Abstract
Microbes have been identified as a major contaminant of water resources. Escherichia coli (E. coli) is a commonly used indicator organism. It is well recognized that the fate of E. coli in surface water systems is governed by multiple physical, chemical, and biological factors. The aim of this work is to provide insight into the physical, chemical, and biological factors along with their interactions that are critical in the estimation of E. coli loads in surface streams. There are various models to predict E. coli loads in streams, but they tend to be system or site specific or overly complex without enhancing our understanding of these factors. Hence, based on available data, a Bayesian Neural Network (BNN) is presented for estimating E. coli loads based on physical, chemical, and biological factors in streams. The BNN has the dual advantage of overcoming the absence of quality data (with regards to consistency in data) and determination of mechanistic model parameters by employing a probabilistic framework. This study evaluates whether the BNN model can be an effective alternative tool to mechanistic models for E. coli loads estimation in streams. For this purpose, a comparison with a traditional model (LOADEST, USGS) is conducted. The models are compared for estimated E. coli loads based on available water quality data in Plum Creek, Texas. All the model efficiency measures suggest that overall E. coli loads estimations by the BNN model are better than the E. coli loads estimations by the LOADEST model on all the three occasions (three-fold cross validation). Thirteen factors were used for estimating E. coli loads with the exhaustive feature selection technique, which indicated that six of thirteen factors are important for estimating E. coli loads. Physical factors included temperature and dissolved oxygen; chemical factors include phosphate and ammonia; biological factors include suspended solids and chlorophyll. The results highlight that the LOADEST model estimates E. coli loads better in the smaller ranges, whereas the BNN model estimates E. coli loads better in the higher ranges. Hence, the BNN model can be used to design targeted monitoring programs and implement regulatory standards through TMDL programs.
Keywords: E. coli, Stream flow, Bayesian Neural Networks, Uncertainty analysis, LOADEST
2. Introduction
Microbes have been identified as a major contaminant (13.2% contamination caused by pathogenic microbes of total impaired water body segments) of water resources in USA (USEPA, 2006). Common bacterial waterborne pathogens include Salmonella sp, Shigella sp., few strains of Escherichia coli (E. coli), Pseudomonas aeruginosa, Aeromonas hydrophila, Mycobacteria, Helicobacter pylori, and various others [Fincher et al., 2009]. The most widely used indicator organisms are the enteric coliform bacteria, which are Gram-negative bacilli that belong to the family Enterobacteriaceae (e.g., Klebsiella spp., Enterobacter spp., Citrobacter spp., Escherichia coli) [Hipsey et al., 2008; Dorner et al., 2006; Mead and Griffin, 1998]. The indicator organisms are mostly harmless as compared to the pathogen(s) of concern. However, the indicator organisms are monitored due to the relative ease and lesser cost involved in their measurements. It is well established that the fate of E. coli in surface water systems is governed by multiple physical (e.g., temperature [Flint, 1987]), chemical (e.g., pH [Sjogren and Gibson, 1981], nutrients [Lessard and Sieburth, 1983], sulfate [Robakis et al., 1983], and nitrate [Noguchi et al., 1997]), and biological (Chlorophyll [Nevers and Whitman, 2005]) factors. The relationship among these factors and E. coli loads gets complicated by flow rate [Whitman et al., 2004; McKergow et al., 2009]. Vidon et al. [2008] have reported that E. coli loads are significantly higher at high flow than at low flow, whereas McKergow et al. [2009] have observed that E. coli peak loads always preceded discharge and turbidity peaks (which had similar timings). Therefore, E. coli evidently has a nonlinear relationship with the flow rate and the turbidity.
It is important to develop an understanding of the relative importance of these physical, chemical, and biological factors in estimating the survival of E. coli in water bodies. However, a direct measurement of E. coli fate is not, in general, easy to implement. Therefore, the degree of impairment of a stream is assessed in terms of Total Maximum Daily Load (TMDL). Load duration curves are often used to estimate the reduction of contaminant loads in a watershed, especially in TMDL programs [Babbar-Sebens and Karthikeyan, 2009]. The load duration curves are measured using the instantaneous “load”. The instantaneous “load” passing through a stream cross-section is the product of the flow rate and the constituent concentration.
