Abstract
The Asteraceae family is at the forefront of the evolution due to frequent hybridization. Hybridization is associated with the induction of widespread genetic and epigenetic changes and has played an important role in the evolution of many plant taxa. We attempted the intergeneric cross Chrysanthemum morifolium × Leucanthemum paludosum. To obtain the success in cross, we have to turn to ovule rescue. DNA profiling of the amphihaploid and amphidiploid was investigated using amplified fragment length polymorphism, sequence-related amplified polymorphism, start codon targeted polymorphism, and methylation-sensitive amplification polymorphism (MSAP). Hybridization induced rapid changes at the genetic and the epigenetic levels. The genetic changes mainly involved loss of parental fragments and gaining of novel fragments, and some eliminated sequences possibly from the noncoding region of L. paludosum. The MSAP analysis indicated that the level of DNA methylation was lower in the amphiploid (∼45%) than in the parental lines (51.5–50.6%), whereas it increased after amphidiploid formation. Events associated with intergeneric genomic shock were a feature of C. morifolium × L. paludosum hybrid, given that the genetic relationship between the parental species is relatively distant. Our results provide genetic and epigenetic evidence for understanding genomic shock in wide crosses between species in Asteraceae and suggest a need to expand our current evolutionary framework to encompass a genetic/epigenetic dimension when seeking to understand wide crosses.
Keywords: intergeneric hybridization, polyploidization, GISH, genome changes, Asteraceae
Introduction
The whole-genome doubling of natural wide hybrids to produce allopolyploids is a common event in the plant kingdom, and their formation has provided a significant means of promoting adaptation and speciation (Kearney 2005; Rapp and Wendel 2005). Plant genomes also have the capacity to spontaneously double their genomes, which in turn produce autopolyploids, whereas autopolyploids are less frequently encountered than allopolyploids (Grant-Downton and Dickinson 2006; Paun et al. 2006). Current estimates suggest that at least 70% of modern plant species are either polyploids or descendants from a polyploid ancestor. Polyploid genomes are fundamentally unstable, tending to decay back to the functional diploid state through a process of pseudogenization and segmental deletion known as diploidization (Wolfe 2001; Ma and Gustafson 2005). Arabidopsis, maize, and rice, traditionally regarded as diploid species, are all good examples of diploidized polyploids (Chen 2007). The first few generations following a wide hybridization event, as has been demonstrated for a diversity of synthetic allopolyploids, are prone to genomic perturbation at both genetic and epigenetic levels, a phenomenon referred to as “genomic shock” (Yu et al. 2005; Jackson and Chen 2010).
The nature of genomic shock has been repeatedly investigated (Matsuoka 2011). Its manifestation includes chromosomal rearrangement, the gain and loss of chromosome segments, gene repression and activation, subfunctionalization, and transposon activation, as well as changes in the epigenome, in particular with respect to patterns of cytosine methylation (Adams et al. 2003; Paun et al. 2006; Otto 2007; Kawakami et al. 2011). Nevertheless, the plant seems well tolerated in many genomic groups by combining traits from two parental species and even potentially benefits from those changes, which promotes growth vigor in response to “genomic shock” stress in their hybrids (McClintock 1984; Paun et al. 2007; Ungerer and Kawakami 2013). Some of these alterations occur very rapidly after the hybridization and/or polyploidization event, as shown in wheat and Tragopogon (Shaked et al. 2001; Tate et al. 2006). Changes in global patterns of cytosine methylation, as detected by methylation-sensitive amplification polymorphism, have been directly associated with the hybridization and/or polyploidization event (Shaked et al. 2001; Salmon et al. 2005; Qi et al. 2010; Hegarty et al. 2011). However, there is little evidence for any rapid methylation changes in polyploidy formation of cotton and ploidy watermelon (Liu et al. 2001; Wang et al. 2009). Thus, polyploidy in the plant kingdom may involve an array of molecular events that are dependent on the genomic context.
In general, the more distantly related are the prospective parents of a wide cross, the harder the cross is to make. In the context of crop improvement, wide crosses such as those involving parents from different genera or/and different ploidy levels are of particular interest because they offer the potentiality of introgressing novel genes into the primary gene pool of the crop (Ozkan et al. 2001; Zhang et al. 2011). The Asteraceae family comprises more than 25,000 geographically widely dispersed species, representing 10% of all angiosperms, and many of these species are polyploid (Bremer and Humphries 1993; Guo et al. 2012). Consequently, the Asteraceae family could provide a unique opportunity to understand the evolutionary genomics of lineage radiation and diversification at various phylogenetic scales (Kane et al. 2011). Although transcriptome shock and cytosine methylation have been studied in interspecific hybrids in Asteraceae (Hegarty et al. 2006; Tate et al. 2006; Hegarty et al. 2008), genetic and epigenetic alterations under intergeneric genomic shock are still poorly understood.
The ornamental species chrysanthemum (Chrysanthemum morifolium) is a hexaploid, whereas the ploidy level of other Chrysanthemum spp. varies from diploid to decaploid (Kondo et al. 1999). The perennial Asteraceae species Leucanthemum paludosum (formerly Mauranthemum paludosum), a native diploid species of the Mediterranean region, is also used for ornamental purposes. Here, we describe the producing of an allopolyploid from the cross C. morifolium × L. paludosum using ovule rescue and detail some of the major genetic and epigenetic changes induced by this intergeneric hybridization and polyploidization using a variety of DNA-based marker technologies.
Materials and Methods
Plant Material, Crossing, and Embryo Rescue
The plant materials used are the C. morifolium × L. paludosum hybrid, chromosome-doubled hybrids derived from the cross combination, its parents C. morifolium cv. “Zhongshanzigui,” and an accession of L. paludosum (fig. 1A–D) and the diploid species C. zawadskii. The materials are all maintained by the Chrysanthemum Germplasm Resource Preserving Centre, Nanjing Agricultural University, China (32°05′N, 118°8′E, 58 m altitude). The plants were propagated by cuttings. All plants were grown in a greenhouse held at 22 °C during the day and above 15 °C during the night, with a relative humidity of 70–75% and under natural light. To make the wide cross, at 9:00–10:00 AM on a sunny day, the bisexual tubular florets of “Zhongshanzigui” were removed using tweezers before anthesis and the inflorescences were enclosed within a paper bag when the stigmas first became visible on the developing receptacle. After 2 or 3 days, fresh pollen from newly blossoming flowers of the L. paludosum was brushed onto the “Zhongshanzigui” pistil (at a Y-shaped stigma stage) using a fine-haired paintbrush. After pollination, the flowers were immediately bagged. Four, 6, 8, 10, 12, 15, 18, and 21 days after pollination (DAP), individual pollinated florets were collected, fixed in FAA (1:1:18, formalin:glacial acetic acid:70% v/v ethanol), and stored at room temperature. Before examination, the florets were dehydrated by passing through an alcohol series, infiltrated with xylene, and embedded in paraffin wax, following the methods described by Deng et al. (2010). Sections of thickness 6–10 μm were cut, stained in Heidenhain’s hematoxylin, and then observed by light microscopy. For ovule rescue, ovaries were excised between 10 and 15 DAP, surface sterilized by immersion in 75% v/v ethanol for 35 s, followed by 10% v/v H2O2 for 15 min, and rinsed six times in sterile water. The ovary coats were aseptically removed to extract the ovules; the extracted ovules were then placed on solidified MS medium (Murashige and Skoog 1962) containing 2 mg/l 6-benzyladenine (6-BA) and 0.5 mg/l α-naphthaleneacetic acid (Tang et al. 2009) and cultured at 25 °C under a 16-h photoperiod provided by cool-white fluorescent lamps (30 μmol m−2 s−1). Regenerating plantlets were transferred to fresh solidified MS medium supplemented with 1.0% w/v sucrose (pH 5.7) until they had developed roots 1 cm long (Murashige and Skoog 1962), and the humidity was gradually lowered over a period of 1 month, after which they were potted into a 2:2:1 mixture of perlite, vermiculite, and leaf mold, and grown in a greenhouse.
