of positions analyzed, the monophyly of the two clusters was strongly supported, each displaying robust monophyletic groups corresponding to the major archaeal phyla and orders (fig. 3A). In particular, when two copies are present in a given taxon, they generally correspond to either one cluster or the other. For instance, this is the case of the two core paralogs of Sulfolobales; one corresponds to the first cluster (Cdc6/Orc1-1) and the other to the second cluster (Cdc6-Orc1-2). The same is true for Halobacteriales, where only two core paralogs belonging to each of the two clusters could be identified. This suggests that Cdc6/Orc1-1 and Cdc6/Orc1-2 are ancient paralogs that arose from gene duplication and were both likely present in the LACA. Therefore, the absence of one of the two copies in present day genomes must be interpreted as the consequence of gene loss (fig. 3B). This trend of gene loss is observed across the whole archaeal tree, with different lineages having lost either one paralog or the other. For example, we can infer loss of Cdc6/Orc1-2 in the ancestor of Thaumarchaeota and in the ancestor of Thermococcales, and loss of Cdc6/Orc1-1 in the ancestor of Thermoproteales and Korarchaeota (fig. 3B). Methanococcales and Methanopyrales have pushed this trend to the extreme by losing both copies, likely in parallel to replacement by a nonorthologous protein (Zhang RR and Zhang C-CT 2004; Berthon et al. 2008). The Cdc6/Orc1-2 cluster appears to evolve faster than the Cdc6/Orc1-1 cluster and exhibits a few inconsistencies with the archaeal phylogeny, such as the branching of Korarchaeota and Aigarchaeota within Thermoproteales, and of Thermoplasmatales/uncultured marine group II at the base of Crenarchaeota (fig. 3A). More data from these lineages will be necessary to clarify whether these taxa acquired their Cdc6/Orc1-2 via horizontal gene transfer from Crenarchaeota, or if these placements are the result of a tree artifact. Indeed, a number of horizontal gene transfers from Crenarchaeota are known to have occurred during adaptation of Thermoplasmatales to thermoacidic environments (Fütterer et al. 2004). Finally, Halobacteriales have kept both Cdc6-Orc1 and Cdc6/Orc1-2 paralogs, but most genomes have acquired multiple extra copies arising from integration of mobile elements (fig. 2). It has to be noted that Cdc6/Orc1-1 coincides with one of the three origins of replication identified in H. volcanii (Hawkins et al. 2013), but Cdc6-Orc1-2 does not. The same is true for Sulfolobus solfataricus, where only Cdc6/Orc1-1 coincides with one of the three origins of replication (Samson et al. 2013).
Fig. 3.—
(A) Maximum likelihood phylogeny of Cdc6/Orc1 core components. The tree was calculated by Treefinder (MIX model + gamma4) based on 261 unambiguously aligned amino acid positions. The scale bar represents the average number of substitutions per site. Dots represent bootstrap values (BV) based on 100 replicates of the original alignment. For clarity, supports are shown for major lineages only: black dots indicate BV > 90%, gray dots BV 80–90%, and white dots BV <80%. (B) Evolutionary scenario for Cdc6/Orc1. The two Cdc6/Orc1 paralogs 1 (red) and 2 (green) arose from ancestral gene duplication in the Last Common Archaeal Ancestor. Independent gene losses occurred subsequently in a number of lineages, involving either one paralog (red crosses) or the other (green crosses), and in some cases both. See text for details.
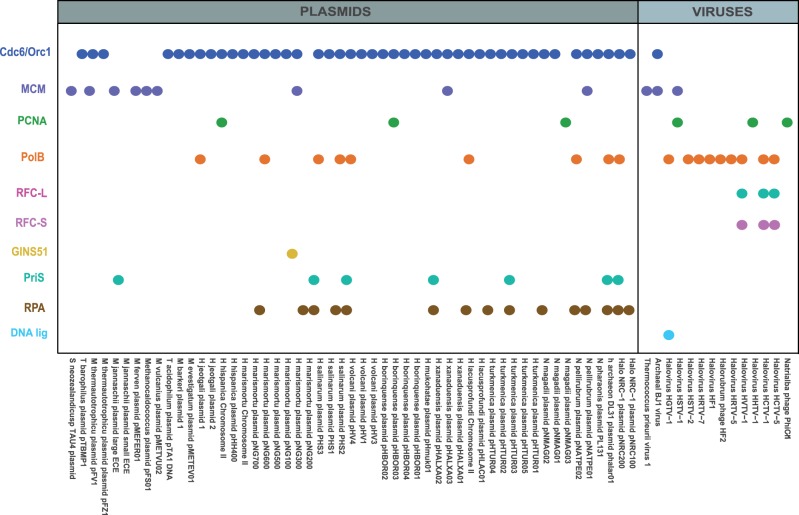
The Cdc6/Orc1 case is not unique. By using the same approach, we identified shell copies for most replication components, with an apparent preference for Cdc6/Orc1, MCM, PCNA, and PolB (fig. 2). Remarkably, the components that appear enriched in shell copies are also specifically present in plasmid and viral sequences, particularly from Halobacteriales (fig. 4; supplementary table S2, Supplementary Material online). This suggests that the shell replication homologs may come predominantly from extrachromosomal elements. In addition, it appears that extrachromosomal entities are enriched with different replication proteins, for example, Cdc6/Orc1 is more abundant in plasmids and PolB is particularly present in viruses (fig. 4). Although the current taxonomic covering of viral and plasmid sequences from archaea is narrow (supplementary table S3, Supplementary Material online), these data suggest that replication proteins are frequently exchanged between extrachromosomal elements and cellular genomes.
Fig. 4.—
Homologs of DNA replication proteins found in archaeal plasmids and viruses. Colors correspond to those used in figure 1. Accession numbers are given in supplementary table S2, Supplementary Material online.
The precise identification of core and shell replication components can be important for functional studies on archaeal replication, as proteins belonging to the core may have essential roles while shell components may keep functions linked to their extrachromosomal entity. For instance, of the three MCM present in T. kodakarensis, we assigned the gene encoding MCM3 (TK1620) to the core (supplementary table S2, Supplementary Material online); in fact, experimental data have shown that this is the only essential copy and is likely the only MCM involved in genome replication (Pan et al. 2011). Additionally, of the two PCNA homologs in T. kodakarensis, we designated PCNA1 (TK0535) as the core component and PCNA2 (TK0582) as the shell, consistent with the finding that only PCNA1 is required for cell viability (Pan et al. 2013).
The analysis of each replication protein allowed us to precisely reconstruct the global evolutionary history of DNA replication in the Archaea and the dynamics that shaped this key cellular machinery from the LACA throughout the subsequent diversification of this Domain of Life. Some of our results also provide interesting evolutionary and functional information, and are detailed hereafter.
Complex Evolutionary History of SSB and RPA Proteins
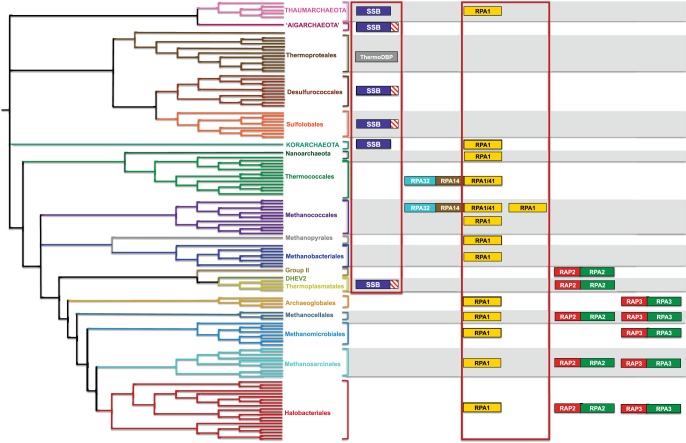
It is commonly assumed that SSB proteins with a single OB fold and a flexible C-terminal tail (SSB) are typical of Crenarchaeota (Wadsworth and White 2001) and that SSB proteins with multiple OB folds (RPA) are typical of Euryarchaeota (Grabowski and Kelman 2003; Kerr et al. 2003). The high degree of sequence divergence among archaeal SSB proteins makes the assignment of homologs particularly challenging. According to sequence similarity and the presence of single or multiple OB folds, we now clarified the distribution of SSB and RPA homologs in all archaeal genomes (fig. 2; supplementary table S1, Supplementary Material online).
