Abstract
Gene clusters encoding accessory or environmentally specialized metabolic pathways likely play a significant role in the evolution of fungal genomes. Two such gene clusters encoding enzymes associated with the tyrosine metabolism pathway (KEGG #00350) have been identified in the filamentous fungus Aspergillus fumigatus. The l-tyrosine degradation (TD) gene cluster encodes a functional module that facilitates breakdown of the phenolic amino acid, l-tyrosine through a homogentisate intermediate, but is also involved in the production of pyomelanin, a fungal pathogenicity factor. The gentisate catabolism (GC) gene cluster encodes a functional module likely involved in phenolic compound degradation, which may enable metabolism of biphenolic stilbenes in multiple lineages. Our investigation of the evolution of the TD and GC gene clusters in 214 fungal genomes revealed spotty distributions partially shaped by gene cluster loss and horizontal gene transfer (HGT). Specifically, a TD gene cluster shows evidence of HGT between the extremophilic, melanized fungi Exophiala dermatitidis and Baudoinia compniacensis, and a GC gene cluster shows evidence of HGT between Sordariomycete and Dothideomycete grass pathogens. These results suggest that the distribution of specialized tyrosine metabolism modules is influenced by both the ecology and phylogeny of fungal species.
Keywords: pathway evolution, phenolic compound, gene cluster, horizontal gene transfer
Introduction
Plants produce a diversity of phenolic compounds that serve as defenses against fungal pathogens. Phenolic compounds like stilbenes and flavonoids can be directly toxic to fungi that are unable to metabolize them (Adrian et al. 1997; Fofana et al. 2002). Phenolic compounds can be degraded through the Tyrosine Metabolism pathway (KEGG #00350) to produce simple sugars. The degradation of tyrosine may carry additional costs, however. For example, two human genetic diseases, alkaptonuria and tyrosinemia, are caused by mutations in tyrosine metabolism genes, leading to the accumulation of toxic metabolic intermediates (Jorquera and Tanguay 2001; Peñalva 2001). Interestingly, homologs of genes involved in two parallel tyrosine degradation (TD) pathways are sometimes tightly linked in fungal genomes. This physical co-location of tyrosine metabolism genes is consistent with the view that metabolic genes cluster because natural selection favors co-inheritance and/or balanced expression of enzyme pairs that handle toxic intermediates (Slot and Rokas 2010; Takos and Rook 2012; McGary et al. 2013).
One of these gene clusters, which is present in the genomes of Aspergillus and Candida species (Fernández-Cañón and Penalva 1995a; Jorquera and Tanguay 2001; Schmaler-Ripcke et al. 2009; Holesova et al. 2011; Keller et al. 2011), is involved in degrading l-tyrosine through a homogentisate intermediate and consists of four enzymatic steps that convert 4-hydroxyphenylpyruvate to acetoacetate and fumarate (fig. 1A). This l-TD gene cluster is also responsible for the formation of pyomelanin in the opportunistic animal pathogen Aspergillus fumigatus (Heinekamp et al. 2012). Pyomelanin contributes to disease persistence in some microbial pathogens (Hunter and Newman 2010; Zheng et al. 2013), although such a role has not been demonstrated in A. fumigatus. The TD gene cluster is considered a model of hereditary diseases resulting from defects in several of its homologous enzymes in humans (Peñalva 2001).
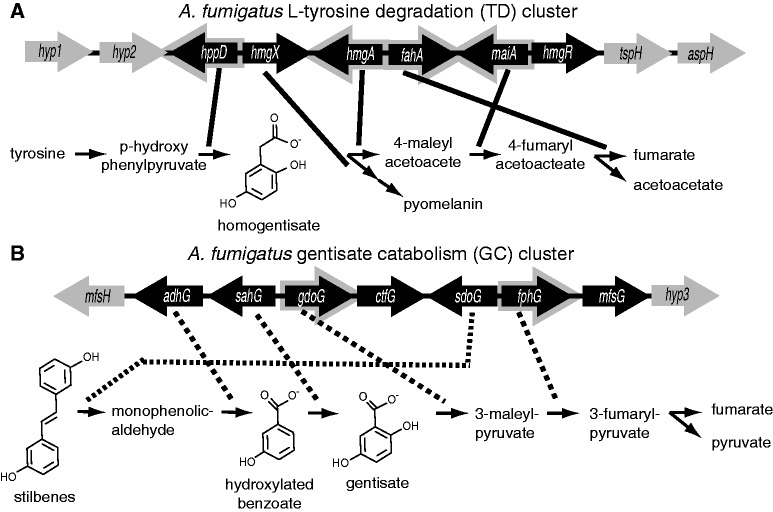
Fig. 1.—
Biochemical pathways for (A) the TD gene cluster and (B) the putative gentisate catabolism (GC) gene cluster in A. fumigatus. TD clusters (indicated by black arrows) include 4-hydroxyphenylpyruvate dioxygenase (hppD; Afu2g04200), homogentisate X factor (hmgX; Afu2g04210), homogentisate 1,2-dioxygenase (hmgA; Afu2g04220), fumarylacetoacetate hydrolase (fahA; Afu2g04230), maleylacetoacetate isomerase (maiA; Afu2g04240), homogentisate regulatory factor (hmgR; Afu2g04262). GC gene clusters include aldehyde dehydrogenase (adhG; Afu4g01550), salicylate hydroxylase, (sahG; Afu4g01530), gentisate 1,2-dioxygenase (gdoG; Afu4g01520), C6 transcription factor (ctfG; Afu4g01510), stilbene alpha-beta-dioxygenase (sdoG; Afu4g01500), fumarylpyruvate hydrolase (fphG; Afu4g01490), and a putative MFS sugar transporter (mfsG; Afu4g01480). Solid gray arrows—hypothetical protein 1 (hyp1; Afu2g04170), hypothetical protein 2 (hyp2;Afu2g04190), TIM17 subunit protein (tspH; Afu2g04270), asparaginase (aspH; Afu2g04280), myo-inositol transporter (mfsH; Afu4g01560), and hypothetical protein 3, a C6 finger domain protein (hyp3; Afu4g01470)—are genes that were included in the evolutionary analysis but are not widely conserved. Gray outlined arrows indicate genes encoding enzymes directly involved in tyrosine catabolism. Genes are connected to pathway functions by either solid lines indicating functional evidence for the specific metabolic role or dashed lines indicating putative functions based on enzyme homology. Gene clusters are not drawn to scale.
Through analysis of fungal genome organization (Zhang et al. 2012), we also noted the presence of a separate cluster of genes in a parallel TD pathway in A. fumigatus. This gene cluster encodes homologs of five enzymes that are collectively expected to degrade phenolic compounds to fumarate and pyruvate through a gentisate intermediate, and additional proteins putatively involved in phenolic metabolism (fig. 1B). We will refer to this as a putative gentisate catabolism (GC) gene cluster.