Fig. 1.—
Morphology of materials. (A) Chrysanthemum morifolium “Zhongshanzigui,” (B) amphihaploid, (C) amphidoploid, and (D) Leucanthemum paludosum). From left to right of each line: plant morphology; floral morphology, bar: 1 cm; ligulate flower, bar: 2 mm; tubular flower, bar: 2 mm; leaf: 0.5 cm.
Cytological Confirmation and Genomic In Situ Hybridization Analysis
Young root tips collected from parental cuttings and from the putative amphihaploid plantlets were held in ice water for 20–24 h, then fixed in 3:1 ethanol:glacial acetic acid (v/v) at 4 °C for 24 h, and squashed in a drop of 45% acetic acid. Mitotic chromosome spreads were observed by phase contrast microscopy. The mitotic chromosomes of the putative amphihaploid were also subjected to genomic in situ hybridization (GISH) by probing with genomic DNA of C. zawadskii extracted from young leaves using the CTAB (cetyltrimethylammonium bromide) method (Stewart and Via 1993) and labeled with biotinylated 16-dUTP (Roche Ltd.). The GISH procedure followed Deng et al. (2010).
Chromosome Doubling
Following the procedure described by Liu et al. (2011), nodal segments from 1-month-old hybrid plantlets were immersed in 600 mg/l colchicine for 48 h, then rinsed three times in sterile water, and placed on hormone-free MS medium for 30 days, at which time the developed lateral buds were transferred onto a rooting medium (Liu et al. 2011).
Genotypic Analysis
DNA was extracted from the fourth and fifth leaves of three individual plants of the two parental lines, the three amphihaploids, and the corresponding amphidiploids using a Nuclei Isolation kit (Solarbio, China). The DNAs were subjected to amplified fragment length polymorphism (AFLP) profiling as described by Vos et al. (1995). Five hundred nanograms of DNA was digested overnight at 37 °C with 10 U each of EcoRI and MseI (New England Biolabs, Beijing, China). Then adaptors were ligated to the ends of the digested fragments at 16 °C for 4 h; the ligation mix contains the digested fragments, 5-pmol EcoRI and 50-pmol MseI adaptors (supplementary table S1, Supplementary Material online), and 4 U T4 DNA polymerase (NEB), after which the enzymes were inactivated by heating to 70 °C for 15 min. The amplicon generated from this template based on EcoRI and MseI primers lacking any selective bases (supplementary table S1, Supplementary Material online) was diluted 1:30 in ddH2O and used as the template for subsequent selective amplifications based on EcoRI and MseI primers carrying three selective bases; in total, the nine primer combinations are EcoRI selective primer #2 plus MseI selective primer #5 (abbreviated “E2 + M5”), E2 + M6, E3 + M2, E4 + M3, E4 + M8, E6 + M7, E7 + M3, E8 + M3, and E8 + M7 (supplementary table S1, Supplementary Material online). The EcoRI primers were labeled with 5-FAM (Carboxyfluorescein). Each polymerase chain reaction (PCR) and electrophoretic separation was repeated twice. The amplicons were separated using an ABI 377 DNA Analyzer device, and only fragments ranging from 80 to 450 bp were considered.
The same genomic DNAs were also subjected to sequence-related amplified polymorphism (SRAP) analysis, a DNA fingerprinting technique that targets open reading frames (Li and Quiros 2001). In all, 24 SRAP primer combinations were used, involving 7 forward and 8 reverse primers (supplementary table S1, Supplementary Material online). The combinations were ME-1 combined with EM-1 (abbreviated “ME1 + EM1”), ME1 + EM7, ME1 + EM14, ME3 + EM2, ME3 + EM5, ME3 + EM10, ME6 + EM1, ME6 + EM2, ME6 + EM5, ME6 + EM14, ME10 + EM1, ME10 + EM7, ME10 + EM11, ME10 + EM14, ME14 + EM2, ME14 + EM5, ME16 + EM2, ME16 + EM10, ME16 + EM11, ME16 + EM14, ME17 + EM1, ME17 + EM5, ME17 + EM7, and ME17 + EM15. Each 25-μl reaction mix comprised 20 ng genomic DNA, 200 μM dNTP, 1.5 mM MgCl2, 0.6 μM of each primer, 2.5 μl 10× buffer, and 2 U Taq polymerase (Takara). The reactions were subjected to a 5-min denaturation at 94 °C, then five cycles of 94 °C/1 min, 35 °C/1 min, 72 °C/2 min, followed by 35 cycles of 94 °C/1 min, 50 °C/1 min, 72 °C/2 min, and finally an extension step of 72 °C/7 min. For the start codon-targeted polymorphism (SCoT) analysis, the primers (supplementary table S1, Supplementary Material online) were derived from a consensus sequence in the flanking regions around the ATG start codon (Joshi et al. 1997). Each 25-μl PCR reaction mix contained 20 ng genomic DNA, 200 μM dNTP, 1.5 mM MgCl2, 0.6 μM of each primer, 2.5 μl 10× buffer, and 2 U Taq polymerase (Takara, Japan), and the cycling regime consisted of an initial denaturation step (94 °C/3 min), followed by 35 cycles of 94 °C/1 min, 50 °C/1 min, 72 °C/2 min, and a final extension step of 72 °C/5 min. Both the SRAP and SCoT amplicons were electrophoresed through 8% nondenaturing polyacrylamide gels run at 300 V for 2.5 h in 1× TBE buffer and were visualized by silver staining.
Isolating and Sequencing Informative AFLP and SRAP Fragments
Informative AFLP amplicons were also separated by electrophoresis through 6% denaturing polyacrylamide gels and silver stained (Vos et al. 1995; Chen et al. 2007). Polymorphic AFLP fragments and polymorphic SRAP fragments were cut from the gel and eluted in 50 μl ddH2O by boiling for 10 min. After centrifugation (12,000 × g, 5 min), an aliquot of the supernatant was collected and served as the template. The fragment was reamplified using the same primer combination and PCR conditions. The resulting amplicon was inserted into the cloning vector PMD19 TA (Takara) and sequenced. The similarity of these sequences to those in public databases was obtained using Blast analysis (http://www.ncbi.nlm.nih.gov/BLAST/, last accessed January 24, 2014).