Euryarchaeal RPAs can display different domain architectures and form various structural conformations. For example, Methanococcus jannaschii encodes a unique SSB protein, homologous to eukaryotic RPA70 that functions as a monomer in solution (Kelly et al. 1998). Methanosarcina acetivorans encodes a homolog of eukaryotic RPA70 called MacRPA1, along with two divergent homologs, MacRPA2 and MacRPA3, each able to self-assemble into a homomultimeric complex (Robbins et al. 2004; Skowyra and MacNeill 2012). In addition, many archaeal genomes encode proteins that are not homologous to RPA but are found close by and therefore were called RPA-associated proteins (Berthon et al. 2008) (hereafter referred to as RAP). In H. volcanii these RPA-associated proteins are thought to be cotranscribed with the adjacent RPA2 and RPA3 genes (Skowyra and MacNeill 2012) and have been shown to interact with them (Stroud et al. 2012). We found that homologs related to Methanosarcina RPA1 are largely distributed in archaeal genomes (in yellow in fig. 5, see also supplementary table S2 [Supplementary Material online] for full accession numbers) and their phylogeny, although not completely resolved, is consistent with the archaeal tree (not shown). Therefore, these likely represent the core RPA component and are likely essential. In fact, among the three RPA copies present in H. volcanii, the copy that we assigned to the core is the only one that is essential (Skowyra and MacNeill 2012).
Fig. 5.—
Taxonomic distribution and diversity of archaeal SSB and RPA homologs plus the associated proteins (RAP2 and RAP3). ThermoDP, the proposed replacement for the native SSB of Thermoproteales, is shown in gray. See text for details.
A number of late emerging euryarchaeal lineages also display one or two additional and divergent RPA homologs that we classified as RPA2 and RPA3 according to their sequence similarity to Methanosarcina acetivorans MacRPA2 and MacRPA3 (in green in fig. 5, see also supplementary table S2 [Supplementary Material online] for full accession numbers). Their specific distribution in late emerging euryarchaeal lineages and phylogenetic analysis (not shown) indicates that RPA2 and RPA3 are paralogs that arose via gene duplication in Euryarchaeota, after the divergence of Thermococcales, Methanococcales, and Methanobacteriales. We found that RPA2 and RPA3 always lie close to RAP2 and RAP3 proteins (in red in fig. 5). RAP2 and RAP3 proteins are homologous and phylogenetic analysis showed a consistent topology to that of RPA2/RPA3 (not shown) suggesting that they also arose by gene duplication in the same ancestor. Such similar evolutionary history and genomic association strongly points to an ancient and important functional linkage of RPA and their associated proteins in these euryarchaeota.
Thermococcales display very peculiar characteristics concerning their SSB proteins. Pyrococcus furiosus harbors three nonhomologous SSB proteins: RPA41, RPA14 (which, despite its name, is not homologous to eukaryotic RPA14), and RPA32. Together these form a stable heterotrimeric complex, and their encoding genes are adjacent in the genome (Komori and Ishino 2001). RPA41 is only distantly related to other archaeal RPA1 homologs, and closely related homologs of RPA32, RPA14, and RPA41 are also found in Methanococcales where they maintain the same genomic arrangement (fig. 5). Because these two orders do not share an exclusive common ancestor according to ribosomal protein trees (Matte-Tailliez et al. 2002; Brochier et al. 2004, 2005; Brochier-Armanet et al. 2011), the presence of such a unique three-protein RPA system may be explained with a horizontal gene transfer, either directly or through a common mobile element, which possibly displaced the original RPA1. In fact, some Methanococcales genomes still harbor an RPA1 homolog that may represent the original protein (fig. 5; supplementary table S2, Supplementary Material online).
In contrast to RPA, SSB homologs have a much more restricted taxonomic distribution and are mostly present in a single copy (fig. 2; supplementary table S2, Supplementary Material online). The presence of an SSB in Thermophilum pendens, an early emerging lineage in the Thermoplasmatales, testifies to the ancestral presence of this protein in this lineage prior to its replacement by the nonhomologous ThermoDPB (Paytubi et al. 2012). The distribution of SSB appears complementary to that of RPA, with the notable exception of Thaumarchaeota, Korarchaeota, Thermoplasmatales/DHEV2, two Nanohaloarchaea, and ARMAN, which harbor both an RPA1 and an SSB homolog (fig. 2). The function of SSB homologs outside the Crenarchaeota is unknown, as is their possible interaction or division of labor in the taxa that harbor an RPA homolog. We noticed that the SSB homologs of Aigarchaeota and Thermoplasmatales/DHEV2 harbor the flexible C-terminal tail typical of crenarchaeal SSB. In Crenarchaeota, this tail appears to be involved in repair and recombination (Cubeddu and White 2005) (schematically represented by a striped box in fig. 5, for a full alignment see supplementary fig. S1, Supplementary Material online). This tail is absent from the SSB of Thaumarchaeota and Korarchaeota, which harbor an RPA1 homolog (fig. 5; supplementary fig. S1, Supplementary Material online). This may hint at a change in function of SSB in these taxa or even a potential interaction with RPA1. Indeed, in the genomes of Candidatus Parvarchaeum acidophilum ARMAN-4 and Candidatus Parvarchaeum acidophilus ARMAN-5′ the gene coding for RPA1 lies next to the gene coding for SSB (supplementary table S1, Supplementary Material online). Phylogenetic analysis of SSB homologs (supplementary fig. S1, Supplementary Material online) suggests that Thermoplasmatales and Aigarchaeota may have acquired their SSB via horizontal gene transfer from Crenarchaeota, an event possibly linked with the loss of the native RPA1 in both lineages. Intriguingly, this putative transfer displays a similar pattern to the one that is likely at the origin of the Cdc6/Orc1-2 of these lineages, as discussed earlier. It is therefore not excluded that both Cdc6/Orc1-2 and RPA1 where transferred together, indicating a possible direct functional linkage of these two components.
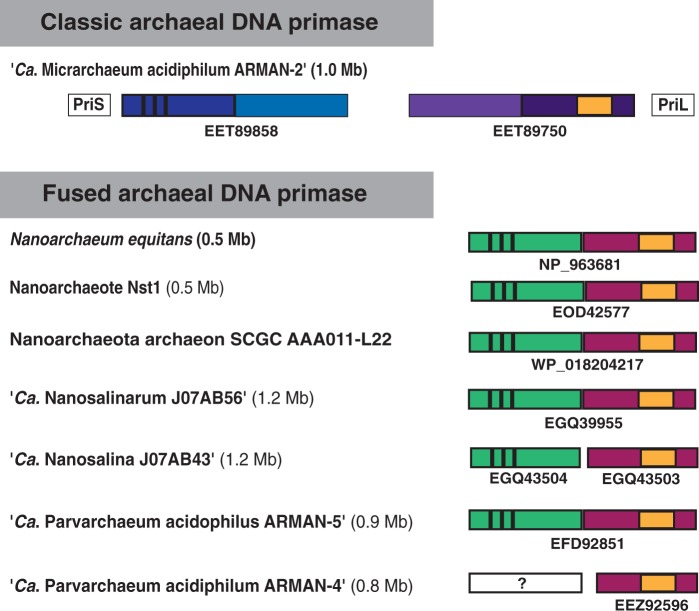
Fused Archaeal DNA Primases: A Shared Derived Character for Nanosized Archaea?