To better understand the evolution of the TD and GC gene clusters, we investigated their gene order and orientation, distribution, and phylogeny in 214 fungal genomes. Our examination revealed that homologs of the TD and GC gene clusters are present in 50 fungal species spanning multiple genera within the phylum Ascomycota and have a spotty distribution shaped by HGT and gene cluster loss that reflects organisms’ ecological specificity. As a consequence of their physical clustering in fungal chromosomes, complex phenolic metabolism modules may be gained, lost, or modified in a lineage in response to shifting environmental conditions and host–pathogen interactions.
Materials and Methods
We used two gene clusters from A. fumigatus (fig. 1) as queries in a computational pipeline constructed to detect homologous clusters in a database of 214 fungal genomes (supplementary table S1, Supplementary Material online).
The TD query (Schmaler-Ripcke et al. 2009) contained the protein sequences of six characterized genes: 4-hydroxyphenylpyruvate dioxygenase (hppD) EC# 1.13.11.27, homogentisate X factor (hmgX), homogentisate 1,2-dioxygenase (hmgA) EC# 1.13.11.5, fumarylacetoacetate hydrolase (fahA) EC# 3.7.1.2, malelylacetoacetate isomerase (maiA) EC# 5.2.1.2, and homogentisate regulatory factor (hmgR). The TD query also contained two additional sequences from each flanking region: two hypothetical proteins (hyp1 and hyp2), a putative TIM17 subunit protein (tspH), and a putative asparaginase (aspH) used to assess the degree of the evolutionary conservation of gene cluster boundaries across fungal species.
The A. fumigatus gene cluster used as the GC query was first identified by genomic co-location of genes encoding for enzymes in the Tyrosine Metabolism pathway (KEGG #00350) using methods previously described by Zhang et al. (2012). The GC query contained the protein sequences of nine genes: fumarylpyruvate hydrolase (fphG) EC# 3.7.2.5, stilbene-alpha, beta-dioxygenase (sdoG) EC# 1.13.11.43, a C6 transcription factor (ctfG), gentisate 1,2-dioxygenase oxidoreductase (a cupin-fold ring-cleaving dioxygenase, gdoG) EC# 1.13.11.4, salicylate hydroxylase (sahG) EC# 1.14.13.1, and two sequences from each flanking region (a hypothetical C6 finger domain protein, hyp3, a putative MFS sugar transporter, mfsG, aldehyde dehydrogenase, adhG, and a hypothetical myo-inositol transporter, mfsH).
Homologous gene clusters in the fungal database were identified as described previously (Slot and Rokas 2010, 2011; Campbell et al. 2012). Briefly, amino acid sequences similar to the query were identified using BlastP (Altschul et al. 1997; Altschul et al. 2005), retaining sequences with an e-value less than 10−4, that had greater than or equal to 50% amino acid sequence identity to the query sequence, and that were between 50% and 150% the length of the query. Sequences retained after the cutoff were considered to be putatively involved in tyrosine metabolism. rpb2 (RNA polymerase II second largest subunit) sequences were also recovered for construction of a species phylogeny. We restricted downstream analyses of clustering to homologous sequences in clades containing orthologs, recent paralogs of the query sequences, and potential xenologs (homologs derived by horizontal gene transfer [HGT]), based on phylogenetic analyses described later. Homologs were considered clustered if they were separated by at most six intervening genes from a homolog of another gene in the query gene cluster. Gene cluster detection was repeated with the Exophiala dermatitidis and Baudoinia compniacensis TD loci as queries in order to more accurately evaluate homology among these genomes. Homology among clusters was evaluated first by phylogenetic concordance among constituent protein phylogenies (described later), then by gene content and conservation of synteny. Conservation of the synteny among closely related species differing in the presence of the GC gene cluster was analyzed by multiple genome alignment of the corresponding loci in Mauve 2.3.1 software (Darling et al. 2010). The absence of genes was confirmed by TBlastN of query protein sequences against nucleotide genome assemblies.
Each protein homolog group was aligned with mafft ver. 6.847 (Fernández-Cañón and Penalva 1995b; Jorquera and Tanguay 2001; Katoh et al. 2002; Katoh and Toh 2008; Heinekamp et al. 2012) under default settings. The resulting alignments were manually curated in MacClade (Maddison and Maddison 2003) to remove poorly aligned taxa and then realigned with mafft. Sites in the alignment missing from greater than 30% of taxa were removed with Trimal ver. 1.2 (Capella-Gutierrez et al. 2009). Maximum likelihood analysis was performed in RAxML ver. 7.3.1 (Stamatakis 2006), with 100 bootstrap replicates under the PROTGAMMADAYHOFF model of amino acid substitution, which was determined to be the best fit of all alignments by the ProtTest (Abascal et al. 2005) script implemented in RAxML.
Analyses that forced gene trees to conform to specific topological constraints were performed in RAxML using the same model parameters. TD and GC phylogenies were constrained to contain clades consisting solely of all Dothideomycetes sequences, all Eurotiomycetes sequences, or all Sordariomycetes sequences (supplementary fig. S1, Supplementary Material online). Likelihood scores of optimal and constrained topologies were compared using the Approximately Unbiased test implemented in the Consel program (Shimodaira 2002). To control for long-branch attraction artifacts, we used the tree-independent method implemented in TIGER (http://bioinf.nuim.ie/tiger/, last accessed January 7, 2014) to classify character evolution rates (Cummins and McInerny 2011) and performed additional maximum likelihood analyses, which excluded the fastest two of ten rate partitions.
Because some homologs of gentisate dioxygenase (gdoG) are known to use salicylate preferentially as a substrate, we sought to better estimate the specificity of fungal gdoG by comparing known function-relevant residues across a set of fungal and a set of bacterial sequences. gdoG amino acid sequences from each set were aligned with mafft and imported into WebLogo 3.3 (Crooks et al. 2004) to determine the extent of conservation and similarity of amino acid residues. Residues of interest (Matera et al. 2008; Ferraroni et al. 2012; Ferraroni et al. 2013) were then compared in the two alignments and with a salicylate-cleaving homolog in Pseudaminobacter salicylatoxidans (Matera et al. 2008).