Methylation-Sensitive Amplification Polymorphism Analysis
Alterations in DNA methylation were investigated by MSAP analysis (Xiong et al. 1999; Benhattar and Clement 2004; Roberts et al. 2007). Aliquots of 500 ng genomic DNA of the parental, amphihaploid, and amphidiploid plants were digested with 10 U EcoRI and 20 U HpaII or 10 U EcoRI and 10 U MspI (NEB) at 37 °C for 12 h, and the digested fragments were ligated to 5 pmol EcoRI and 50 pmol HpaII/MspI adaptors (supplementary table S1, Supplementary Material online), as described for the AFLP method. The amplicons amplified from this template using EcoRI and HpaII/MspI primers lacking any selective bases (supplementary table S1, Supplementary Material online) were diluted 1:30 in ddH2O and used as the template for subsequent selective amplifications, for which fluorescently labeled EcoRI and unlabeled HpaII/MspI primers each carrying three selective bases were used (supplementary table S1, Supplementary Material online). The nine primer combinations used were EcoRI selective primer #2 plus HpaII/MspI selective primer #5 (abbreviated “E2 + HM5”), E4 + HM3, E4 + HM7, E4 + HM8, E6 + HM6, E6 + HM8, E7 + HM1, E8 + HM2, and E8 + HM8. Electrophoresis and fragment detection were carried out in the same way as for the AFLP analysis, and each assay was repeated twice. For statistical analysis of the differences between the midparental value and the hybrids (U values), the following formula was used (Zhao et al. 2007):
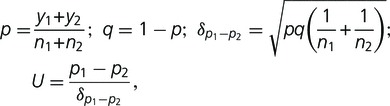 |
where n1 represents the total number of fragments of the midparent values, n2 the total fragments of a hybrid, y1 the total number of methylated and hemimethylated fragments of the midparent values, y2 the total number of methylated and hemimethylated fragments in the hybrid, p1 the proportion (%) of methylated and hemimethylated fragments of the midparent values, and p2 the proportion (%) of methylated and hemimethylated fragments in the hybrid.
Results
Wide Cross and Ovule Rescue
By 12 h after pollination, pollen germination had been initiated, and the L. paludosum pollen tubes had begun to penetrate the C. morifolium stigma (fig. 2A). The hybrid zygote developed normally up to the multicell proembryo stage by 10 DAP. However, by 12 DAP, only two (0.5%) out of 400 ovules formed globular proembryos (fig. 2B and C). No further development of embryos occurred beyond this time point, after which embryo abortion occurred completely (fig. 2D). As a result, to obtain amphihaploid plants, ovules were rescued at 12 DAP. Of the 1,287 ovules cultured in vitro, only 105 (8.2%) formed callus, and of these only three (2.9%) generated buds and roots. The mature plants of amphihaploid and amphidiploid were morphologically homogeneous but were distinct from either of the parents (fig. 1). The plant height, plant canopy, flower diameter, and leaf size of the amphihaploid were smaller than those of the two parental plants. The plant height, plant canopy, and flower diameter were improved in amphidiploid compared with the amphihaploid, while those were still inferior to the two parental plants.
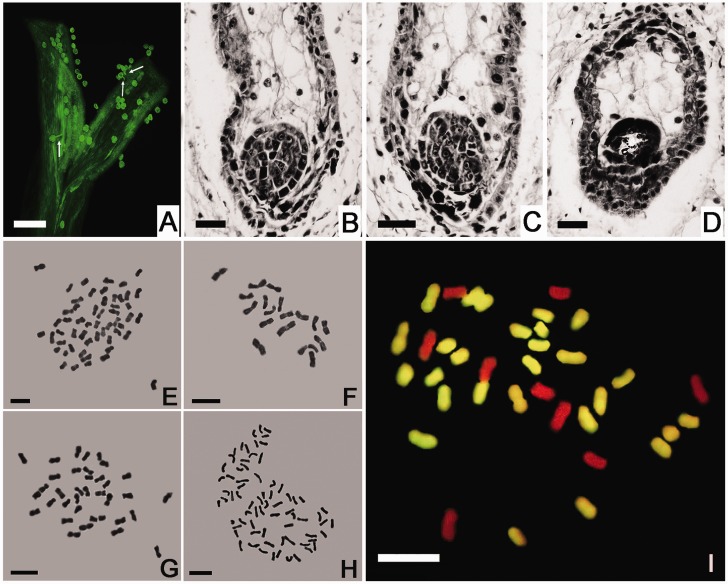
Fig. 2.—
The germination of L. paludosum pollen on the Chrysanthemum morifolium stigma and the development of the hybrid embryo. (A) Pollen germination 12 h postpollination. Bar: 100 μm. (B, C) Two globular stage proembryos with a degenerating endosperm in a 12-DAP ovule. (D) A degenerated globular proembryo in an 18-DAP ovule. Bar: 20 μm. (E–H) The mitotic chromosomes of (E) C. morifolium cv. “Zhongshanzigui” (2n = 54), (F) Leucanthemum paludosum (2n = 18), (G) the C. morifolium × L. paludosum amphihaploid (2n = 36), and (H) the C. morifolium × L. paludosum amphidiploid (2n = 72). Bar: 5 μm. (I) GISH analysis of the C. morifolium × L. paludosum amphihaploid. The chromosomes were probed with labeled C. zawadskii genomic DNA; the 27 yellow fluorescing chromosomes were inherited from C. morifolium, and the nine unlabeled (red) ones from L. paludosum. Bar: 5 μm.
Chromosome Number and GISH Analysis
The somatic chromosome number of C. morifolium is 54 (fig. 2E), whereas that of L. paludosum is 18 (fig. 2F). As expected, the amphihaploid and amphidiploid somatic chromosome numbers were, respectively, 36 and 72 (fig. 2G and H). GISH analysis confirmed the hybridity of the three putative amphihaploids; when probed with labeled C. zawadskii genomic DNA, 27 of the mitotic chromosomes from “Zhongshanzigui” recognized the labeled probes, whereas the other 9 from L. paludosum did not (fig. 2I). There was no evidence for any chromosome elimination.