Archaeal DNA primases (PriS and PriL) show low sequence similarity with their eukaryotic counterparts and even within Archaea. Most archaea contain a classic primase, made of a catalytic subunit PriS and an accessory subunit PriL (fig. 6). The PriL subunit contains a conserved Fe-S cluster-binding domain that plays an important role in primase activity (Klinge et al. 2007) (fig. 6, yellow box). The activity of PriS lies in an N-terminal catalytic domain with a conserved motif (fig. 6, black bars). It has been previously observed that Nanoarchaeum equitans contains a short atypical primase encoded by a single gene, which is composed of a fusion of the catalytic domain of PriS and the Fe–S cluster-binding domain of PriL (Iyer et al. 2005). We identified this same type of primase in the recently sequenced Nanoarchaeote Nst1 (Podar et al. 2012) and in an uncultured nanoarchaeon from a recent single cell genomics survey (Rinke et al. 2013).
Fig. 6.—
Schematic representation of the classic archaeal DNA primase genes encoding for the two subunits PriS and PriL, as opposed to the single genes encoding for fused archaeal primases that we found in some nanosized lineages. The presence of a PriS in Ca. Parvarchaeum acidophilum ARMAN-4 is unknown (question mark). The genome sizes are given in parentheses. See text for details.
Besides Nanoarchaeota, two novel uncultured archaeal lineages characterized by reduced genomes and very small cell sizes have been highlighted recently: a candidate class called Nanohaloarchaea represented by three metagenomic assemblies isolated from a highly saline lake in Australia (Narasingarao et al. 2012), and the Archaeal Richmond Mine Acidophilic Nanoorganisms or ARMAN lineage represented by three metagenomic assemblies isolated from an acidic iron-rich mine in the United States (Baker et al. 2010). Interestingly, we found that Candidatus Parvarchaeum acidophilus ARMAN 5 and the nanohaloarchaeon Candidatus Nanosalinarum sp. J07AB56 contain a single gene encoding a fused PriS/PriL whose sequences are closely related to that of N. equitans but are very divergent in comparison to other archaeal primases. The second available nanohaloarchaeum Candidatus Nanosalina sp. J07AB43 harbors two adjacent genes encoding for a short primase that clearly align with the other fused primases (fig. 6). Candidatus Parvarchaeum acidiphilum ARMAN 4 has a PriL homolog that aligns well with the C-terminal metal binding domain of the short PriL, but appears to lack the N-terminal catalytic PriS domain (fig. 6). However, it is located at the end of a contig in this nonassembled genome, and therefore the presence of the PriS domain cannot be excluded at present. In contrast, Candidatus Micrarchaeum acidiphilum ARMAN 2 possesses a classic primase (fig. 6).
It could be argued that these peculiar fused primases arose from evolutionary convergence following genome streamlining in these nanosized lineages. However, the hypothesis of convergence can be excluded because they are related at the sequence level. This leaves two possibilities: either the lineages harboring a fused primase share a common ancestor or the fused primases have replaced the original primases via horizontal gene transfer. Based on phylogenetic analysis of 38 universal protein markers, Rinke et al. (2013) have proposed the existence of a monophyletic superphylum called DPANN whose members would be characterized by small cell and genome sizes and would include the ARMANS, Nanohaloarchaea, and Nanoarchaeota. The sharing of fused primases may appear consistent with the existence of a DPANN clade. However, it is not consistent with Ca. Micrarchaeum acidiphilum ARMAN 2 harboring a classical primase. Moreover, the grouping of nanosized archaeal lineages in phylogenetic trees should be interpreted with caution given that robust clustering of fast evolving lineages is a well-known artifact of phylogenetic reconstruction (Gribaldo and Philippe 2002). Indeed, recent ribosomal protein trees support the clustering of Ca. Parvarchaeum acidiphilum ARMAN 4, Ca. Parvarchaeum acidophilus ARMAN 5 and Nanoarchaeota to the exclusion of Ca. Micrarchaeum acidiphilum ARMAN 2 (Brochier-Armanet et al. 2011), and the grouping of Nanohalobacteria with Halobacteriales (Narasingarao et al. 2012).
Alternatively, it may be hypothesized that these fused primases have replaced the original primase via horizontal gene transfer among these lineages, possibly through related integrative elements. Fused DNA primases might be frequent in integrative elements, as suggested by the DNA polymerase/primase recently highlighted in the plasmid pTN2 from Thermococcus nautilus (Soler et al. 2010) that harbors a similar PriS/PriL fusion. However, we observe that this fused primase displays no sequence similarity with the primases of nanosized archaea, indicating an independent origin. Moreover, organisms belonging to nanosized lineages thrive in very different environments (hyperthermophilic [Huber et al. 2002], extreme halophilic [Narasingarao et al. 2012], or extreme acidic [Baker et al. 2006]), making the hypothesis of a horizontal gene transfer puzzling. Undoubtedly, more data are needed to clarify the issue and further understand the diversity and evolutionary history of these fascinating lineages.
Acquisition of Bacterial DNA Gyrase: When and How Many Times?
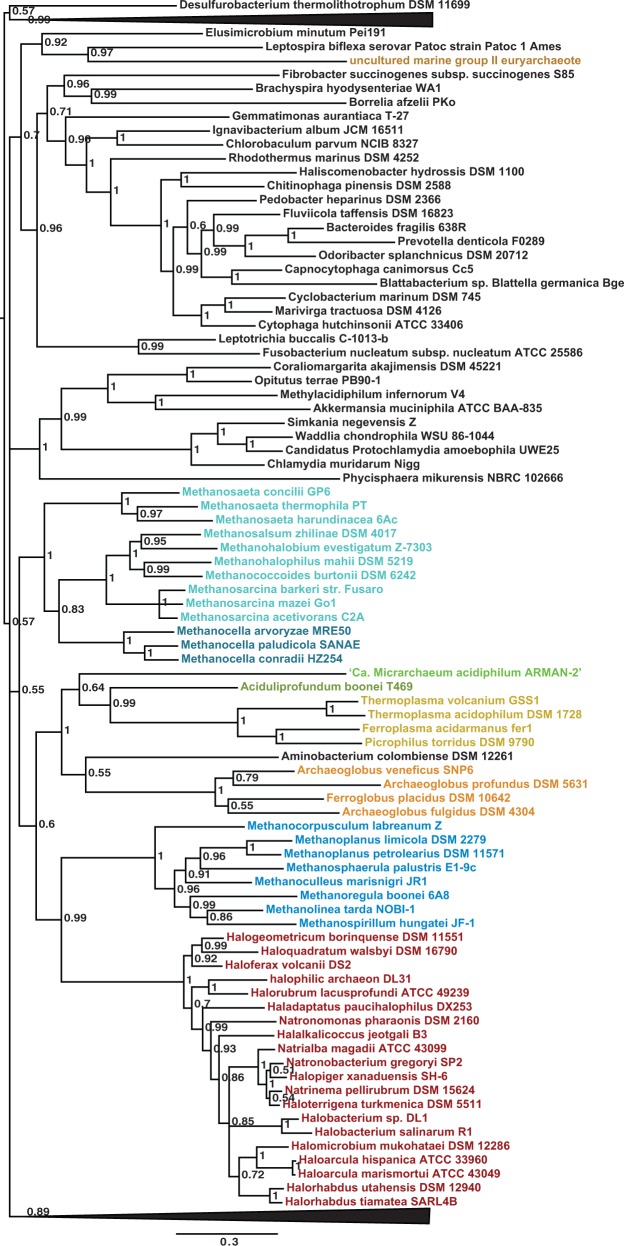
To resolve topological conflicts arising during replication, archaea use a TopoVI that relaxes both positive and negative supercoils. Previous phylogenetic analysis has indicated that bacterial-like DNA gyrases were acquired in a number of euryarchaeota through horizontal gene transfer (Forterre et al. 2007). Because bacterial DNA gyrases actively introduce negative DNA supercoiling, this transfer event likely had a significant impact, changing the overall genome topology and all associated cellular processes, such as the pattern of gene expression (Forterre et al. 2007; Forterre and Gadelle 2009). In most of these euryarchaea, DNA gyrase now coexists with the endogenous TopoVI. In contrast, Thermoplasmatales have lost their original TopoVI and now must solely rely on DNA gyrase for replication and chromosome decatenation (Forterre et al. 2007; Forterre and Gadelle 2009). With the availability of an expanded taxonomic sampling covering more euryarchaeal diversity, we sought to address the timing and number of events that introduced DNA gyrase into this phylum. Consistent with previous reports, we found both DNA gyrase subunits in all genomes from the orders Archaeoglobales, Methanosarcinales, and Halobacteriales (Bergerat et al. 1997; Forterre et al. 2007; Berthon et al. 2008; Forterre and Gadelle 2009). We also identified both subunits in all analyzed genomes of the orders Methanomicrobiales and Methanocellales (which together with Methanosarcinales form the methanogen class II), as well as in DHEV2, uncultured marine group II, and Ca. Micrarchaeum acidiphilum ARMAN 2 (fig. 2; supplementary table S1, Supplementary Material online).