Various models have been developed that use complex mechanistic and empirical relationships to predict the loads of E. coli in surface water systems e.g, Soil and Water Assessment Tool (SWAT) [Arnold and Fohrer, 2005 and Pachepsky et al., 2006], Hydrological Simulation Program—Fortran (HSPF) [Benham et al. 2006], and a watershed model developed by Tian et al. [2002]. However, overly complex mechanistic relationships and requirement of detailed descriptions of stream geometry and capacity, detailed information about sources within the watershed, sedimentation and re-suspension characteristics, and bacteria die-off rates limit the utility of these models. Input parameter approximation and simplification in describing transport processes result in significant uncertainties in E .coli loads in streams. Other models have been developed that use statistical modeling framework to predict the loads of E. coli in surface water systems. For instance, Nevers and Whitman (2005) used regression modeling to determine E. coli using the wave height, lake chlorophyll, and turbidity for individual beaches of southern Lake Michigan. Furthermore, Money et al. (2009) estimated E. coli concentrations using turbidity, where E. coli data were not available, to assess fecal contamination along the Raritan River in New Jersey. Different models are relevant for different surface water environments, such as freshwater lakes and reservoirs [Auer and Niehaus, 1993; Walker and Stedinger, 1999; Jin et al., 2003; Hipsey et al., 2008], streams and rivers [Wilkinson et al., 1995; Medema and Schijven, 2001], and estuaries and coastal lagoons [Steets and Holden, 2003; McCorquodale et al., 2004]. It is also difficult for users to confidently implement these models, since models tend to be system or site specific.
In comparison to these mechanistic and statistical models, a Bayesian Neural Network (BNN) provides a Bayesian modeling framework for estimating E. coli loads by utilizing routinely monitored flow rate and water quality data. The input data will comprise of water quality data (physical, chemical, and biological factors) that will provide a functional framework for the BNN. In case of sparse datasets, the Bayesian framework helps in representing input parameters as random variables emphasizing the statistical strength of the available data. Also, the uncertainty in input data sets is reflected through the probabilistic prediction of E. coli loads. The graphical structure of the BNN represents a cause-and-effect relationship between system variables (water quality data) and E. coli loads, as shown in Fig. 1. One can use various basis functions in the formulation of the BNN such as multi-layer perceptron (MLP), or radial basis functions (RBF). BNN models with radial basis functions (RBF) have been used in this study as they have an ability to deal with sparse datasets and parameter over-fitting [Cilek and Yilmazer, 2003].
Figure 1.
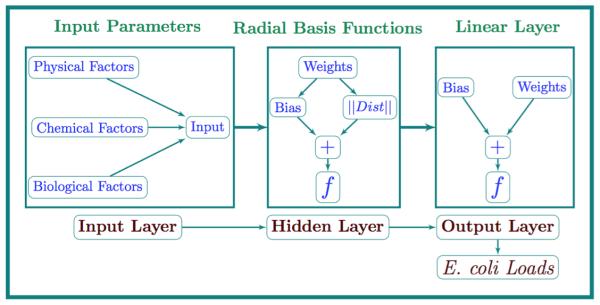
The graphical structure of BNN representing cause-and-effect relationship between system variables (water quality parameters) and the E. coli loads. The Radial Basis layer is the hidden layer, which uses the transfer function f (thin plate spline); the output layer is a linear layer, which uses the transfer function f (linear function). The transfer function f establishes a relationship between inputs and outputs, in case of estimation of E. coli loads, thin plate splines works better than other transfer functions (Gaussian or r4 functions).
The specific objective of this study is to identify the key water quality factors for estimating the E. coli loads in streams. Based on identified water quality factors, E. coli loads will be estimated in streams along with characterization of possible uncertainties.
3. Study Area Description and Data Availability
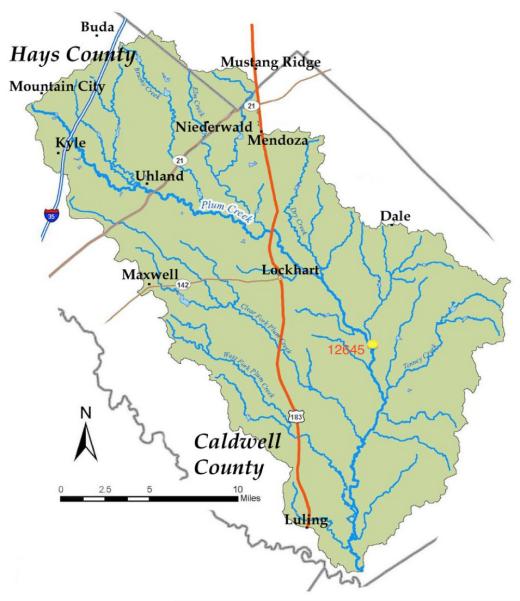
This study is conducted at a station (Station ID: 12645; Latitude 29°40′02″ and Longitude 97°39′14″) in Plum Creek (Fig. 2), which is monitored by the Texas Commission on Environmental Quality (TCEQ). The Plum Creek watershed is a part of the Guadalupe River basin and is located in east central Texas. It surrounds a drainage area of 1028 km2 in the counties of Hays, Caldwell, and Travis. According to the 2008 Texas water quality inventory and 303(d) list of impaired water bodies, Plum Creek is impaired for bacteria throughout the entire segment [http://www.gbra.org/CRP]. Plum Creek is a shallow, intermittent fifth-order stream. It is 83 km long and joins the San Marcos River that in turn connects with the Guadalupe River. The watershed has several rapidly growing towns such as Lockhart, Kyle, and Luling. The watershed has a diversified land use from urban to agriculture and oil field activities. The watershed encompasses 38% rangeland, 17% pasture, 11% cultivated cropland, 18% forest, 8% developed land, 6% near riparian forest, and 2% open water and barren land. The landscape is characterized as rolling hills of pasture and cropland surrounded by scrub oak forest. Plum Creek lies in a semi-humid subtropical climate zone and is heavily influenced by its proximity to the Gulf of Mexico (http://www.gbra.org/CRP).