Genomic Changes Detected under Intergeneric Hybridization and Polyploidization Cycle
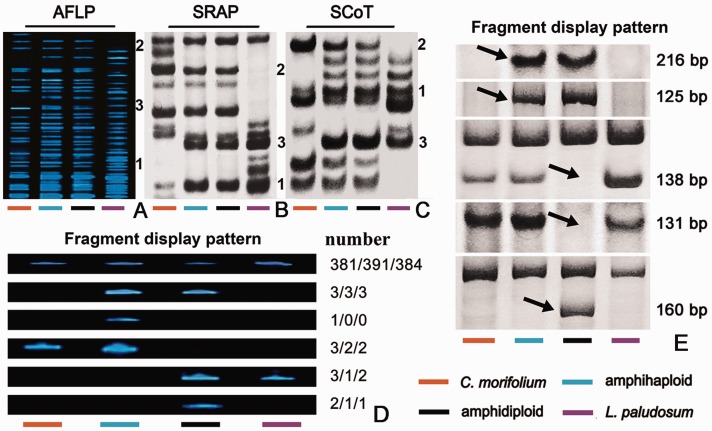
All three amphihaploids appeared to be cytologically uniform and shared a similar plant phenotype; hence, the investigation into genomic perturbation was conducted on these three plants. The nine AFLP primer combinations amplified 375 fragments from the C. morifolium and 354 from the L. paludosum template. The AFLP profiles of amphihaploid lines 1/2/3 consisted of 388/396/389 fragments, respectively, of which 4/3/3 were not present in either parent’s profile (table 1, supplementary table S2, Supplementary Material online), whereas 118/115/115 (30.4%/29.0%/29.6%) were present in both parental profiles (fig. 3A1), 180/188/182 (46.4%/47.5%/46.8%) were inherited from C. morifolium (fig. 3A2), and 86/90/89 (22.2%/22.7%/22.9%) from L. paludosum (fig. 3A3). The amphidiploid’s AFLP profile comprised 389/396/390 fragments, 381/391/384 of which were inherited from one of the parental lines. Of the 4/3/3 nonparental fragments detected in the amphihaploid, 3/3/3 were also present in the amphidiploid’s profile, but only one was absent in the amphidiploid line 1. The 3/2/2 parental fragments present in the amphihaploid’s profile were absent from the amphidiploid’s profile, whereas a further 3/1/2 paternal parent fragments were present in the amphidiploid’s profile but not in the amphihaploid’s. A further 2/1/1 fragments were unique to the amphidiploid’s profile (fig. 3D). Of the 27 nonparental fragments, two fragments yielded poor sequence quality and 25 fragments were successfully sequenced. Fifteen out of 25 fragments showed no homology to any GenBank entries, two matched hypothetical or uncharacterized genes, and the remaining eight sequences hit microsatellite sequence and a sequence encoding an NBS-LRR resistance-like protein (table 1, supplementary table S2, Supplementary Material online). Remarkably, three fragments were present twice in different amphihaploid lines, that is, one fragment was present in lines 1 and 3, and the remaining two were present in lines 2 and 3 (supplementary table S2, Supplementary Material online).
Table 1.
Nine Annotated Novel AFLP Fragments Affected by Either Hybridization or Polyploidization
| Fragment Size (bp) | Fragment Display Pattern |
Sequence Similaritya | |||
|---|---|---|---|---|---|
| C. m | Hy | Am | L. p | ||
| 189 | − | + | + | − | Ricinus communis hypothetical protein, mRNA |
| 122 | − | + | − | − | Acrossocheilus monticola microsatellite sequence |
| 124 | +b | + | − | +b | Helianthus annuus NBS-LRR resistance-like protein |
| 188 | +b | − | + | +b | Linum usitatissimum microsatellite sequence |
| 173 | − | − | + | − | Linum usitatissimum microsatellite sequence |
| 92c | +b | + | − | +b | Linum usitatissimum microsatellite sequence |
| 125 | − | − | + | − | PREDICTED: Strongylocentrotus purpuratus uncharacterized, mRNA |
| 211 | +b | + | − | +b | Linum usitatissimum microsatellite sequence |
| 188 | +b | − | + | +b | Linum usitatissimum microsatellite sequence |
Note.—C. m, C. morifolium; Hy, amphihaploid; Am, amphidiploid; L. p, L. paludosum; +, parental fragment present; –, parental fragment absent. Detailed information displayed in supplementary table S2, Supplementary Material online.
aBlastN or BlastX threshold E values <0.05.
bNonparental fragments.
cAppeared twice in different amphihaploid lines.
Fig. 3.—
Representative DNA profiling of the Chrysanthemum morifolium × Leucanthemum paludosum amphihaploid, amphidiploid, and its parents. (A) AFLP. (B) SRAP, (C) ScoT: 1, fragments present in both parental profiles; 2, fragments inherited from C. morifolium; and 3, fragments inherited from L. paludosum. (D) Variation in AFLP profiles, from top to bottom: most of the parental fragments were inherited by both the amphihaploid and the amphidiploid; four nonparental fragments present in the amphihaploid were also present in the amphidiploid; a nonparental fragment detected in the amphihaploid but not transmitted to the amphidiploid; three parental fragments present in the amphihaploids but not in the amphidiploid; three parental fragments absent in the amphihaploid but present in the amphidiploid; and two nonparental fragments present in the amphidiploid but not in the amphihaploid. (E) Variation in SRAP profiles (line 1), from top to bottom: two nonparental fragments present in both the amphihaploid and the amphidiploid; two parental fragments not present in either the amphihaploid or the amphidiploid; and one nonparental fragment present in the amphidiploid but not in the amphihaploid.
The 24 SRAP primer combinations amplified 318/322/326 fragments from the three amphihaploid templates, of which 101/103/106 (31.8%/32.0%/32.5%) were present in both parents’ profiles (fig. 3B1), 120/121/117 (37.7%/37.6%/35.9%) were inherited from C. morifolium (fig. 3B2), and 95/95/101 (29.9%/29.1%/31.0%) from L. paludosum (fig. 3B3). The 2/3/2 (0.6%/0.9%/0.6%) novel fragments were detected (table 2, supplementary table S2, Supplementary Material online), all of which were transmitted to the amphidiploid. The amphiploid’s profiles also showed the loss of 2/1/1 parental fragments and the gain of 1/1/1 novel fragment. However, only a few fragments can find sequence similarity with accessions deposited in the GenBank, and no repeat sequences between amplified fragments have been found (supplementary table S2, Supplementary Material online). For example, in amphihaploid line 1, only three nonparental fragments showed homology to Ciona intestinalis HyTSR1 protein mRNA, Cynara cardunculus cardosin B gene for aspartic proteinase, and a clone of Brassica rapa subsp. pekinensis sequence (fig. 3E and table 2). The ScoT profiles (12 primers) comprised 200/210/202 fragments (the amphihaploid) and 199/207/202 (the amphidiploid). These fragments were classifiable as 1/2/2 (0.5%/1.0%/1.0%) nonparental, 65/70/67 (32.5%/33.3%/33.2%) inherited from either parent (fig. 3C1), 77/80/75 (38.5%/38.1%/37.1%) from C. morifolium (fig. 3C2), and 57/58/58 (28.5%/27.6%/28.7%) from L. paludosum (fig. 3C3); the difference in fragment number between the amphihaploid and amphidiploid profiles showed a loss of 2/4/2 and a gain of 1/1/2 fragments in amphidiploid.
Table 2.