Given that these lineages form a late emerging monophyletic cluster in the archaeal phylogeny, and that DNA gyrase is most likely rarely acquired because of its biological consequences, we speculated that this horizontal gene transfer occurred only once at the base of this group. Albeit not completely resolved, a phylogenetic tree of concatenated large and small DNA gyrase subunits shows that archaeal sequences form a monophyletic cluster (fig. 7) supporting a single acquisition of DNA gyrase in these archaea via horizontal gene transfer from an unidentified bacterium. The uncultured marine group II is an exception and likely represents an independent horizontal transfer. However, the weak phylogenetic signal makes this monophyletic group very unstable, as it can be broken up in two clusters depending on the bacterial taxonomic sampling used (not shown). In this case, one cluster corresponds to Halobacteriales and Methanogens class II, and the other to Thermoplasma/DHEV2/Archaeoglobales/ARMAN-2. This would indicate that two independent horizontal gene transfers from bacteria are at the origin of DNA gyrases in the two groups of archaea. However, we speculate that the second transfer would have been possible only because the newly introduced DNA gyrase replaced an already present bacterial-type enzyme. The two alternative scenarios (a single transfer or two successive transfers) remain possible, as statistical tests showed that the data do not reject either of the two topologies (P > 0.48 for all tests, see Materials and Methods for details).
Fig. 7.—
Bayesian phylogeny of a concatenation of archaeal DNA gyrase small and large subunits and a selection of bacterial homologs (1,083 amino acid positions). The tree was calculated by MrBayes (MIX model + gamma4). The scale bar represents the average number of substitutions per site. Supports at nodes indicate posterior probabilities. Colors correspond to archaeal orders according to those used in figure 2. The tree is collapsed for clarity. See supplementary table S1 (Supplementary Material online) for accession numbers and taxonomic information.
DNA gyrase is likely essential in all species that harbor it, suggesting that it may be difficult to lose this enzyme once acquired. We could not find any homologs of DNA gyrase in the genomes of Nanohaloarchaea nor of ARMAN-4 and ARMAN-5 (fig. 2). This may be consistent with an emergence of these lineages prior to the alleged first horizontal gene transfer introducing DNA gyrase in the Thermoplasma/DHEV2/Archaeoglobales/ARMAN-2.
DNA Replication Proteins Harbor a Robust Signal for Archaeal Phylogeny
Fourteen core DNA replication orthologs present in more than 60% of the taxa (PriS, MCM, PCNA, Cdc6/Orc1, DPL, DPS, PolB, TopoVI-A, TopoVI-B, RFC-s, RFC-l, RNaseH, DNA ligase, and FEN-1) were concatenated into a large supermatrix of 4,295 amino acid positions from 129 complete or nearly complete archaeal genomes (keeping only one genome per species, see Materials and Methods). The amount of missing data from the concatenation was analyzed, and except for phyla or orders displaying specific losses or absences (e.g., both small and large subunits of PolD absent in all Crenarchaeota) there are no specific species that are underrepresented (supplementary fig. S2, Supplementary Material online). The phylogeny obtained from this supermatrix (fig. 8) is highly consistent with the previous archaeal phylogenies inferred from transcription and translation components (Matte-Tailliez et al. 2002; Brochier et al. 2004, 2005; Brochier-Armanet et al. 2011). The monophylies of Crenarchaeota, Euryarchaeota, Korarchaeota, and Thaumarcheaota are all recovered with strong support as well as those of all major orders. The phylogeny solidifies the clustering of uncultured marine group II and the DHEV2 representative with the Thermoplasmatales (Brochier-Armanet et al. 2011) and the monophyly of Methanogens class I (i.e., Methanopyrus kandleri + Methanobacteriales + Methanococcales) (Bapteste et al. 2005). The robust monophly of Thaumarchaeota and Aigarchaeota observed in the replication tree is in agreement with the proposal that Aigarchaeota represent an early emerging thaumarchaeotal lineage (Brochier-Armanet et al. 2011). Other important points that should be underlined are 1) the emergence of Acidilobus within Desulfurococcales, which refutes the recent proposal of the new order Acidilobales (Prokofeva et al. 2009); 2) the clustering of Halobacteriales with Methanogens class II, with a specific grouping of Methanomicrobiales and Halobacteriales; 3) the grouping of Methanogens class II + Halobacteriales with Archaeaoglobales and Thermoplasmatales (fig. 8).
Fig. 8.—
Bayesian phylogeny of a concatenated data set of 14 replication components (4,295 amino acid positions). The tree was calculated by Phylobayes (CAT + GTR + gamma4). The scale bar represents the average number of substitutions per site. Values at nodes represent posterior probabilities and BV based on 100 resamplings of the original data set calculated by PhyML (LG model + gamma4), when the same node was recovered.
A few differences were observed between the replication phylogeny and the previous trees based on ribosomal proteins (Brochier-Armanet et al. 2011). For example, the robust monophyly of Methanogens class I and Thermococcales, the grouping of Korarchaeota with Thaumarchaeota, and the early emergence of Methanocellales within Methanogens class II (fig. 8). Finally, all of the nanosized archaea (Nanoarchaeota, ARMAN-5, ARMAN-4, and the three Nanohaloarchaea), except for ARMAN-2, form a monophyletic clade that emerges after the divergence of Thermococcales and Methanogens class I (fig. 8). Considering the very fast evolutionary rate of these lineages, it cannot be excluded that this grouping is due to a tree reconstruction artifact. To test this possibility, we created several versions of the concatenated dataset containing different combinations of taxa (i.e., we removed all nanosized lineages from the concatenation and reintroduced them one by one) and we recoded the amino acid supermatrix using Dayhoff6 and Dayhoff4 recoding schemes, a procedure known to alleviate certain artifacts due to fast evolutionary rates (Delsuc et al. 2005). However, no major differences were observed.
Discussion
Dynamic History of a Key Cellular System
Through our precise identification and phylogenetic analysis of core replication components, we reconstructed the global evolutionary history of the DNA replication machinery in Archaea. In particular, we inferred the presence of a complete and modern type machinery in the LACA (table 1). The LACA would have harbored two Cdc6/Orc1 paralogs, two GINS paralogs (GIN23 and GIN51), and one homolog each of the MCM helicase, the sliding clamp PCNA and its loader RFC with both subunits, the polymerase PolB, the archaeal primase with both subunits, the Okazaki fragment processing flap endonuclease Fen1 and RNaseH II, the ATP-dependent DNA ligase, and the topoisomerase Topo VI with both subunits. Although the involvement of DnaG in replication is dubious, this protein must have an important and conserved role because it is universally present in archaea. Moreover, the phylogeny is robustly supported and is strikingly consistent with the archaeal species tree (not shown). This indicates that the presence of DnaG in archaea is not due to horizontal gene transfer from bacteria but instead was harbored by the LACA and was subsequently strictly vertically inherited up to present. For the few remaining components (PolD, SSB, and RPA1), their presence in the LACA strongly depends on the root of the archaeal tree, which is presently unclear (Brochier-Armanet et al. 2011; table 1). TopoIB represents a special case because its presence in LACA relies on whether Archaea and Eukaryotes are sister lineages, a currently unsettled matter (see below).