Figure 2.
Map showing the Guadalupe River basin in east central Texas and the station ID 12645 in Plum Creek (Map modified from http://www.gbra.org/CRP).
Two US Geological Survey (USGS) gage stations are located on Plum Creek to monitor stream flows: one north of Lockhart (Station 08172400) and one near Luling (Station 08173000). Near Lockhart, periods of no flow have occurred almost every year on record. Southern reaches of Plum Creek, particularly south of Lockhart, are fed by a number of small springs and are usually perennial. Based on routine water quality sampling, the TCEQ initially listed portions of Plum Creek for bacteria impairment for contact recreation use in 2002. The possible sources of E. coli contamination in the creek are cows¸ livestock, wildlife, wastewater treatment plants, septic systems, and pet sources [Teague et al., 2009]. By 2004, bacterial contamination level in Plum Creek was elevated, and it was included in the list of impaired waters of Texas prohibiting wading and swimming. The E. coli criteria for designated use of a stream specified in water quality standards (e.g., recreational uses, irrigation, and navigation etc.) require a geometric mean (GM) concentration of E. coli less than 126 cfu/100 mL of water with no sample exceeding 235 cfu/100 mL of water. E. coli and water quality data at the monitoring sites were available from October 1996 to December 2008 [http://www.gbra.org/CRP]. Water quality data were collected monthly by the TCEQ. The available water quality data include thirteen factors, wherein physical factors include turbidity (NTU), temperature (°C), conductivity (µmhos/cm)), and dissolved oxygen (mg/L) (DO); chemical factors include pH, phosphate (mg/L), nitrate-N (mg/L), chloride (mg/L), sulfate (mg/L), total hardness (mg/L), and ammonia (mg/L); biological factors include suspended solids (mg/L) (SS) and chlorophyll (mg/m3).
4. Methodology
In order to identify the key water quality factors responsible for E. coli loads in streams, BNN models are run in conjunction with the exhaustive feature selection technique. We use the thirteen physical, chemical, and biological factors described above. The exhaustive feature selection technique provides the best set of water quality factors for estimating E. coli loads. A principal component analysis (PCA) is also conducted to get insight into the relative importance of the factors identified by the exhaustive feature selection. These selected factors are subsequently utilized in estimating E. coli loads by the BNN model in Plum Creek. The BNN model results are also compared with the LOADEST [Runkel et al., 2004] model. For a better decision making, uncertainty analysis is also conducted. In the subsequent sections, we provide a description of BNN and LOADSEST models, exhaustive feature selection, PCA, and uncertainty analysis.
4.1 Bayesian Neural Networks
The application of the Bayesian learning paradigm to neural networks results in a flexible and powerful nonlinear modeling framework that can be used for regression, density estimation, prediction and classification supporting adaptive decision-making, and accounting for uncertainties [Andrieu et al., 2001, Reckhow, 1999]. The regression of a target variable Y on an input set of covariates X given the data D = {(x1, y1),(x2, y2 ),….}:
| (1) |
where M is the model and ξi are independent and identically distributed errors (i.i.d.) ~ N(0,σ2).
The E. coli loads for each point (i) in time are estimated as follows:
| (2) |
The central process of the Bayesian framework is the calculation of a probability distribution on the unknown parameter (weight) vector w. Prior knowledge that we might have, say for small weights, is updated using the data. These posterior distributions are used in model predictions, with point forecasts given as expectations [Holmes and Mallick, 1998]:
| (3) |
where, E[Y| x, D] represents the posterior probability of the parameters of the model m(x, w) given the training data D.
BNN generates a probability distribution of the layer weights, which is dependent on the given input data:
| (4) |
where P(Y | X ) = ∫ P(Y | w, X)P(w)dw is the marginal distribution of Y, P(w) is the prior distribution of weights, and P(Y|w, χ) is the likelihood function [Gelman et al., 1995]. Artificial Neural Network (ANN) combined with Monte Carlo Markov Chain (MCMC) generates multiple samples from a continuous target density [Bates and Campbell, 2001]. A flat prior can be assumed here, as we do not have any concrete prior knowledge of weights [Sims and Ƶha, 1998].