Results
Three Types of TD Gene Clusters Are Found in Dothideomycetes and Eurotiomycetes
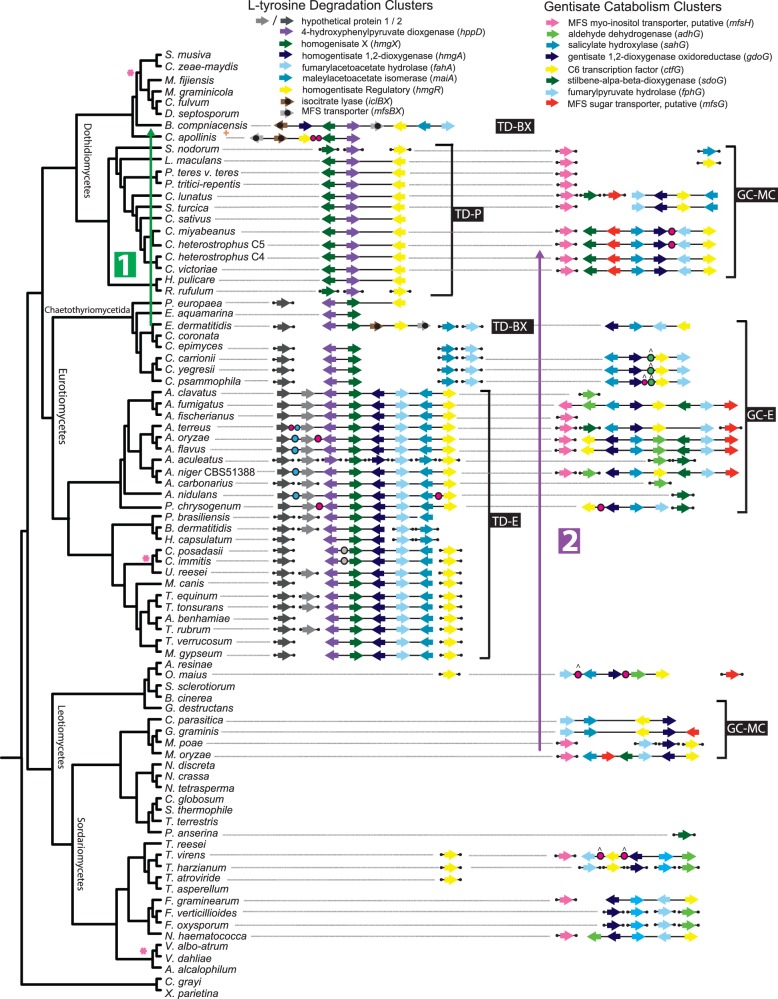
We identified 38 TDs (fig. 2) distributed among Dothideomycete and Eurotiomycete genomes. Examination of these clusters enables their division into three general types (TD-E, TD-P, and TD-BX) defined by overall concordance of specific clades in the protein phylogenies (fig. 3A and supplementary fig. S2A–H, Supplementary Material online) that correspond to the gene content and order of these types. The TD-E type consists of gene clusters that closely match the query gene cluster in gene content and order and which are found in Eurotiomycetes (excluding E. dermatitidis and other Chaetothyriomycetida). Clustering of hppD, hmgX, hmgA, fahA, and maiA in this order is highly conserved in Penicillium and Aspergillus, and clustering of hmgR is additionally well conserved in Aspergillus. hmgR orthologs are present within most genomes in Eurotiomycetes even when not clustered. The TD-P type consists of gene clusters that contain three genes (hmgX, hppD, and hmgR) differently ordered than their counterparts in TD-E gene clusters and is found exclusively in the Pleosporomycetidae (Dothideomycetes). The TD-BX type also consists of gene clusters that contain hmgX, hppD, and hmgR, but in a different order, and is found only in one Eurotiomycete, E. dermatitidis, and one Dothideomycete, B. compniacensis. TD-BX type gene clusters share two additional proteins not otherwise associated with TD gene clusters, isocitrate lyase (iclBX) and an MFS transporter (mfsBX). Furthermore, fahA and maiA orthologs are not part of the TD-BX gene cluster found in E. dermatitidis. hppD, hmgX, and sometimes hmgR are found clustered in the greater Chaetothyriomycetida (Eurotiomycetes), which contains E. dermatitidis. A similar TD cluster including iclBX and mfsBX in Coniosporium apollinis has similar gene content to the E. dermatitidis TD-BX cluster. Gene order is not highly conserved between the two TD-BX clusters.
Fig. 2.—
Distribution of two specialized tyrosine metabolism gene clusters in Ascomycete fungi. The rpb2 phylogeny (left panel) depicts species relationships among the lineages beside homologs of the l-tyrosine degradation (TD) gene clusters (middle panel) and gentisate catabolism (GC) gene clusters (right panel). Gene colors indicate homology with the query genes in A. fumigatus; absence of gene arrows for taxa denotes absence of these orthologs from their genomes. Clustering is indicated by solid lines connecting genes. Homologs that are not clustered are indicated by detached line segments. Specific gene cluster types defined first by clades in gene trees then by shared synteny are indicated with brackets. Genes with black dots in B. compniacensis and E. dermatitidis TD gene clusters were not part of the initial query. The first of these (brown) is isocitrate lyase (iclBX), and the second (gray) is an MFS transporter gene (mfsBX). The pink asterisk denotes that a homolog of MFS myo-inositol transporter of the GC gene cluster was detected for all species in the indicated clade. Dots in clusters represent other intervening genes not found in the A. fumigatus query: magenta dots represent unique intervening genes, and all other colored dots represent homologs (i.e., the blue dots in Aspergillus clade are homologous). Dots with carets above them indicate the presence of 2–6 intervening genes. Labeled arrow 1 indicates a horizontal transfer of a TD-BX gene cluster from a relative of E. dermatitidis (Eurotiomycetes) to B. compniacensis (Dothideomycetes) and labeled arrow 2 indicates a horizontal transfer of a GC-MC gene cluster from a relative of Magnaporthe spp. (Sordariomycetes) to Pleosporineae (Dothideomycetes).
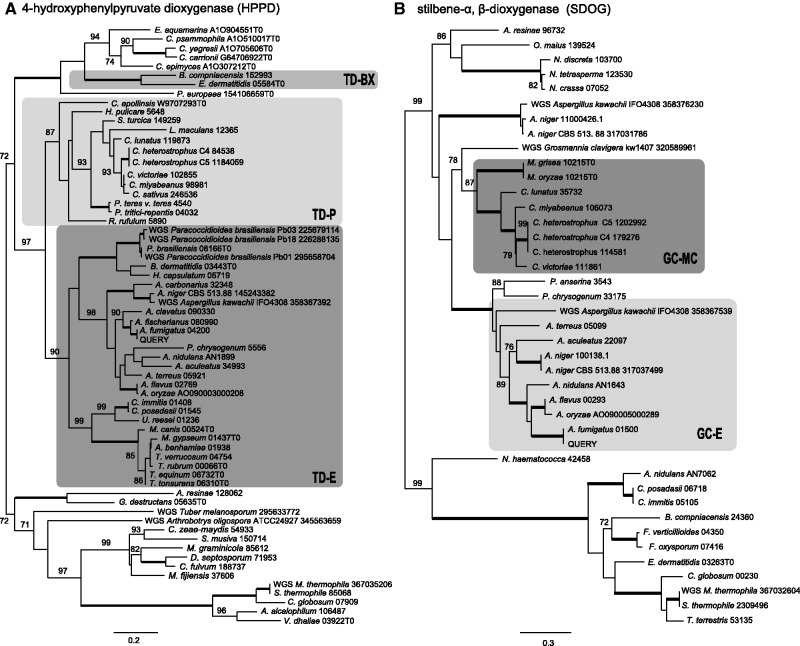
Fig. 3.—
Phylogenetic trees of (A) 4-hydroxyphenylpyruvate dioxgenase (hppD) encoded in the TD gene cluster and (B) stilbene-α, β-dioxygenase (sdoG) encoded in the gentisate catabolism (GC) gene cluster. The ortholog group for each cluster type is bounded by a shaded box. Support values represent percentage bootstrap support (out of 1000) under maximum likelihood. Only values greater than or equal to 70% are shown. Thickened black lines indicate nodes with 100% support.