Table 1.
Inferred Components of DNA Replication in the LACA and in the Ancestor of Each Major Phylum
| LACA | Thaumarchaeota/Aigarchaeota | Korarchaeaota | Crenarchaeota | Euryarchaeota |
|---|---|---|---|---|
| Cdc6/Orc1-1 | Cdc6/Orc1-1 | Cdc6/Orc1-1 | Cdc6/Orc1-1 | |
| Cdc6/Orc1-2 | Cdc6/Orc1-2 | Cdc6/Orc1-2 | Cdc6/Orc1-2 | Cdc6/Orc1-2 |
| MCM | MCM | MCM | MCM | MCM |
| GINS51 | GINS51 | GINS51 | GINS51 | GINS51 |
| GINS23 | GINS23 | GINS23 | GINS23 | GINS23 |
| RPA1 | RPA1 | RPA1 | RPA1 | |
| SSB | SSB | SSB | SSB | |
| PolB | PolB | PolB (X2) | PolB (X2) | PolB |
| PolD-L/S | PolD-L/S | PolD-L/S | DP-L/S | |
| RFC-S/L | RFC-S/L | RFC-S/L | RFC-S/L | RFC-S/L |
| PCNA | PCNA | PCNA | PCNA (X2) | PCNA |
| Pri-S/L | Pri-S/L | Pri-S/L | Pri-S/L | Pri-S/L |
| RNaseH II | RNaseH II | RNaseH II | RNaseH II | RNaseH II |
| FEN-1 | FEN-1 | FEN-1 | FEN-1 | FEN-1 |
| ATP DNA ligase | ATP DNA ligase | ATP DNA ligase | ATP DNA ligase | ATP DNA ligase |
| TopoIV-A/B | TopoVI-A/B | TopoVI-A/B | TopoVI-A/B | TopoVI-A/B |
| TopoIB | TopoIB | |||
| Root-dependent components | ||||
| Thaumarchaeota/“Aigarchaeota” → PolD-L/S, RPA, SSB, TopoIB | ||||
| Korarchaeota → PolD-L/S, RPA, SSB | ||||
| Crenarchaeota → SSB | ||||
| Euryarchaeota → PolD-L/S, RPA | ||||
Note.—Additional components that would have been present in the LACA according to a rooting in each of the four major phyla are indicated. Components shown in bold have homologs in eukaryotes and those shown in gray are root dependent.
The core components inferred in the ancestor of each phylum are overall very similar (table 1). Major differences appear most evident in the ancestor of Crenarchaeota, with a number of specific characters such as the presence of at least two PCNA and PolB paralogs, the absence of PolD, and the presence of SSB but not RPA. The subsequent evolutionary history of the DNA replication machinery appears very dynamic. In particular, the absence in any present day lineage of a component inferred to have been present in the LACA has to be interpreted as a consequence of gene loss. We observed many independent gene losses frequently involving one of two ancestral paralogs, for example, Cdc6/Orc1 and GINS. A similar phenomenon of gene loss has been observed in archaeal ribosomes, which appear to have experienced independent losses of components in different lineages (Desmond et al. 2010; Yutin et al. 2012), as well as on a global genomic scale (Csuros and Miklos 2009). Our results are therefore consistent with a growing consensus on a complex LACA (Makarova et al. 2007; Csuros and Miklos 2009; Wolf et al. 2011).
However, there is not a unique trend toward gene loss in regard to the replication machinery. We highlighted the occurrence of a number of component accretions throughout archaeal diversification. Examples are the multiplication of RPA copies in Euryarchaeota and the expansion of the MCM family in Methanococcales. These are both due to gene duplication of core components and acquisition of additional shell components from extrachromosomal elements. Some of these events also led to increased complexity of multiprotein machineries involved in replication. For example, whereas most archaeal RFC are composed of four identical RFC small subunits (RFC-S) and one RFC large subunit (RFC-L) (Barry and Bell 2006), some species contain two RFC-S homologs (RFC-S1 and RFC-S2). In these cases, three RFC-S1 subunits and one RFC-S2 subunit assemble with RFC-L to form the pentameric RFC complex (Chen et al. 2005). Similarly, Crenarchaeota contain two or three copies of PCNA that have arisen from gene duplication and form a heterotrimeric structure in which each subunit has specific binding functions to different replication proteins (Grabowski and Kelman 2003; Barry and Bell 2006). It is noteworthy that, according to current knowledge, these accretions of components in multisubunit complexes appear to be due to gene duplication rather than integration of shell components or horizontal gene transfer. However, it will be very interesting to study if extra copies arising from integrative elements may, in some instances, replace the native component or integrate complexes made of core components.
As opposed to the high dynamics of shell components, horizontal gene transfers involving core components appear to be relatively rare. A few cases can been seen which are consistent with known exchanges amongst archaea thriving in the same environments such as from Crenarchaeota to Thermoplasmatales. Moreover, we show that horizontal gene transfer events involving bacterial replication components, albeit rare, have occurred during archaeal diversification. For example, other than the previously discussed case of DNA gyrase, we observed a single horizontal gene transfer introducing a bacterial-type NAD + -dependent DNA ligase in the ancestor of Halobacteriales (not shown), which may have in some cases replaced the native archaeal/eukaryal ATP- dependent DNA ligase (fig. 2; supplementary table S1, Supplementary Material online).
Why So Many DNA Replication Components in Extracellular Elements?
An evident phenomenon affecting archaeal DNA replication is the presence of many divergent extra copies particularly those involved in the first steps of replication, such as Cdc6/Orc1, MCM, RPA1, and PolB (fig. 2). Moreover, different archaeal viruses, proviruses, and plasmids are known to encode homologs of Cdc6/Orc1 and MCM (Pagaling et al. 2006; Yamashiro et al. 2006; Krupovic, Forterre, et al. 2010). Similarly, an archaeal homolog of eukaryotic Ctd1 called WhiP was recently identified in the integrative element that contributed the third origin of replication in Sulfolobales (Robinson and Bell 2007). Precise identification of all extra copies of replication components that reside in integrative elements in archaeal genomes requires extensive work and is beyond the scope of this article. Nevertheless, our study strongly suggests that extrachromosomal elements have had an impact on the evolution of the archaeal DNA replication machinery and actively modeled its composition, both by picking up and transferring components to and from cellular genomes. Considering the small number and taxonomic coverage of viral sequences presently available in public databases (supplementary table S3, Supplementary Material online) our analysis suggests that the world of archaeal extrachromosomal entities may be particularly enriched in genes encoding for replication proteins. Moreover, the presence of highly divergent and related components in Thermococcales and Methanococcales, such as their DNA primase and the RPA three-gene cluster, may indicate potential avenues of gene sharing through a common pool of plasmids and viruses (Soler et al. 2010).
Archaeal plasmids and viruses rarely encode components of the transcription machinery and, to our knowledge, no translation components. The targeting of DNA replication by virus/plasmid entities to hijack the host machinery provides a strong advantage and is a well-known phenomenon. However, it is much less known that, upon viral/plasmid integration, many DNA replication proteins of extrachromosomal origin became residents (either transient or permanent) of cellular genomes. This can confuse the phylogeny of these proteins if the difference between real and false cellular genes is not correctly assessed. Finally, it will be interesting to carry out a similar global analysis in Bacteria and Eukaryotes to understand whether this phenomenon is particularly evident in the Archaea or is a more general trend.
An Archaeon at the Origin of Eukaryotes?