SRAP Fragments Affected by Either Hybridization or Polyploidization in Amphihaploid Line 1
| Fragment Size (bp) | Fragment Display Pattern |
Sequence Similaritya | |||
|---|---|---|---|---|---|
| C. m | Hy | Am | L. p | ||
| 216 | − | + | + | − | PREDICTED: Ciona intestinalis similar to HyTSR1 protein mRNA |
| 125 | − | + | + | − | None |
| 138 | +b | + | − | +b | None |
| 131 | +b | + | − | +b | Cynara cardunculus cardosin B gene for aspartic proteinase, exons 1–14 |
| 160 | − | − | + | − | Brassica rapa subsp. pekinensis clone KBrB084K02, complete sequence |
Note.—C. m, C. morifolium; Hy, amphihaploid; Am, amphidiploid; L. p, L. paludosum; +, parental fragment present; –, parental fragment absent. Detailed information displayed in supplementary table S2, Supplementary Material online.
aBlastN or BlastX threshold E values <0.05.
bNonparental fragments
Epigenetic Changes Induced by Allopolyploidization
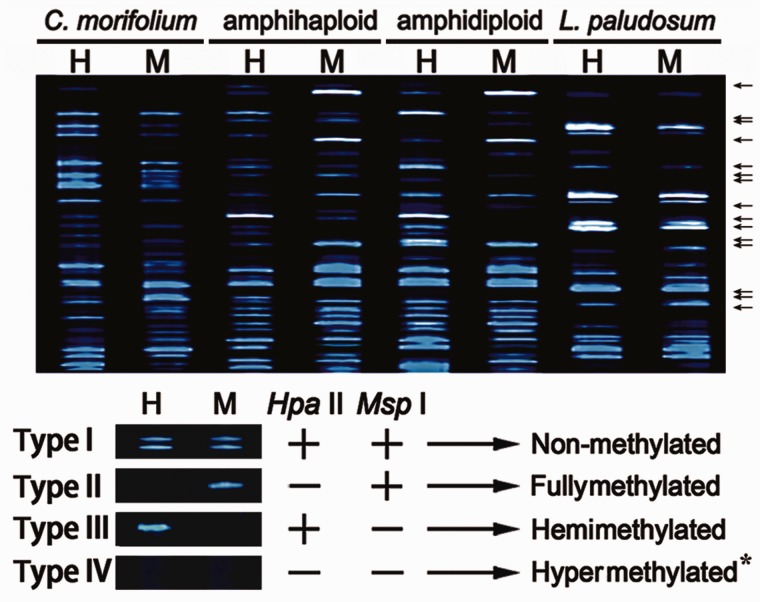
Changes in methylation status were investigated using MSAP profiling, based on nine combinations of EcoRI + HpaII/MspI primer combinations (fig. 4). Four types of fragment were revealed: type I fragments are nonmethylated and were present in both the H and M digests; type II are fully methylated and only appeared in the M digests; type III are hemimethylated and appeared in the H digests; finally, type IV were fragments absent from both H and M lanes in amphihaploid but present in either H or M lane of amphidiploid, inferring decreased methylation, or those absent from both H and M lanes in amphidiploid but present in either H or M lane of amphihaploid, indicating increased methylation.
Fig. 4.—
Representative variation in MSAP profiles. “←,” Variation of DNA methylation between amphihaploid and amphidiploid; “+,” fragments obtained after digestion with EcoRI or HpaII. “−,” fragments not digested by EcoRI or HpaII. Note: hypermethylation (type IV) is detectable from the comparison between the amphihaploid and amphidiploid profiles. For example, fragments absent from amphihaploid (both H and M lanes) but present in amphidiploid (either H or M lane), and vice versa.
The C. morifolium profile included 267 type I, 169 type II, and 114 type III fragments, whereas the corresponding numbers in L. paludosum were 264, 154, and 116. The proportion of methylated fragments was 51.5% in C. morifolium and 50.6% in L. paludosum. In the amphihaploid lines, the proportion of cytosine methylated fragments was 45.1–45.2%, a level that is inconsistent with the midparent value (table 3). The amphihaploids included a significantly lower proportion of fully methylated fragments (23.4–23.8%) than the midparent level (the resulting U values were 2.02–2.23, with U0.05 = 1.96); however, with respect to the proportion of hemimethylated fragments present, the proportion of amphihaploid templates (20.8–21.3%, with a U value was 0.01–0.13) equaled the midparent value. The mean number of fragments that showed changes in methylation state between the amphihaploid to amphidiploid profiles was 161.3 on average, of which 57.3 showed a decrease in methylation, whereas the remainder exhibited an increase in methylation (table 3). It showed that 21.3 of these sites involved a shift from type III to type I and 14.3 from type IV to type II (decreased methylation), whereas 39.0 shifted from type I to type III and 23.0 from type I to type II (increased methylation) (table 4).
Table 3.
Levels of Cytosine Methylation Detected in the Amphihaploid and Parental Plants
| Plant Lines | Total Sites | Nonmethylated | Methylated |
||
|---|---|---|---|---|---|
| Type I | Total (II + III) | Type II | Type III | ||
| C. m | 550 | 267 (48.5%) | 283 (51.5%) | 169 (30.8%) | 114 (20.7%) |
| L. p | 534 | 264 (49.4%) | 270 (50.6%) | 154 (28.8%) | 116 (21.7%) |
| Midparent value | 100% | 49.0% | 51.0% | 29.8% | 21.2% |
| Hy1 | 554 | 304 (54.9%) | 250 (45.1%) | 132 (23.8%)a | 118 (21.3%) |
| Hy2 | 556 | 306 (55.0%) | 251 (45.0%) | 136 (24.5%)a | 114 (20.5%) |
| Hy3 | 575 | 311(54.1%) | 264 (45.9%) | 141 (24.5%)a | 123 (21.4%) |
aValues significantly lower than the corresponding midparent value at the U0.05 threshold.
Table 4.
MSAP Fragments Affected by Either Hybridization or Polyploidization
| Fragment Type |
Fragment Display Pattern in MSAP Gel |
Number of Sites | Status | ||||
|---|---|---|---|---|---|---|---|
| Hy | Am | Hy H Lane | Hy M Lane | AM H Lane | AM M Lane | ||
| Type IV | Type I | − | − | + | + | 5.0 ± 0.9 | ↓ |
| Type I | Type IV | + | + | − | − | 14.7 ± 0.5 | ↑ |
| Type IV | Type III | − | − | + | − | 11.0 ± 1.4 | ↓ |
| Type III | Type IV | + | − | − | − | 20.7 ± 1.0 | ↑ |
| Type IV | Type II | − | − | − | + | 14.3 ± 1.2 | ↓ |
| Type II | Type IV | − | + | − | − | 7.0 ± 0.9 | ↑ |
| Type II | Type I | − | + | + | + | 5.3 ± 0.3 | ↓ |
| Type I | Type II | + | + | − | + | 23.0 ± 1.9 | ↑ |
| Type III | Type I | + | − | + | + | 21.3 ± 1.7 | ↓ |
| Type I | Type III | + | + | + | − | 39.0 ± 2.8 | ↑ |
Note.—↓, decreased methylation; ↑, increased methylation; Hy, amphihaploid; Am, amphidiploid.
Discussion
Intergeneric Hybridization between C. morifolium and L. paludosum
Embryo rescue is a frequently used technique in wide cross, which has allowed for the successful production of many interspecific and intergeneric hybrids (Van Tuyl et al. 1991; Sharma 1995; Tang et al. 2009). Although many C. morifolium × L. paludosum hybrid zygotes were able to develop the multicellular proembryo, further development of embryo did not occur in vivo. Therefore, we resorted to ovary culture, which contributed to successful hybrid zygotes rescue before hybrid embryo abortion occurred. However, only a small number of true hybrids could be regenerated to form viable amphihaploid seedlings. The amphihaploid and amphidiploid are able to flower normally, but the meiotic pairing behavior of the chromosomes is disordered in the amphihaploid.