GC Gene Clusters Have a Spotty Distribution in Three Classes of Fungi
We identified 23 GC gene clusters (fig. 2) spottily distributed among genomes from Dothideomycetes, Eurotiomycetes, and Sordariomycetes through the concordance of topologies among protein phylogenies (fig. 3B and supplementary fig. S2J–T, Supplementary Material online). We were able to identify two general types (GC-E and GC-MC) defined by general concordance of specific clades in the protein phylogenies that correspond to the gene content. In protein phylogenies, these clades are usually nested among paraphyletic Sordariomycete sequences and sequences from Oidiodendron maius and E. dermatitidis, which have inconsistent branching patterns. Gene order is not well conserved among these clusters. Four fully conserved genes in these gene clusters are fphG, ctfG, gdoG, and sahG. The GC-E type consists of gene clusters most closely matched in gene content to the query cluster and is found only in Aspergillus and Penicillium. sdoG, adhG, and mfsG are variably clustered in GC-E. The GC-MC type consists of gene clusters found in divergent lineages in Pleosporaceae (Dothideomycetes) and Sordariomycetidae (Sordariomycetes). sdoG and mfsG are variably found in Pleosporaceae and Sordariomycetidae GC-MC gene clusters, but adhG is not found in gene clusters in these groups. Exophiala dermatitidis and O. maius display wide variation in their placement in tree topology. The presence of closely related paralogous gene clusters (supplementary fig. S3, Supplementary Material online) in O. maius and E. dermatitidis suggest that gene cluster duplication has also played a role in diversification of GC gene clusters. Comparisons of gdoG amino acid residues among fungi and bacteria (supplementary table S2 and fig. S4, Supplementary Material online) indicate structural diversity, which may allow relaxed substrate specificity and cleavage of salicylate and other benzoate derivatives in fungi.
Phylogenetic Analyses Reveal Conflict between Gene Clusters and the Species Phylogeny
The gene trees from hmgR, hppD, hmgA, fahA, and hmgX recover the TD-E and TD-P clades (those of maiA, hyp1, and hyp2 lack TD-P clade sequences), and within these two clades, branching order is consistent with the species phylogeny. The recovery of a B. compniacensis and E. dermatitidis (BX) clade in four TD gene trees, however, conflicts with a phylogeny of Ascomycota. fahA and hmgA trees do not recover a BX clade but are consistent with a hypothesis that B. compniacensis homologs are monophyletic with the greater Chaetothyriomycetida. In addition to three TD genes inferred by comparison with the query gene cluster, these genomes share two other genes in their TDs, iclBX and mfsBX. The conflict between these gene trees and the species phylogeny is confounded by sequences from a cluster containing both genes in Rhytidhysteron rufulum, which show alternative branching orders. Baudoinia compniacensis is the only Dothideomycete in which hmgA and maiA orthologs or xenologs were detected. TD cluster sequences from C. apollinis, a related Dothideomycete, group with other Dothideomycetes and are not in conflict with a species phylogeny. Synteny of hppD, hmgX, and hmgA and of fahA and maiA is shared between B. compniacensis and gene clusters of the TD-E type but not of the TD-P type. Approximately Unbiased (AU) tests, in which constrained topologies (enforcing either a Dothideomycete clade or a Eurotiomycete clade) were compared with the optimum topology (BX clade), rejected monophyly of both classes (P < 0.05) in hppD, hmgR, hmgX, and maiA (table 1). The TD-BX clade was also supported by analyses of hmgR, iclBX, and hppD and B. compniacensis grouped with Chaetothyriomycetida in analyses of maiA after exclusion of the fastest evolving characters, suggesting that these results are not long-branch attraction artifacts.
Table 1.
Topology Tests Fail to Reject Horizontal Transfer
| Protein | Optimum Clade | Clade Enforced | P Value (AU Test) |
|---|---|---|---|
| hppD | TD-BX | Dothideomycetes | 3e−06 |
| Eurotiomycetes | 1e−11 | ||
| hmgR | TD-BX | Dothideomycetes | 1e−05 |
| Eurotiomycetes | 4e−04 | ||
| hmgX | TD-BX | Dothideomycetes | 1e−03 |
| Eurotiomycetes | 7e−03 | ||
| fahA | TD-BX | Dothideomycetes | >5e−02 |
| Eurotiomycetes | 5e−02 | ||
| maiA | TD-BX | Eurotiomycetes | 3e−05 |
| sdoG | GC-MC | Sordariomycetes | 3e−33 |
| sahG | GC-MC | Sordariomycetes | 3e−48 |
| gdoG | GC-MC | Sordariomycetes | 3e−03 |
| ctfG | GC-MC | Sordariomycetes | 4e−20 |
| fphG | GC-MC | Sordariomycetes | 5e−09 |
Protein phylogenies of six genes in the GC cluster support a GC-MC clade. Branching order supports monophyly of Magnaporthe and Pleosporineae GC-MC gene clusters. The placement of a sahG ortholog from Stagonospora nodorum and a ctfG ortholog from Leptosphaeria maculans as outgroups of the clustered orthologs suggests that the GC-MC gene cluster was originally found more broadly in Pleosporineae but was subsequently subject to repeated degradation and loss events. Five protein phylogenies (sbdG, sahG, gdxG, ctfG, and fphG) had the required taxon sampling to make them amenable to constrained analyses, which forced a Sordariomycetes clade, and all of these rejected monophyly. There was additional support for a GC-MC cluster clade, albeit very weak, from shared synteny between the two lineages, which included the conserved convergent transcription of gdoG and fphG among GC-MC type gene clusters. The presence of transposable elements in the flanks of the Cochliobolus species was notable due to the ability of these elements to mobilize DNA, which raises the hypothesis that the unexpected distribution of these genes may be explained by HGT.