A recent analysis inferred the core DNA replication components in the last eukaryotic common ancestor (Aves et al. 2012). Aves et al. predicted that LECA (the Last Eukaryotic Common Ancestor) would have possessed all of the components that we have inferred in the archaeal ancestor, with the exclusion of PolD (table 1). This is coherent with the classical scenario indicated by ancient paralogous protein pairs where Archaea are a sister lineage to Eukaryotes (Gogarten et al. 1989; Iwabe et al. 1989; Gribaldo and Cammarano 1998). In contrast, recent analyses support the emergence of Eukaryotes from within the archaeal radiation (Cox et al. 2008; Foster et al. 2009; Guy and Ettema 2011; Williams et al. 2012; Alvarez-Ponce et al. 2013; Lasek-Nesselquist and Gogarten 2013). In particular, a deep branching within a cluster composed of Thaumarchaeota, Aigarcharchaeota, Korarchaeota, and Crenarchaeota seems to be predominant, and would be consistent with an apparent enrichment of eukaryotic-like characters in these phyla with respect to Euryarchaeota (Guy and Ettema 2011).
Unfortunately, archaeal DNA replication components are very divergent from their eukaryotic homologs, preventing the reconstruction of reliable phylogenies to test the evolutionary relationship between these two domains of life. Nonetheless, our reconstruction of the evolution of the DNA replication machinery along archaeal diversification sheds new light on this issue. The absence of eukaryotic core components from the replication machinery of the ancestor of a given archaeal lineage would exclude the emergence of eukaryotes from one of its members (unless invoking an extremely unparsimonious scenario where the component was independently lost in all members of the lineage but only kept in the one that would have given rise to eukaryotes). By this rationale, we can exclude the emergence of eukaryotes from within the radiation of any of the major archaeal phyla. For example, the lack of GINS 23 and SSB in the ancestor of Euryarchaeota (table 1) would exclude an emergence of Eukaryotes from within this phylum. Similarly, the absence of RPA in the ancestor of Crenarchaeota would also exclude an emergence of Eukaryotes from within the radiation of this phylum. Furthermore, an origin of Eukaryotes from within Crenarchaeota also seems unlikely given the presence of a peculiar heterotrimeric PCNA derived from an ancestral homotrimeric structure. In this situation, the complex would have reverted back into the homo-trimeric form observed in present day eukaryotes, an improbable scenario. Among the four major archaeal phyla, none seem to be particularly enriched in characters shared with Eukaryotes, perhaps with the exception of Thaumarchaeota (table 1). However, this kind of argument should not be used to infer a specific evolutionary link between Eukaryotes and Thaumarchaeota. In fact, gene loss appears to be a common process that has substantially affected DNA replication, along with many other cellular processes during the diversification of Archaea.
Irrespective of the different evolutionary scenarios for the origin of eukaryotes, our study indicates that the ancestral replication machinery of these two domains of life was very similar (table 1). Therefore, our analysis provides a key starting point for understanding the subsequent evolutionary history of the eukaryotic DNA replication machinery. For example, specific gene duplications would have occurred in the eukaryotic ancestor giving rise to paralogous components such as MCM(2-7) and GINS (Sld5, Psf1, Psf2, and Psf3), or the addition of multiple nonhomologous subunits like ORC(1-6), RPA(70, 34, 14), and RNaseH2 (A, B, C). A few components with homology to archaea are not involved in replication in eukaryotes, and it can be speculated that they were reassigned to other cellular functions. For example, most eukaryotes encode a homolog of the A subunit of archaeal TopoVI called Spo11 (Bergerat et al. 1997), which is not involved in replication but instead induces the double stand breaks that initiate meiotic recombination (Bergerat et al. 1997; Martini and Keeney 2002). In contrast, members of the Archaeplastida (land plants and green, red, and glaucocystophyte algae) possess homologs of both subunits (A and B) of archaeal TopoVI, where they combine into a functional enzyme that appears to play a role in DNA endoreduplication, a process required for polyploidization (Hartung and Puchta 2001). The presence of both subunits in some protist lineages such as Kinetoplastids opens up the possibility that a functional TopoVI was present in the ancestor of Eukaryotes (Malik et al. 2007), and was subsequently lost in most lineages. The same logic applies to the archaeal-like SSB that we identified in representatives of most eukaryotic phyla (supplementary table S4, Supplementary Material online), where it may have an important and possibly ancestral role in (Robbins et al. 2005; Richard et al. 2008; Shi et al. 2012).
On the other hand, a few of the core components of eukaryotic DNA replication are not present in Archaea and therefore would have arisen specifically in the lineage leading to Eukaryotes. This is the case of DNA pol-α and the B-subunit of the primase complex, topoisomerase IIA, and the FACT complex (Aves et al. 2012). The emergence of DNA pol-α is particularly fascinating. In Bacteria and Archaea the RNA primer is directly extended by the main replicative DNA polymerase, but in Eukaryotes Pol-α adds 10-30 nt DNA stretches to the RNA primer, and only then does the complex hand-off to the main replicative DNA polymerase (DePamphilis and Bell 2010). These 10–30 nucleotides therefore need to be removed during Okazaki fragment maturation (Stillman 2008), raising the question of the origin of this polymerase (Forterre 2013). The future availability of both genomic and experimental data from a larger fraction of eukaryotic diversity will surely allow a better understanding of the diversity and evolutionary history of DNA replication in this Domain of Life.
Finally, further exploration of diversity and function of archaeal replication may uncover unsuspected links with their eukaryotic cousins. It is not excluded that some of these components/functions were ancestrally present in the Archaea and subsequently lost.
Increasing the Conserved Phylogenomic Core for Archaea
In the past, we have shown that the components of the transcription and translation machineries contain a consistent and robust phylogenetic signal that reflects the history of archaeal diversification (Brochier et al. 2005; Gribaldo and Brochier-Armanet 2006; Gribaldo and Brochier 2009). The third major informational system that remained to be analyzed was the DNA replication machinery. However, the complex evolutionary history of DNA replication components and the occurrence of multiple highly divergent copies of unclear origin rendered the application of phylogenomic approaches to this cellular machinery particularly challenging. Our precise identification of orthologs has now made it possible to perform such analysis, and indeed, archaeal DNA replication carries a robust phylogenetic signal that is largely consistent with that of the two other informational systems. Moreover, reconstructing the evolution of DNA replication brings novel information to the archaeal phylogeny. It consolidates important relationships such as Aigarchaeota as a sister lineage of Thaumarchaeota, and the monophyly of Methanogens class I. The clustering of Thermococcales and Methanococcales merits further study, because it is not apparent in trees based on ribosomal proteins or transcription components (Matte-Tailliez et al. 2002; Brochier et al. 2004, 2005; Brochier-Armanet et al. 2011) but is in agreement with some common peculiarities in their replication machinery (see above). Therefore, this relationship in the tree based on replication components may reflect a bias introduced by undetected independent transfers from related mobile elements, viruses, and/or plasmids. The phylogenetic placement of nanosized archaea remains unclear. Their grouping in our trees may indicate common ancestry, but only partially supports the recently proposed DPANN cluster (Rinke et al. 2013). In fact, one member of the ARMANS (Ca. Micrarchaeum acidiphilum ARMAN-2) does not cluster with the other nanosized lineages, consistent with the analysis of ribosomal proteins (Brochier-Armanet et al. 2011). This is congruous with a number of additional observations: the absence of a fused primases (figs. 2 and 6), the presence of bacterial DNA gyrase (figs. 2 and 5), the presence of an SSB with an N-terminal tail (figs. 2 and 5; supplementary fig. S1, Supplementary Material online), and the absence of RPA. Targeted phylogenomic analyses combined with novel genomic data from these peculiar lineages will bring important insights into this issue.