Genetic Relationship between Chrysanthemum and Leucanthemum
The meiotic pairing behavior of the chromosomes in a wide hybrid is arguably the most reliable means to deduce the taxonomic relationship between the parental species; however, the taxonomic relationships remain as yet not fully resolved within Chrysanthemum sensu lato (Teixeira da Silva 2003). In the absence of such data, GISH analysis can provide a strong basis for assessing the level of distinctness between pairs of genomes, and it has been widely used to characterize hybrids generated from species of the genera Chrysanthemum, Ajania, Tanacetum, Brachanthemum, Elachanthemum, Leucanthemella, and Nipponanthemum (El-Twab et al. 1999; El-Twab and Kondo 2001; Zhao et al. 2008). Genomes that are closely related to one another require the incorporation of blocking (unlabeled) DNA to achieve successful discrimination (El-Twab and Kondo 2004; Deng et al. 2010). Here, GISH, in which the probe DNA was obtained from the diploid C. zawadskii, was able to distinguish the 27 C. morifolium chromosomes from the 9 L. paludosum ones in mitotic spreads of the amphihaploid plant without any blocking DNA. Chrysanthemum zawadskii has been proposed as a potential ancestor of C. morifolium (Yang et al. 2006); here, our data again suggested a relatively close relationship between the two species within Chrysanthemum, whereas the relationship between Chrysanthemum and Leucanthemum spp. in the tribe Anthemideae is distant, which is consistent with the observation of hybrid embryo abortion.
Genome Perturbation in the Synthetic Amphihaploids and Amphidiploids
Plants tolerate hybridization and polyploidization much more easily than do animals. Newly synthesized plant polyploids have been observed to undergo a range of chromosomal rearrangements, the activation of dormant transposable elements, the elimination of DNA sequences, and epigenetic silencing (Barton 2001; Chen 2007; Xiong et al. 2011). Commonly, wide hybrids express certain characteristics (at either the morphological or molecular level) clearly inherited from one of the parents, some from the other parent, and a number of de novo ones that are apparently not inherited from either parent (Soltis and Soltis 1999; Richards 2003; Bell et al. 2013). Synthetic allopolyploids between wheat and its near relatives suffer a measurable degree of sequence elimination (Shaked et al. 2001), as also do those generated in Cucumis spp. (Chen et al. 2007), Brassica spp. (Song et al. 1995), and Tragopogon spp. (Tate et al. 2006). Here, AFLP profiling showed that DNA diversification had probably occurred in the C. morifolium × L. paludosum amphidiploid, as some fragments were lost, whereas others were gained in amphidiploid. Several novel fragments were also detected in the amphihaploid, showing that genome changes are also induced by the intergeneric hybridization event itself. New fragments can also form via point mutation if the mutations remove (or create) a restriction site. Genomic shock can activate quiescent transposons/retrotransposon and some mutation (Liu et al. 1998; Shaked et al. 2001; Kashkush et al. 2002). We did not find any new fragments corresponding to retrotransposon or DNA transposons, however, some fragments contained microsatellite sequence, suggesting whole-genome doubling may be accompanied by the modification of microsatellite sequence.
Although marker-based analysis has become the method for many studies on genetics, evolution, and ecology, genotyping error rates are not explicitly calculated in most empirical studies (Pompanon et al. 2005). These errors cause confusion in the signal-to-noise ratio and hence a loss of resolving power; fortunately, they might not bias all the results of the analysis (Bonin et al. 2004). The number of amplification fragments in each primer combination is decided by the template quality, primer sequence, PCR reaction conditions, competition between potential amplicons, and base mismatching between primer and template (Smith and Williams 1994; Bussell et al. 2005). In this study, we attempt to minimize error rates by employing optimized DNA extraction, reaction mix, PCR reaction conditions, and primer screening. Theoretically, the rate of genetic diversity for genotypes was nearly the same using AFLP, SRAP, or SCoT markers (Schuler et al. 1996; Arabidopsis Genome Initiative 2000; Li and Quiros 2001; Meudt and Clarke 2007). In this study, however, the proportion of each parent’s contribution to the amphihaploid profiles differed when AFLP and SRAP or ScoT markers were used, and only 46.4% of the amphihaploid’s AFLP fragments were inherited from C. morifolium, whereas 22.2% were inherited from L. paludosum; in contrast, the relative contributions with respect to the SRAP fragments were 37.7% and 29.9%, and for the SCoT fragments were 38.5% and 28.5%. The difference between these contributions implies a difference in the nature of the fragments detected by the three profiling methods; a plausible explanation could relate to the random nature of AFLP fragments, which reflect genome-wide variation in DNA sequence (largely consisting of noncoding DNA), whereas both SRAP and SCoT fragments are derived mostly from coding sequence. The reason may be that some “redundant” sequence possibly from the noncoding region of L. paludosum was eliminated in the amphihaploid and the change may be nonrandom during intergeneric hybridization. Sequence elimination is one of the earliest responses of the genome to wide hybridization or allopolyploidy (Shaked et al. 2001). The identity of the sequences eliminated has been shown in wheat (Shaked et al. 2001; Kashkush et al. 2002), Tragopogon spp. (Tate et al. 2006; Koh et al. 2010), Cucumis spp. (Chen et al. 2007), and Brassica spp. (Song et al. 1995). Our present data support previous observation that sequence elimination after wide hybrid is reproducible (McClintock 1984).
Cytosine methylation is the most common covalent modification of DNA in eukaryotes, and it provides a ubiquitous epigenetic control (Zilberman et al. 2006). In this study, Chrysanthemum and Leucanthemum methylated sites are up to 51.6% and 50.6%, respectively (table 3). A number of epigenetic phenomena have been associated with wide hybridization and whole-genome doubling (Lee and Chen 2001; Wang et al. 2006). For example, >13% of the site alterations in cytosine methylation occurred either in the F1 hybrid or in the allopolyploid in wheat (Shaked et al. 2001). After hybridization, the global level of DNA methylation, as measured by MSAP, was somewhat lower in the amphihaploid than in the parental genome, which suggests that one effect of the wide hybridization may be to reduce global DNA methylation. Similar epigenetic phenomena have been observed in other newly synthesized triticale (wheat×rye hybrids), which tended to be rather unstable in the first few generations, probably owing to a reduced methylation in the hybrids, but eventually became stable (Ma and Gustafson 2008). It should be noted that a drop in DNA methylation could also be induced by the callus phase that the primary hybrids go through (Miguel and Marum 2011). However, it has been reported that in spite of the different kind of methylation changes in individual regenerants of Codonopsis lanceolata, overall tissue culture did not cause significant global alteration in the DNA methylation (Guo et al. 2007). In oil palm, the highest number of polymorphic bands had occurred during tissue culture, and most frequently, the HpaII recognition was only 0.3% (Matthes et al. 2001). Further investigation using whole-genome bisulfite sequencing revealed that these changes resulting from tissue culture accounted for a relatively small proportion of the rice genome (Stroud et al. 2013). We suggested that wide hybridization may be the major driver of global DNA methylation reduction.