Presence of GC Gene Clusters Is Variable among Closely Related Species
In the Aspergillus section Nigri (Eurotiomycetes), A. aculeatus and A. carbonarius lack a GC-E type gene cluster, while such a gene cluster is present in A. niger. Similarly, A. clavatus (section Clavati) and its close relative Aspergillus fischerianus (section Fumigati) lack a GC-E gene cluster, while A. fumigatus (section Fumigati), the sister taxon of A. fischerianus, has a full GC-E gene cluster. Similarly, A. nidulans (section Nidulantes) lacks a GC-E gene cluster, but Penicillium chrysogenum has a partial (4-gene) GC-E gene cluster and two additional unclustered homologs, suggesting that these genes are ancestral in mitosporic Trichocomaceae but were later lost. Mauve alignments of the GC locus in A. fischerianus and A. fumigatus (supplementary fig. S5, Supplementary Material online) reveal alternative sets of genes totaling a similar length of ∼12 kb. Orthologs of the alternative gene sets are reciprocally absent in the paired species. Four genomes, F. oxysporum, F. verticillioides, T. harzianum, and Magnaporthe poae, do not retain clustering of GC gene cluster homologs but do retain constituent enzymes that bracket putative toxic intermediates in the pathway.
Discussion
Conservation of Gene Content and Synteny Suggest Metabolic Phenotypes Conferred by Tyrosine Metabolism Gene Clusters
Genes involved in tyrosine metabolism are widely distributed throughout Ascomycete fungi, a subset of which is found in metabolic gene clusters (supplementary fig. S6, Supplementary Material online). Here, we trace the evolution of two types of gene clusters, which encode processes that overlap background functions of tyrosine metabolism (i.e., they are functionally semi-redundant with noncluster paralogs). The TD gene cluster (Schmaler-Ripcke et al. 2009; Keller et al. 2011) overlaps tyrosine metabolism in the path for homogentisate degradation, and the GC gene cluster overlaps tyrosine metabolism in the path for gentisate degradation. Both of these clusters contain genes conferring specialized or accessory functions beyond those conferred by the core degradation pathways.
Conservation of gene content of these clusters despite extensive general genomic rearrangement circumscribes modular functions (Muto et al. 2013) that are retained by natural selection in fungal lineages. Using these conserved gene cluster constituents as a guide, we propose testable models for the net biochemical functions of these gene clusters (fig. 1). For example, the TD-E gene cluster in A. fumigatus has already been characterized to produce pyomelanin as a shunt in the breakdown of tyrosine and also to allow the use of l-tyrosine as a sole carbon source (Schmaler-Ripcke et al. 2009). The TD-BX gene cluster contains many of the same genes as the TD-E gene cluster but also contains isocitrate lyase, which could support downstream carbon retention by these fungi and suggests a role for TD-BX in fungal pathogenesis (Dunn et al. 2009). TD-BX also includes an MFS transporter, which could be involved in transport of simple sugars across the mitochondrial membrane. By analogy with the characterized gene cluster in A. fumigatus, the TD-P gene cluster in Dothideomycetes may be involved in pyomelanin production but not in degradation of tyrosine as an energy source. Melanins are pathogenicity factors in fungi, and therefore TD-P could indicate selection focused on this component of the pathway for evasion of host oxidative defenses (Williamson et al. 1998; Langfelder et al. 2003; Karkowska-Kuleta et al. 2009; Youngchim et al. 2011).
In GC gene clusters, we propose a model in which multiple genes are required to generate and degrade gentisate and other benzoate derivatives. In our model of the core GC pathway, a hydroxylated benzoate is converted to gentisate by sahG. sahG is homologous with 3-hydroxybenzoate 6-hydroxylase (mnx2), which is encoded in a GC-like gene cluster in Candida parapsilosis where it hydroxylates a benzoate derivative for degradation via gentisate (Holesova et al. 2011). Gentisate is then converted to 3-maleyl pyruvate by gdoG, 3-maleyl pyruvate spontaneously isomerizes to 3-fumarylpyruvate, and finally 3-fumarylpyruvate is converted to fumarate and pyruvate by fphG. A stilbene dioxygenase gene, (sdoG) is incompletely but broadly conserved in GC gene clusters. Stilbene dioxygenases convert ethylene-linked phenolic dimers such as pinosylvin and resveratrol into monophenolic aldehydes (Marasco and Schmidt-Dannert 2008). sdoG is similar in sequence to lignostilbene dioxygenase, which preferentially cleaves trans-4-hydroxy-3-methoxystilbene (Kamoda et al. 2003) into the monophenolic aldehyde, vanillin. Notably, one of the genes that is conserved across divergent lineages, adhG, is predicted to encode an aldehyde dehydrogenase, which may complete the path from stilbene to citric acid cycle intermediates. Relaxed specificities for the 2,5 hydroxylation pattern in gentisate (Fetzner 2012) could allow a broader range of stilbenes or benzoate derivatives to be metabolized by GC pathways (Ferraroni et al. 2012; Ferraroni et al. 2013). The conserved sugar transporter in the GC gene cluster may then usher simple sugars into mitochondria for energy production. Similar clusters of homologs of sahG, gdoG, fphG, and maleylpyruvate isomerase are spottily distributed among bacteria expected to be under phenolic stress, such as Acinetobacter sp. Strain DR1 (YP_003732082-YP_003732085) (Jung et al. 2010), Polaromonas naphthalenivorans strain CJ2 (YP_983363–YP_983366) (Yagi et al. 2009), Azoarcus sp. strain BH72 (YP_933924–YP_933927) (Krause et al. 2006), and Cupriavidus necator JMP134 (YP_300048–YP_300051) (Lykidis et al. 2010), further supporting the functional association of these genes.
A Spotty Distribution of Specialized Tyrosine Pathway Clusters Is Explained by Overlapping Ecology
The spotty distribution of specialized tyrosine metabolism clusters is inconsistent with fungal phylogeny. Distantly related species share similar clusters, while close relatives with different ecologies differ in the presence of clusters. This spotty distribution is best exemplified by the fact that the locus containing the GC gene cluster in A. fumigatus is occupied by an alternative set of genes in its very close relative A. fischerianus. The genes in this locus are reciprocally absent in A. fumigatus and A. fischerianus suggesting either recent replacement of the GC gene cluster by HGT or a gene cluster polymorphism in this locus (Gibbons et al. 2012; Zhang et al. 2012). Aspergillus fischerianus does not readily decay grasses like its close relative A. fumigatus, which is often a primary agent of grass silage spoilage (Aragon et al. 2011), suggesting that the difference in content at this locus might reflect the different selective pressures exerted on the two species.