It is important to highlight that a detailed analysis such as ours allows for the identification of novel phylogenetic markers that would most likely be discarded by more automated analyses. A commonly used approach to build concatenated data sets for phylogenetic analysis is to choose genes present in a single copy in all (or nearly all) genomes to avoid problems arising from a mixture of orthologs and paralogs. Such a strategy drastically reduces the number of usable markers, especially when dealing with deep evolutionary relationships. In addition, this type of strategy biases our understanding of prokaryotic evolution, by underrepresenting vertical inheritance (tree-like process) with respect to horizontal gene transfers (net or forest-like process) (Dagan and William Martin 2006). Had we applied such strategy, we would have essentially discarded all replication components. Instead, we have shown that reliable phylogenetic information can be extracted even from proteins that are not universally distributed or exist in multiple paralogs—allowing the tree to appear from the forest. Even if a strict core of vertically inherited genes might be limited, our results clearly demonstrate the existence of a soft core of cellular components involved in different processes whose genes have similar histories and can therefore be used to trace back the evolutionary relationships among the organisms that carry them (Gribaldo and Brochier-Armanet 2006; Gribaldo and Brochier 2009). It is likely that this soft phylogenomic core is much richer than usually assumed.
Conclusions
The emergence of novel techniques grants rapid access to an ever-wider fraction of microbial diversity, both from a genomic and functional point of view. In this context, the integration of evolutionary studies will be of primary importance, not only to provide key information for experimental work but also to uncover general trends in the global evolutionary history of the largest fraction of the biosphere.
Supplementary Material
Supplementary figures S1 and S2 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge the article by Makarova and Koonin (2013) that was published while this manuscript was under review and which presents a comparison of DNA replication in Archaea and Eukaryotes. The authors thank the PRABI (Pôle Rhône-Alpes de Bioinformatique) for providing computing facilities. K.R. is a scholar from the Pasteur–Paris University (PPU) International PhD program and received a stipend from the Paul W. Zuccaire Foundation. C.B.A. is member of the Institut Universitaire de France. This work was supported by the Investissement d’Avenir grant “Ancestrome” (ANR-10- BINF-01-01).
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Lopez P, Bapteste E, McInerney JO. Gene similarity networks provide tools for understanding eukaryote origins and evolution. Proc Natl Acad Sci U S A. 2013;110:E1594–E1603. doi: 10.1073/pnas.1211371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aves SJ, Liu Y, Richards TA. Evolutionary diversification of eukaryotic DNA replication machinery. Subcell Biochem. 2012;62:19–35. doi: 10.1007/978-94-007-4572-8_2. [DOI] [PubMed] [Google Scholar]
- Baker BJ, et al. Lineages of Acidophilic Archaea Revealed by Community Genomic Analysis. Science. 2006;314:1933–1935. doi: 10.1126/science.1132690. [DOI] [PubMed] [Google Scholar]
- Baker BJ, et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci U S A. 2010;107:8806–8811. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E, Brochier CL, Boucher Y. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea. 2005;1:353–363. doi: 10.1155/2005/859728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry ER, Bell SD. DNA replication in the archaea. Microbiol Mol Biol Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer RJ, Graham BW, Trakselis MA. Novel interaction of the bacterial-Like DnaG primase with the MCM helicase in archaea. J Mol Biol. 2013;425:1259–1273. doi: 10.1016/j.jmb.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Beattie TR, Bell SD. Molecular machines in archaeal DNA replication. Curr Opin Chem Biol. 2011;15:614–619. doi: 10.1016/j.cbpa.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Bell SD. DNA replication: archaeal oriGINS. BMC Biol. 2011;9:36. doi: 10.1186/1741-7007-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A, et al. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Berthon J, Cortez D, Forterre P. Genomic context analysis in Archaea suggests previously unrecognized links between DNA replication and translation. Genome Biol. 2008;9:R71. doi: 10.1186/gb-2008-9-4-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Forterre P, Gribaldo S. Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox. Genome Biol. 2004;5:R17. doi: 10.1186/gb-2004-5-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Forterre P, Gribaldo S. An emerging phylogenetic core of Archaea: phylogenies of transcription and translation machineries converge following addition of new genome sequences. BMC Evol Biol. 2005;5:36. doi: 10.1186/1471-2148-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier-Armanet C, Forterre P, Gribaldo S. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol. 2011;14:274–281. doi: 10.1016/j.mib.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Buckley TRT, Simon CC, Shimodaira HH, Chambers GKG. Evaluating hypotheses on the origin and evolution of the New Zealand alpine cicadas (Maoricicada) using multiple-comparison tests of tree topology. Mol Biol Evol. 2001;18:223–234. doi: 10.1093/oxfordjournals.molbev.a003796. [DOI] [PubMed] [Google Scholar]
- Cann I, Komori K, Toh H, Kanai S, Ishino Y. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc Natl Acad Sci U S A. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, et al. Biochemical and mutational analyses of a unique clamp loader complex in the archaeon Methanosarcina acetivorans. J Biol Chem. 2005;280:41852–41863. doi: 10.1074/jbc.M508684200. [DOI] [PubMed] [Google Scholar]
- Chia N, Cann I, Olsen GJ. Evolution of DNA replication protein complexes in eukaryotes and Archaea. PLoS One. 2010;5:e10866. doi: 10.1371/journal.pone.0010866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJC, Foster PGP, Hirt RPR, Harris SRS, Embley TMT. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci U S A. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csuros M, Miklos I. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol. 2009;26:2087–2095. doi: 10.1093/molbev/msp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeddu L, White MF. DNA damage detection by an archaeal single-stranded DNA-binding protein. J Mol Biol. 2005;353:10–10. doi: 10.1016/j.jmb.2005.08.050. [DOI] [PubMed] [Google Scholar]
- Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc FF, Brinkmann HH, Philippe HH. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML, Bell SD. Genome duplication. London and New York: Garland Publications; 2010. [Google Scholar]
- Desmond E, Brochier-Armanet C, Forterre P, Gribaldo S. On the last common ancestor and early evolution of eukaryotes: reconstructing the history of mitochondrial ribosomes. Res Microbiol. 2010;162:18–18. doi: 10.1016/j.resmic.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Forterre P. Why are there so many diverse replication machineries? J Mol Biol. 2013;425:4714–4726. doi: 10.1016/j.jmb.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D, Serre M-C. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Forterre PP, Gadelle DD. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PG, Cox CJ, Embley TM. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Philos Trans R Soc Lond B Biol Sci. 2009;364:2197–2207. doi: 10.1098/rstb.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, et al. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005;15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer OO, et al. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc Natl Acad Sci U S A. 2004;101:9091–9096. doi: 10.1073/pnas.0401356101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, et al. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci U S A. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Anderson JP, Rodrigo AG. Likelihood-based tests of topologies in phylogenetics. Syst Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- Grabowski B, Kelman Z. Archeal DNA replication: eukaryal proteins in a bacterial context. Annu Rev Microbiol. 2003;57:487–516. doi: 10.1146/annurev.micro.57.030502.090709. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Brochier C. Phylogeny of prokaryotes: does it exist and why should we care? Res Microbiol. 2009;160:513–521. doi: 10.1016/j.resmic.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Brochier-Armanet C. The origin and evolution of Archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci. 2006;361:1007–1022. doi: 10.1098/rstb.2006.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Cammarano P. The root of the universal tree of life inferred from anciently duplicated genes encoding components of the protein-targeting machinery. J Mol Evol. 1998;47:508–516. doi: 10.1007/pl00006407. [DOI] [PubMed] [Google Scholar]