DNA methylation is generally recognized to suppress gene expression as regulatory factors, and changes in cytosine methylation resulting from wide hybridization and/or polyploidization can be expected to have a major effect on gene expression in polyploids. It has long been recognized that genes can be silenced by methylation and turned on by demethylation (Martienssen and Colot 2001). Here, the global pattern of methylation in the amphihaploid was noticeably different than those of the two parental lines (table 3). Although it was reduced by the wide hybridization event, it was restored somewhat by whole-genome doubling, and the fragments of increased DNA methylation in the amphihaploid are 1.8 (104/57.3) times those of decreased methylation. Similarly, in the allotetraploid of Cucumis × hytivus, 68.2% (15 from 22 sites) of methylation loci changes showed an increase in cytosine methylation, whereas only 31.8% displayed demethylation (Belzile et al. 2008). Taken together, it suggests that DNA methylation partly functions epigenetically and dynamically to control and compromise with unbalanced gene expressions caused under certain circumstances. Although the underlying mechanisms for these phenomena are not yet determined, it is possible that the cytosine methylation machinery responds as a “genome defense” system by uniting divergent genomes into one nucleus (Yoder et al. 1997; Osborn et al. 2003).
Conclusion
The taxonomic relationship between Chrysanthemum and Leucanthemum is relatively distant. Combining such diverse genomes into a single nucleus can be expected to cause genomic shock in the sense of McClintock: “a highly programmed sequence of events within the cell that serves to cushion the effects of the shock.” The induced changes to the genome must occur very early in the development of the hybrid zygote and continue when the amphihaploid is subjected to whole-genome doubling. A further manifestation of the shock is the alteration in the pattern of DNA methylation. These rapid and massive changes in the genome of hybrids may serve as a tolerance of genome shock and might contribute to rapid genetic diploidization of the newly formed allopolyploid. An important question is whether the genomic and epigenomic events observed here are common in wide hybrids within the Asteraceae family. Understanding these genomic changes and epigenetic modifications is a worthy goal to increase understanding of the evolutionary process of the Asteraceae family, which will require the production and analysis of other wide hybridization combinations between members of the Asteraceae family.
Supplementary Material
Supplementary tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31071820, 31071825, 31272203, 31272196); 863 project (2011AA100208); the Fundamental Research Funds for the Central Universities (KYZ201112, KYZ201147); the University Program for New Century Excellent Talents in of Chinese Ministry of Education (Grant No. NCET-10-0492, NCET-12-0890); Youth Science and Technology Innovation Fund (KJ2011009); and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Literature Cited
- Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci U S A. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Barton NH. The role of hybridization in evolution. Mol Ecol. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- Bell GD, Kane NC, Rieseberg LH, Adams KL. RNA-seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol Evol. 2013;5:1309–1323. doi: 10.1093/gbe/evt072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzile F, Chen L, Chen J. Changes of cytosine methylation induced by wide hybridization and allopolyploidy in Cucumis. Genome. 2008;51:789–799. doi: 10.1139/G08-063. [DOI] [PubMed] [Google Scholar]
- Benhattar J, Clement G. Methylation-sensitive single-strand conformation analysis: a rapid method to screen for and analyze DNA methylation. Methods Mol Biol. 2004;287:181–193. doi: 10.1385/1-59259-828-5:181. [DOI] [PubMed] [Google Scholar]
- Bonin A, et al. How to track and assess genotyping errors in population genetics studies. Mol Ecol. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Bremer K, Humphries CJ. Generic monograph of the Asteraceae-Anthemideae. Bull Nat Hist Mus London (Bot). 1993;23:71–177. [Google Scholar]
- Bussell JD, Waycott M, Chappill JA. Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspect Plant Ecol. 2005;7:3–26. [Google Scholar]
- Chen L, et al. Cytological diploidization and rapid genome changes of the newly synthesized allotetraploids Cucumis x hytivus. Planta. 2007;225:603–614. doi: 10.1007/s00425-006-0381-2. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, et al. Production and characterisation of the intergeneric hybrids between Dendranthema morifolium and Artemisia vulgaris exhibiting enhanced resistance to chrysanthemum aphid (Macrosiphoniellasanbourni) Planta. 2010;231:693–703. doi: 10.1007/s00425-009-1081-5. [DOI] [PubMed] [Google Scholar]
- El-Twab A, Kondo K. Identification of parental chromosomes and changes of artificial, intergeneric F1 hybrid between Dendranthema horaimontana and Nipponanthemum nipponicum by fluorescence genomic in situ hybridization (FISH) and fluorescence in situ hybridization (GISH) Chrom Sci. 2004;8:71–79. [Google Scholar]
- El-Twab A, Kondo K, Hong D. Isolation of a particular chromosome of Ajania remotipinna in a chromosome complement of an artificial F1 hybrid of Dendranthema lavandulifolia x Ajania remotipinna by use of genomic in situ hybridization. Chrom Sci. 1999;3:21–28. [Google Scholar]
- El-Twab MHA, Kondo K. Molecular cytogenetic identification of the parental genomes in the intergeneric hybrid between Leucanthemella linearis and Nipponanthemum nipponicum during meiosis and mitosis. Caryologia Firenze. 2001;54:109–114. [Google Scholar]
- Grant-Downton RT, Dickinson HG. Epigenetics and its implications for plant biology 2. The “epigenetic epiphany”: epigenetics, evolution and beyond. Ann Bot. 2006;97:11–27. doi: 10.1093/aob/mcj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, et al. Tissue culture-induced locus-specific alteration in DNA methylation and its correlation with genetic variation in Codonopsis lanceolata Benth. et Hook. f. Plant Cell Rep. 2007;26:1297–1307. doi: 10.1007/s00299-007-0320-0. [DOI] [PubMed] [Google Scholar]
- Guo YP, Wang SZ, Vogl C, Ehrendorfer F. Nuclear and plastid haplotypes suggest rapid diploid and polyploid speciation in the N Hemisphere Achillea millefolium complex (Asteraceae) BMC Evol Biol. 2012;12:2. doi: 10.1186/1471-2148-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty MJ, et al. Changes to gene expression associated with hybrid speciation in plants: further insights from transcriptomic studies in Senecio. Philos Trans R Soc Lond B Biol Sci. 2008;363:3055–3069. doi: 10.1098/rstb.2008.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty MJ, et al. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, et al. Nonadditive changes to cytosine methylation as a consequence of hybridization and genome duplication in Senecio (Asteraceae) Mol Ecol. 2011;20:105–113. doi: 10.1111/j.1365-294X.2010.04926.x. [DOI] [PubMed] [Google Scholar]
- Jackson S, Chen ZJ. Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol. 2010;13:153–159. doi: 10.1016/j.pbi.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Mol Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- Kane NC, Barker MS, Zhan SH, Rieseberg LH. Molecular evolution across the Asteraceae: micro- and macroevolutionary processes. Mol Biol Evol. 2011;28:3225–3235. doi: 10.1093/molbev/msr166. [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Dhakal P, Katterhenry AN, Heatherington CA, Ungerer MC. Transposable element proliferation and genome expansion are rare in contemporary sunflower hybrid populations despite widespread transcriptional activity of LTR retrotransposons. Genome Biol Evol. 2011;3:156–167. doi: 10.1093/gbe/evr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M. Hybridization, glaciation and geographical parthenogenesis. Trends Ecol Evol. 2005;20:495–502. doi: 10.1016/j.tree.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Koh J, Soltis PS, Soltis DE. Homeolog loss and expression changes in natural populations of the recently and repeatedly formed allotetraploid Tragopogon mirus (Asteraceae) BMC Genomics. 2010;11:97. doi: 10.1186/1471-2164-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, El-Twab A, Tanaka R. Fluorescence genomic in situ hybridization identifies reciprocal translocation of somatic chromosomes and origin of extra chromosome by an artificial, intergeneric hybrid between Dendranthema japonica and Tanacetum vulgare. Chrom Sci. 1999;3:15–20. [Google Scholar]