In general, the robustly supported observed discordances between gene tree topologies and the species phylogeny can be explained by either HGT of gene clusters or by the independent maintenance of an ancient duplicate gene cluster in specific lineages and its loss in all others. For example, the clade formed by genes in TD-BX gene clusters suggests either HGT from Chaetothyriomycetida to an ancestor of B. compniacensis or retention of an ancient duplicate cluster by B. compniacensis and E. dermatitidis, but not by other Dothideomycetes or Chaetothyriomycetida genomes. We favor the hypothesis of HGT in this case due to the high number of gene cluster losses required to reconcile the gene trees with the conflicting species phylogeny under the alternative hypothesis (see also Khaldi et al. 2008). Similarly, the phylogeny of GC genes is best explained by HGT of the GC-MC cluster between Magnaporthe and the clade containing Cochliobolus spp. There is an intriguing phenotypic overlap between divergent species that is not found among all species of their respective classes present in our data set. Both E. dermatitidis (like other Chaetothyriomycetida) and B. compniacensis have constitutively melanized cell walls and thrive in extreme environments. Exophiala dermatitidis, although most known as a human pathogen acquired in bathhouses in Southeast Asia, is most readily cultured from creosote-treated railroad ties (Sudhadham et al. 2008). The preservative effect of creosote on exposed wood is due in part to antifungal phenolic compounds (Kim et al. 2010). Baudoinia compniacensis is most commonly isolated from exterior surfaces near alcohol distilleries (Scott et al. 2007). Both of these species are thermotolerant and resistant to environmental exposure. Aside from human environments, fallen fruit has been postulated as a natural reservoir for both of these fungi (Scott et al. 2007; Sudhadham et al. 2008). Sharing niches under oxidative stress may have provided an opportunity for the ancestors of B. compniacensis and E. dermatitidis to engage in HGT.
Acquisition of the TD-BX gene cluster may have benefitted B. compniacensis by facilitating additional protective melanization and assimilation of carbon. Two genes (hmgR and hmgX) that may have been transferred with the TD-BX gene cluster facilitate a metabolic shunt for the production of pyomelanin (Heinekamp et al. 2012) and appear to be ancestrally present in TD gene clusters. This shunt may benefit B. compniacensis by converting high concentrations of toxic phenolic compounds to pyomelanin, which protects cells from oxidation damage (Rodriguez-Rojas et al. 2009). The inclusion of isocitrate lyase in the transferred gene cluster suggests these species are able to directly assimilate fixed carbon from the breakdown of phenolics (Dunn et al. 2009).
The GC gene cluster variants in this study are likely to facilitate metabolic specialization by prepending one or two additional metabolic steps upstream of ring cleavage. These clusters all encode sahG, which may transform benzoate derivatives for degradation by gdoG. Salicylate and gentisate are plant pathogen-response hormones with antifungal activity (Dempsey et al. 2011; Qi et al. 2012). Biotrophic and endophytic fungi may require resistances to these compounds to maintain stable infections of plant hosts (Ambrose and Belanger 2012). The fungi in this analysis that can live endophytically in plants are often found with GC gene clusters that are expected to degrade benzoate derivatives such as gentisate and salicylate. One of them, Fusarium graminearum, has also been shown to use salicylate as a carbon source (Qi et al. 2012). These monophenolic compounds are likely to be produced throughout the life history of many plants and consequently favor biotrophs with a mechanism to cope with them. GC gene clusters with sahG are found in distantly related endophytic fungi, such as Fusarium graminearum, Trichoderma virens, and Oidiodendron maius, but not in their close relatives with different ecological preferences (T. reesei—a wood decayer [Kubicek et al. 2011] and Amorphotheca resinae—a hydrocarbon degrader [Seifert et al. 2007]). We hypothesize that this reflects their use in symbiosis-related metabolism.
Stilbenes are antifungal compounds produced by sorghum and other grasses in response to multiple types of pathogenic fungal infections (Brinker and Seigler 1991; Yu et al. 2005, 2008). Consequently, grass pathogens may benefit from alpha-beta dioxygenases, such as sdoG, to degrade stilbenes to monophenolic aldehydes (Adrian et al. 1997; Hipskind and Paiva 2000; Schulze et al. 2005). GC-MC and GC-E types are two GC gene cluster variants that contain sdoG orthologs and that are found in grass pathogens (Magnaporthe spp. and Cochliobolus spp.) and common spoilage agents of grain (Aspergillus spp.), respectively. Aspergillus species containing GC-E also elicit production of stilbenes during spoilage of peanut (Sobolev 2013). The distribution of sdoG-containing GC cluster variants is more strongly associated with grass colonization than simple linear descent through the species phylogeny, which further suggests that shared ecology provides both the opportunity and selection pressure to drive HGT of gene clusters (Slot and Hibbett 2007; Slot and Rokas 2010; Slot and Rokas 2011; Campbell et al. 2012).
The ecological pattern to the distribution of specialized tyrosine metabolism gene clusters is facilitated by the physical linkage of the pathways they encode. The core genes involved in tyrosine metabolism are commonly dispersed in the genome, while more specialized genes that function in contexts such as toxin degradation and resistance to host defenses appear to be frequently clustered. Decoupling of specialized functions from core metabolism through gene duplication-induced redundancy (fig. 4) frees specialized genes and gene clusters to adapt to novel usage, while feeding into established metabolic pathways (Zhang et al. 2012). Selection for the maintenance of specialized versions of these pathways may be sporadic and variable due to changing environments and host–pathogen co-evolution, even though the non-clustered core metabolic pathways remain relatively stable. Relaxation of selection from environmental factors explains why specialized pathways are prone to loss during vertical transmission. Gradual loss of the constituent genes may be prevented by selection against the accumulation of toxic intermediate compounds produced by incomplete pathways (McGary et al. 2013), but isolation of these pathways to a specific location weakens their link to the genome by facilitating complete loss in a single event (Slot and Rokas 2010). HGT may counteract the loss of genes that are not under constant selection if they can provide benefit to the organisms that receive them (Lawrence and Roth 1996; Walton 2000). The net benefit of these gene clusters to fungi may be enhanced by additional conserved genes, such as transcription factors and transporters, which contribute to a complete, modular function. In these ways, genomic modularity of complex specialized metabolic pathways enhances the adaptability of receptive fungal genomes to shifts in ecological landscapes.