- Gribaldo SS, Philippe HH. Ancient phylogenetic relationships. Theor Popul Biol. 2002;61:391–408. doi: 10.1006/tpbi.2002.1593. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guy L, Ettema TJ. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:8–8. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Hartman ALA, et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2009;5:e9605–e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung FF, Puchta HH. Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene. 2001;271:81–86. doi: 10.1016/s0378-1119(01)00496-6. [DOI] [PubMed] [Google Scholar]
- Hawkins M, Malla S, Blythe MJ, Nieduszynski CA, Allers T. Accelerated growth in the absence of DNA replication origins. Nature. 2013:1–16. doi: 10.1038/nature12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashibata H, Kikuchi H, Kawarabayasi Y, Matsui I. Helicase and nuclease activities of hyperthermophile Pyrococcus horikoshii Dna2 inhibited by substrates with RNA segments at 5′-end. J Biol Chem. 2003;278:15983–15990. doi: 10.1074/jbc.M207748200. [DOI] [PubMed] [Google Scholar]
- Hou L, Klug G, Evguenieva-Hackenberg E. The archaeal DnaG protein needs Csl4 for binding to the exosome and enhances its interaction with adenine-rich RNAs. RNA Biol. 2013;10:415–424. doi: 10.4161/rna.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- Hurvich CM, Tsai C-L. Regression and time series model selection in small samples. Biometrika. 1989;72:297–307. [Google Scholar]
- Iwabe NN, Kuma KK, Hasegawa MM, Osawa SS, Miyata TT. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobb G, Haeseler von A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJT, Simancek PP, Brush GSG. Identification and characterization of a single-stranded DNA-binding protein from the archaeon Methanococcus jannaschii. Proc Natl Acad Sci U S A. 1998;95:14634–14639. doi: 10.1073/pnas.95.25.14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoven R, van Enckevort FHJ, Boekhorst J, Molenaar D, Siezen RJ. Visualization for genomics: the Microbial Genome Viewer. Bioinformatics. 2004;20:1812–1814. doi: 10.1093/bioinformatics/bth159. [DOI] [PubMed] [Google Scholar]
- Kerr ID, et al. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H, Takashi M, Hasegawat M. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J Mol Evol. 1990;31:151–160. [Google Scholar]
- Kishino HH, Hasegawa MM. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol. 2007;14:875–877. doi: 10.1038/nsmb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori KK, Ishino YY. Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J Biol Chem. 2001;276:25654–25660. doi: 10.1074/jbc.M102423200. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Forterre P, Bamford DH. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J Mol Biol. 2010;397:17–17. doi: 10.1016/j.jmb.2010.01.037. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Gribaldo S, Bamford DH, Forterre P. The evolutionary history of archaeal MCM helicases: a case study of vertical evolution combined with hitchhiking of mobile genetic elements. Mol Biol Evol. 2010;27:2716–2732. doi: 10.1093/molbev/msq161. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E, Gogarten JP. The effects of model choice and mitigating bias on the ribosomal tree of life. Mol Phylogenet Evol. 2013;69:17–38. doi: 10.1016/j.ympev.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. Archaeology of eukaryotic DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a012963. doi: 10.1101/cshperspect.a012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV, Kelman Z. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol Direct. 2012;7:7. doi: 10.1186/1745-6150-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S-BS, Ramesh MAM, Hulstrand AMA, Logsdon JMJ. Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol Biol Evol. 2007;24:2827–2841. doi: 10.1093/molbev/msm217. [DOI] [PubMed] [Google Scholar]
- Martin IV, MacNeill SA. ATP-dependent DNA ligases. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-4-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini EE, Keeney SS. Sex and the single (double-strand) break. Mol Cell. 2002;9:700–702. doi: 10.1016/s1097-2765(02)00512-9. [DOI] [PubMed] [Google Scholar]
- Matte-Tailliez O, Brochier C, Forterre P, Philippe H. Archaeal phylogeny based on ribosomal proteins. Mol Biol Evol. 2002;19:631–639. doi: 10.1093/oxfordjournals.molbev.a004122. [DOI] [PubMed] [Google Scholar]
- McGeoch AT, Bell SD. Extra-chromosomal elements and the evolution of cellular DNA replication machineries. Nat Rev Mol Cell Biol. 2008;9:569–574. doi: 10.1038/nrm2426. [DOI] [PubMed] [Google Scholar]
- Meselson M, Stahl FW. The replication of DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1958;44:671–682. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasingarao P, et al. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 2012;6:81–93. doi: 10.1038/ismej.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagaling E, et al. Sequence analysis of an Archaeal virus isolated from a hypersaline lake in Inner Mongolia, China. BMC Genomics. 2006;8:410. doi: 10.1186/1471-2164-8-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, et al. Thermococcus kodakarensis has two functional PCNA homologs but only one is required for viability. Extremophiles. 2013;17:453–461. doi: 10.1007/s00792-013-0526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Santangelo TJ, Li Z, Reeve JN, Kelman Z. Thermococcus kodakarensis encodes three MCM homologs but only one is essential. Nucleic Acids Res. 2011;39:9671–9680. doi: 10.1093/nar/gkr624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paytubi S, et al. Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales. Proc Natl Acad Sci U S A. 2012;109:E398–E405. doi: 10.1073/pnas.1113277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, et al. Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol Direct. 2012;8:9. doi: 10.1186/1745-6150-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokofeva MI, et al. Isolation of the anaerobic thermoacidophilic crenarchaeote Acidilobus saccharovorans sp. nov. and proposal of Acidilobales ord. nov., including Acidilobaceae fam. nov. and Caldisphaeraceae fam. nov. Int J Syst Evol Microbiol. 2009;59:3116–3122. doi: 10.1099/ijs.0.010355-0. [DOI] [PubMed] [Google Scholar]
- Richard DJD, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Robbins JB, et al. The euryarchaeota, nature’s medium for engineering of single-stranded DNA-binding proteins. J Biol Chem. 2005;280:15325–15339. doi: 10.1074/jbc.M412870200. [DOI] [PubMed] [Google Scholar]
- Robbins JBJ, et al. Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase BI. J Biol Chem. 2004;279:6315–6326. doi: 10.1074/jbc.M304491200. [DOI] [PubMed] [Google Scholar]
- Robinson NP, Bell SD. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc Natl Acad Sci U S A. 2007;104:5806–5811. doi: 10.1073/pnas.0700206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, et al. Specificity and functionof archaeal DNA replication initiator proteins. Cell Rep. 2013;3:485–496. doi: 10.1016/j.celrep.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, et al. Essential developmental, genomic stability, and tumour suppressor functions of the mouse orthologue of hSSB1/NABP2. PLoS Genet. 2012;9:e1003298–e1003298. doi: 10.1371/journal.pgen.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Skowyra A, MacNeill SA. Identification of essential and non-essential single-stranded DNA-binding proteins in a model archaeal organism. Nucleic Acids Res. 2012;40:1077–1090. doi: 10.1093/nar/gkr838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler N, et al. Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins. Nucleic Acids Res. 2010;38:5088–5104. doi: 10.1093/nar/gkq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. DNA polymerases at the replication fork in eukaryotes. Mol Cell. 2008;30:259–260. doi: 10.1016/j.molcel.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud A, Liddell S, Allers T. Genetic and biochemical identification of a novel single-stranded DNA-binding complex in Haloferax volcanii. Front Microbiol. 2012;3:224–224. doi: 10.3389/fmicb.2012.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2005;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth RIR, White MFM. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 2001;29:914–920. doi: 10.1093/nar/29.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol Microbiol. 2001;40:1241–1248. doi: 10.1046/j.1365-2958.2001.02479.x. [DOI] [PubMed] [Google Scholar]
- Williams TAT, Foster PGP, Nye TMWT, Cox CJC, Embley TMT. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci. 2012;279:4870–4879. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YIY, Makarova KSK, Yutin NN, Koonin EVE. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct. 2011;7:46–46. doi: 10.1186/1745-6150-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro KK, Yokobori S-IS, Oshima TT, Yamagishi AA. Structural analysis of the plasmid pTA1 isolated from the thermoacidophilic archaeon Thermoplasma acidophilum. Extremophiles. 2006;10:327–335. doi: 10.1007/s00792-005-0502-z. [DOI] [PubMed] [Google Scholar]
- Yutin N, Puigbò P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RR, Zhang C-TC. Identification of replication origins in the genome of the methanogenic archaeon, Methanocaldococcus jannaschii. Extremophiles. 2004;8:253–258. doi: 10.1007/s00792-004-0385-4. [DOI] [PubMed] [Google Scholar]
- Zhao A, Gray FC, MacNeill SA. ATP- and NAD+-dependent DNA ligases share an essential function in the halophilic archaeon Haloferax volcanii. Mol Microbiol. 2006;59:743–752. doi: 10.1111/j.1365-2958.2005.04975.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.