- Lee HS, Chen ZJ. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci U S A. 2001;98:6753–6758. doi: 10.1073/pnas.121064698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Quiros CF. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet. 2001;103:455–461. [Google Scholar]
- Liu B, Vega J, Feldman M. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome. 1998;41:535–542. doi: 10.1139/g98-052. [DOI] [PubMed] [Google Scholar]
- Liu B, Brubaker CL, Mergeai G, Cronn RC, Wendel JF. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome. 2001;44:321–330. [PubMed] [Google Scholar]
- Liu SY, et al. In vitro induced tetraploid of Dendranthema nankingense (Nakai) Tzvel. shows an improved level of abiotic stress tolerance. Sci Hortic. 2011;127:411–419. [Google Scholar]
- Ma XF, Gustafson J. Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenet Genome Res. 2005;109:236–249. doi: 10.1159/000082406. [DOI] [PubMed] [Google Scholar]
- Ma XF, Gustafson JP. Allopolyploidization-accommodated genomic sequence changes in triticale. Ann Bot. 2008;101:825–832. doi: 10.1093/aob/mcm331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- Matthes M, Singh R, Cheah S-C, Karp A. Variation in oil palm (Elaeis guineensis Jacq.) tissue culture-derived regenerants revealed by AFLPs with methylation-sensitive enzymes. Theor Appl Genet. 2001;102:971–979. [Google Scholar]
- Matsuoka Y. Evolution of polyploid triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011;52:750–764. doi: 10.1093/pcp/pcr018. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Meudt HM, Clarke AC. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007;12:106–117. doi: 10.1016/j.tplants.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Miguel C, Marum L. An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J Exp Bot. 2011;62:3713–3725. doi: 10.1093/jxb/err155. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Osborn TC, et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–147. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Ozkan H, Levy AA, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O, Fay MF, Soltis DE, Chase MW. Genetic and epigenetic alterations after hybridization and genome doubling. Taxon. 2007;56:649–656. [PMC free article] [PubMed] [Google Scholar]
- Paun O, Stuessy TF, Horandl E. The role of hybridization, polyploidization and glaciation in the origin and evolution of the apomictic Ranunculus cassubicus complex. New Phytol. 2006;171:223–236. doi: 10.1111/j.1469-8137.2006.01738.x. [DOI] [PubMed] [Google Scholar]
- Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: causes, consequences and solutions. Nat Rev Genet. 2005;6:847–846. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- Qi B, et al. Generality and characteristics of genetic and epigenetic changes in newly synthesized allotetraploid wheat lines. J Genet Genomics. 2010;37:737–748. doi: 10.1016/S1673-8527(09)60091-6. [DOI] [PubMed] [Google Scholar]
- Rapp RA, Wendel JF. Epigenetics and plant evolution. New Phytol. 2005;168:81–91. doi: 10.1111/j.1469-8137.2005.01491.x. [DOI] [PubMed] [Google Scholar]
- Richards AJ. Apomixis in flowering plants: an overview. Philos Trans R Soc Lond B Biol Sci. 2003;358:1085–1093. doi: 10.1098/rstb.2003.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon A, Ainouche ML, Wendel JF. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae) Mol Ecol. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- Schuler GD, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell. 2001;13:1749–1759. doi: 10.1105/TPC.010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma H. How wide can a wide cross be? Euphytica. 1995;(82):43–64. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci U S A. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CN, Jr, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques. 1993;14:748–750. [PubMed] [Google Scholar]
- Stroud H, et al. Plants regenerated from tissue culture contain stable epigenome changes in rice. Elife. 2013;2:e00354. doi: 10.7554/eLife.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Williams J. Arbitrary primer mediated fingerprinting in plants: case studies in plant breeding, taxonomy and phylogeny. In: Schierwater B, Streit B, Wagner GP, DeSalle R, editors. Molecular ecology and evolution: approaches and applications. 1994. Experientia supplements Vol. 69. Basel (Switzerland): Birkhauser Verlag. p. 5–16. [Google Scholar]
- Tang F, Chen F, Chen S, Teng N, Fang W. Intergeneric hybridization and relationship of genera within the tribe Anthemideae Cass.(I. Dendranthema crassum (kitam.) kitam.× Crossostephium chinense (L.) Makino) Euphytica. 2009;169:133–140. [Google Scholar]
- Tate JA, et al. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics. 2006;173:1599–1611. doi: 10.1534/genetics.106.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira da Silva JA. Chrysanthemum: advances in tissue culture, cryopreservation, postharvest technology, genetics and transgenic biotechnology. Biotechnol Adv. 2003;21:715–766. doi: 10.1016/s0734-9750(03)00117-4. [DOI] [PubMed] [Google Scholar]
- Ungerer MC, Kawakami T. Transcriptional dynamics of LTR retrotransposons in early generation and ancient sunflower hybrids. Genome Biol Evol. 2013;5:329–337. doi: 10.1093/gbe/evt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tuyl JM, et al. Application of in vitro pollination, ovary culture, ovule culture and embryo rescue for overcoming incongruity barriers in interspecific Lilium crosses. Plant Sci. 1991;74:115–126. [Google Scholar]
- Vos P, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-G, et al. Marker-based analysis of genome structure and DNA methylation in a watermelon (Citrullus lanatus) ploidy series. Bot Stu. 2009;50:389–402. [Google Scholar]
- Wang J, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH. Yesterday's polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- Xiong L, Xu C, Maroof MS, Zhang Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet. 1999;261:439–446. doi: 10.1007/s004380050986. [DOI] [PubMed] [Google Scholar]
- Xiong Z, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci U S A. 2011;108:7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Glover BJ, Rao GY, Yang J. Molecular evidence for multiple polyploidization and lineage recombination in the Chrysanthemum indicum polyploid complex (Asteraceae) New Phytol. 2006;171:875–886. doi: 10.1111/j.1469-8137.2006.01779.x. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Yu J, et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SZ, Wang YL, He ZC, Ejder E. Genome differentiation in Magonoliaceae as revealed from meiotic pairing in interspecific and intergeneric hybrids. J Syst Evol. 2011;49:518–527. [Google Scholar]
- Zhao H, Chen F, Fang W, Guo W, Xie W. Creating novel germplasms of chrysanthemum by employing the Ajania pacifica. Sci Agric Sin. 2008;41:2077–2084. [Google Scholar]
- Zhao X, Chai Y, Liu B. Epigenetic inheritance and variation of DNA methylation level and pattern in maize intra-specific hybrids. Plant Sci. 2007;172:930–938. [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2006;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.