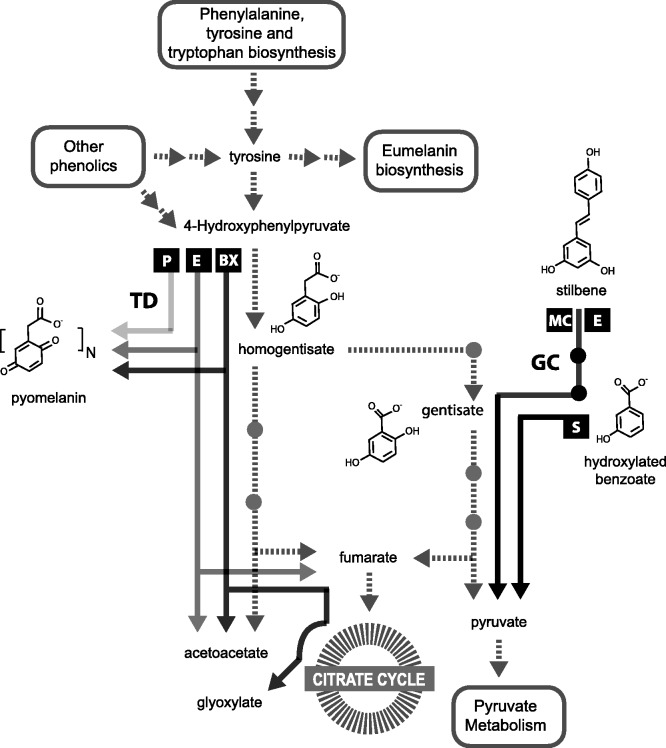
Fig. 4.—
Modular evolution and specialization of pathways in tyrosine metabolism. Basic TD via homogentisate and gentisate is depicted by dashed lines. Intermediate metabolites are indicated by names (and compound structure) or solid points. Metabolic modules and putative metabolic modules encoded by specialized gene clusters that parallel the basic pathways are depicted with solid lines of different shades. The TD-P, TD-E, TD-BX, GC-MC/GC-E, and TD-S (where TD-S is a nonmonophyletic assemblage of clusters expected to degrade benzoate derivative compounds by way of gentisic acid) are indicated from lightest to darkest grayscale.
Supplementary Material
Supplementary figures S1–S6 and tables S1–S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. This work was partially supported by funds provided by a Vanderbilt Undergraduate Summer Research Program Fellowship (G.H.G.) and the National Science Foundation (DBI-0805625 to J.C.S. and DEB-0844968 to A.R.).
Literature Cited
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adrian M, Jeandet P, Veneau J, Weston LA, Bessis R. Biological activity of resveratrol, a stilbenic compound from grapevines, against Botrytis cinerea, the causal agent for gray mold. J Chemic Ecol. 1997;23:1689–1702. [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose KV, Belanger FC. SOLiD-SAGE of endophyte-infected red fescue reveals numerous effects on host transcriptome and an abundance of highly expressed fungal secreted proteins. PLoS One. 2012;7:e53214. doi: 10.1371/journal.pone.0053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon YA, Rodrigues I, Hofstetter U, Binder EM. Mycotoxins in silages: occurrence and prevention. Iran J Appl Anim Sci. 2011;1:1–10. [Google Scholar]
- Brinker AM, Seigler DS. Isolation and identification of piceatannol as a phytoalexin from sugarcane. Phytochemistry. 1991;30:3229–3232. [Google Scholar]
- Campbell MA, Rokas A, Slot JC. Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol Evol. 2012;4:289–293. doi: 10.1093/gbe/evs011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins CA, Mcinerney JO. A method for inferring the rate of evolution of homologous characters that can potentially improve phylogenetic inference, resolve deep divergence and correct systematic biases. System Biol. 2011;60(6):833–844. doi: 10.1093/sysbio/syr064. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF. Salicylic acid biosynthesis and metabolism. Arabidopsis Book. 2011;9:e0156. doi: 10.1199/tab.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn MF, Ramirez-Trujillo JA, Hernandez-Lucas I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology. 2009;155:3166–3175. doi: 10.1099/mic.0.030858-0. [DOI] [PubMed] [Google Scholar]
- Fernández-Cañón JM, Penalva MA. Molecular characterization of a gene encoding a homogentisate dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J Biol Chem. 1995a;270:21199–21205. doi: 10.1074/jbc.270.36.21199. [DOI] [PubMed] [Google Scholar]
- Fernández-Cañón JM, Penalva MA. Fungal metabolic model for human type I hereditary tyrosinaemia. Proc Natl Acad Sci U S A. 1995b;92: 9132–9136. doi: 10.1073/pnas.92.20.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraroni M, et al. Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: a step towards the comprehension of the structural Basis for substrate selection in class III ring cleaving dioxygenases. J Struct Biol. 2012;177:431–438. doi: 10.1016/j.jsb.2011.11.026. [DOI] [PubMed] [Google Scholar]
- Ferraroni M, et al. The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate. FEBS J. 2013;280:1643–1652. doi: 10.1111/febs.12173. [DOI] [PubMed] [Google Scholar]
- Fetzner S. Ring-cleaving dioxygenases with a cupin fold. Appl Environ Microbiol. 2012;78:2505–2514. doi: 10.1128/AEM.07651-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofana B, et al. Milsana-Induced resistance in powdery mildew-infected cucumber plants correlates with the induction of chalcone synthase and chalcone isomerase. Physiol Mol Plant Pathol. 2002;61:121–132. [Google Scholar]
- Gibbons JG, et al. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr Biol. 2012;22:1403–1409. doi: 10.1016/j.cub.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinekamp T, et al. Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence. Front Microbiol. 2012;3:440. doi: 10.3389/fmicb.2012.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipskind JD, Paiva NL. Constitutive accumulation of a resveratrol-glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol Plant Microbe Interact. 2000;13:551–562. doi: 10.1094/MPMI.2000.13.5.551. [DOI] [PubMed] [Google Scholar]
- Holesova Z, et al. Gentisate and 3-oxoadipate pathways in the yeast Candida parapsilosis: identification and functional analysis of the genes coding for 3-hydroxybenzoate 6-hydroxylase and 4-hydroxybenzoate 1-hydroxylase. Microbiology. 2011;157:2152–2163. doi: 10.1099/mic.0.048215-0. [DOI] [PubMed] [Google Scholar]
- Hunter RC, Newman DK. A putative ABC transporter, HatABCDE, Is among molecular determinants of pyomelanin production in Pseudomonas aeruginosa. J Bacteriol. 2010;192:5962–5971. doi: 10.1128/JB.01021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorquera R, Tanguay RM. Fumarylacetoacetate, the metabolite accumulating in hereditary tyrosinemia, activates the ERK pathway and induces mitotic abnormalities and genomic instability. Hum Mol Genet. 2001;10:1741–1752. doi: 10.1093/hmg/10.17.1741. [DOI] [PubMed] [Google Scholar]
- Jung J, Baek J, Park W. Complete genome sequence of the diesel-degrading Acinetobacter sp. strain DR1. J Bacteriol. 2010;192:4794–4795. doi: 10.1128/JB.00722-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoda S, Terada T, Saburi Y. A common structure of substrate shared by lignostilbenedioxygenase isozymes from Sphingomonas paucimobilis TMY1009. Biosci Biotechnol Biochem. 2003;67:1394–1396. doi: 10.1271/bbb.67.1394. [DOI] [PubMed] [Google Scholar]
- Karkowska-Kuleta J, Rapala-Kozik M, Kozik A. Fungi pathogenic to humans: molecular bases of virulence of Candida albicans, Cryptococcus neoformans and Aspergillus fumigatus. Acta Biochim Pol. 2009;56:211–224. [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Keller S, et al. Pyomelanin formation in Aspergillus fumigatus requires HmgX and the transcriptional activator HmgR but is dispensable for virulence. PLoS One. 2011;6:e26604. doi: 10.1371/journal.pone.0026604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N, Collemare J, Lebrun MH, Wolfe KH. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol. 2008;9(1):R18. doi: 10.1186/gb-2008-9-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, et al. Diversity of fungi in creosote-treated crosstie wastes and their resistance to polycyclic aromatic hydrocarbons. Antonie Van Leeuwenhoek. 2010;97:377–387. doi: 10.1007/s10482-010-9416-6. [DOI] [PubMed] [Google Scholar]
- Krause A, et al. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat Biotechnol. 2006;24:1385–1391. doi: 10.1038/nbt1243. [DOI] [PubMed] [Google Scholar]
- Kubicek CP, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12:R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder K, Streibel M, Jahn B, Haase G, Brakhage AA. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet Biol. 2003;38:143–158. doi: 10.1016/s1087-1845(02)00526-1. [DOI] [PubMed] [Google Scholar]
- Lawrence J, Roth J. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykidis A, et al. The complete multipartite genome sequence of Cupriavidus necator JMP134, a versatile pollutant degrader. PLoS One. 2010;5:e9729. doi: 10.1371/journal.pone.0009729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. MacCLADE: analysis of phylogeny and character evolution, Version 4.06. Sunderland (MA): Sinauer Associates; 2003. [Google Scholar]
- Marasco EK, Schmidt-Dannert C. Identification of bacterial carotenoid cleavage dioxygenase homologues that cleave the interphenyl α,β double bond of stilbene derivatives via a monooxygenase reaction. Chembiochem. 2008;9:1450–1461. doi: 10.1002/cbic.200700724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera I, et al. Salicylate 1,2-dioxygenase from Pseudaminobacter salicylatoxidans: crystal structure of a peculiar ring-cleaving dioxygenase. J Mol Biol. 2008;380:856–868. doi: 10.1016/j.jmb.2008.05.041. [DOI] [PubMed] [Google Scholar]
- McGary KL, Slot JC, Rokas A. Physical linkage of metabolic genes in fungi is an adaptation against the accumulation of toxic intermediate compounds. Proc Natl Acad Sci U S A. 2013;110:11481–11486. doi: 10.1073/pnas.1304461110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, et al. Modular architecture of metabolic pathways revealed by conserved sequences of reactions. J Chem Info Model. 2013;53:613–622. doi: 10.1021/ci3005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva MÁ. A fungal perspective on human inborn errors of metabolism: alkaptonuria and beyond. Fungal Genet Biol. 2001;34:1–10. doi: 10.1006/fgbi.2001.1284. [DOI] [PubMed] [Google Scholar]
- Qi P, et al. Effect of salicylic acid on Fusarium graminearum, the major causal agent of fusarium head blight in wheat. Fungal Biol. 2012;116:413–426. doi: 10.1016/j.funbio.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rojas A, et al. Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology. 2009;155:1050–1057. doi: 10.1099/mic.0.024745-0. [DOI] [PubMed] [Google Scholar]
- Schmaler-Ripcke J, et al. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl Environ Microbiol. 2009;75:493–503. doi: 10.1128/AEM.02077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze K, Schreiber L, Szankowski I. Inhibiting effects of resveratrol and its glucoside piceid against Venturia inaequalis, the causal agent of apple scab. J Agric Food Chem. 2005;53:356–362. doi: 10.1021/jf048375h. [DOI] [PubMed] [Google Scholar]
- Scott JA, Untereiner WA, Ewaze JO, Wong B, Doyle D. Baudoinia, a new genus to accommodate Torula compniacensis. Mycologia. 2007;99:592–601. doi: 10.3852/mycologia.99.4.592. [DOI] [PubMed] [Google Scholar]
- Seifert KA, Hughes SJ, Boulay H, Louis-Seize G. Taxonomy, nomenclature and phylogeny of three cladosporium-like hyphomycetes, Sorocybe resinae, Seifertia azaleae and the Hormoconis anamorph of Amorphotheca resinae. Stud Mycol. 2007;58:235–245. doi: 10.3114/sim.2007.58.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Slot JC, Hibbett DS. Horizontal transfer of a nitrate assimilation gene cluster and ecological transitions in fungi: a phylogenetic study. PLoS One. 2007;2:e1097. doi: 10.1371/journal.pone.0001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JC, Rokas A. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc Natl Acad Sci U S A. 2010;107:10136–10141. doi: 10.1073/pnas.0914418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JC, Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr Biol. 2011;21:134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Sobolev VS. Production of phytoalexins in peanut (Arachis hypogaea) seed elicited by selected microorganisms. J Agric Food Chem. 2013;61:1850–1858. doi: 10.1021/jf3054752. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sudhadham M, et al. The neurotropic black yeast Exophiala dermatitidis has a possible origin in the tropical rain forest. Stud Mycol. 2008;61:145–155. doi: 10.3114/sim.2008.61.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Rook F. Why biosynthetic genes for chemical defense compounds cluster. Trends Plant Sci. 2012;17:383–388. doi: 10.1016/j.tplants.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Walton J. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet Biol. 2000;30:167–171. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- Williamson PR, Wakamatsu K, Ito S. Melanin biosynthesis in Cryptococcus neoformans. J Bacteriol. 1998;180:1570–1572. doi: 10.1128/jb.180.6.1570-1572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi JM, Sims D, Brettin T, Bruce D, Madsen EL. The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ Microbiol. 2009;11:2253–2270. doi: 10.1111/j.1462-2920.2009.01947.x. [DOI] [PubMed] [Google Scholar]
- Youngchim S, Pornsuwan S, Nosanchuk JD, Dankai W, Vanittanakom N. Melanogenesis in dermatophyte species in vitro and during infection. Microbiology. 2011;157:2348–2356. doi: 10.1099/mic.0.047928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CK, Shih CH, Chu IK, Lo C. Accumulation of trans-piceid in sorghum seedlings infected with Colletotrichum sublineolum. Phytochemistry. 2008;69:700–706. doi: 10.1016/j.phytochem.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Yu CK, et al. A stilbene synthase gene (SbSTS1) is involved in host and nonhost defense responses in sorghum. Plant Physiol. 2005;138:393–401. doi: 10.1104/pp.105.059337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rokas A, Slot JC. Two different secondary metabolism gene clusters occupied the same ancestral locus in fungal dermatophytes of the Arthrodermataceae. PLoS One. 2012;7:e41903. doi: 10.1371/journal.pone.0041903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chatfield CH, Liles MR, Cianciotto NP. Secreted pyomelanin of Legionella pneumophila promotes bacterial iron uptake and growth under iron-limiting conditions. Infect Immun. 2013;81:4182–4191. doi: 10.1128/IAI.00858-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.