Predictive distribution of Yn+1 is given by:
| (5) |
where n+1 denotes the next realization.
We considered Radial Basis Functions (RBF) architecture in BNN, which has an ability of closely approximating any nonlinear multidimensional mapping [Ciocoiu, 2002]. A brief summary of RBF is provided below for completeness.
Radial basis function (RBF)
RBF networks are one of the most commonly used types of feed forward networks. The feed forward neural network is most widely used to solve engineering problems. It is a simple nonlinear model that maps the input vector onto the output vector [Lanouette et al., 1999]. The architecture of a RBF network consists of three layers: an input layer, a hidden layer, and an output layer. The transformation from input space to hidden unit space is nonlinear, whereas transformation from hidden unit space to output space is linear [Ciocoiu, 2002]. During the training stage, a known set of input and output data pairs are delivered to the RBF network to select the centers and compute the output layer weights. The models have radial functions, where each basis is parameterized by a knot or position vector located in the d-dimensional covariate space x. The hidden layer provides a set of functions that constitute an arbitrary basis for the input patterns. The hidden units are known as radial centers and represented by the vectors (C1; C2; …, Ch). Conventionally, there are as many basis functions (h) as data points to be approximated with the position vectors set to the data values. The model output m(x) is given by a linear combination of the basis functions response and a low-order polynomial term:
| (6) |
where ∥·∥ denotes a distance metric, usually Euclidean or Mahalanobis, and qm(x) represents a polynomial of degree m. The coefficients w and a are calculated by least squares where the constraint is imposed to ensure the uniqueness of the solution [Holmes and Mallick, 1998]. Different radial functions (e.g., Gaussian, quadratic, thin plate spline, inverse quadratic functions) are used for different problems. We used the thin plate spline (TPS) for estimating E. coli loads. The TPS is given as:
| (7) |
| (8) |
RBF networks enlarge the dimensionality of the input data in order to increase the probability that originally nonlinearly separable classes become linearly separable (Cover’s theorem) [Ciocoiu, 2002]. For modeling a system with limited experimental data, RBF has an advantage over the other techniques. One of the problems that may occur during neural network training is over-fitting. A frequently used method for improving network generalization is to use an adequate-sized network, which is just large enough to provide an adequate fit [Cilek and Yilmazer, 2003]. Over fitting happens when the model has too many degrees of freedom, which is the result of including too many hidden neurons. The neurons in the hidden layer contain transfer functions whose outputs are inversely proportional to the distance from the center of the neuron. With small data sets used in this study, we ensured model accuracy by running multiple simulations by randomizing data sets. We split all the valid data into three randomly distributed groups. Three random sets are selected using randperm function in MATLAB. Initially, random split 1 is set aside for testing, while the models are parameterized on the basis of random splits 2 and 3. The fitted models are then used to test/predict E. coli loads by using input data from the random split 1. Next, random split 2 is set aside for testing, while random splits 1 and 3 are used for training. This pattern is also repeated for the random split 3. We use the same random splits (1, 2, and 3) for estimating E. coli loads by using the LOADEST Model.
4.2 Load Estimator (LOADEST)
Load estimator (LOADEST) is a regression-based model for estimating constituent loads in streams and rivers [Runkel et al., 2004]. Given a time series of streamflow and constituent concentration (E. coli), LOADEST facilitates users in developing a regression model for the estimation of constituent loads [Cohn, 2005]. Explanatory variables within the regression model include multiple functions of flow, time, and additional data variables. The developed regression model is then used to estimate loads over a user-specified time interval. Mean loads, standard errors, and 95% confidence intervals are also estimated on a monthly and/or seasonal basis. There are three statistical methods used for calibration and validation (estimation) of LOADEST including Adjusted Maximum Likelihood Estimation (AMLE), Maximum Likelihood Estimation (MLE), and Least Absolute Deviation (LAD). AMLE and MLE are appropriate when the calibration model errors (residuals) are normally distributed, whereas LAD is appropriate when model errors (residuals) are not normally distributed. In our case, calibration model errors are normally distributed, so we used AMLE for estimating E. coli loads. The detailed mathematical formulation of LOADEST is provided elsewhere [Cohn, 2005]. In general, total mass loading over an arbitrary time period, τ, is given by:
| (9) |
| (10) |
where C is concentration [M/L3], L is total load [M], 𝒬 is instantaneous stream flow [L3/T], t is time [T], and NP is number of discrete points in time. The hats on 𝒬, and Lτ and L denotes the instantaneous values of the respective variables. E. coli loads estimated by the LOADEST model are compared with the E. coli loads estimated by the BNN model using the key water quality factors. The key water quality factors are identified using the exhaustive feature selection technique.
4.3 Exhaustive Feature Selection
The BNN models are run multiple times with all possible combinations of the thirteen water quality factors (13C1+13C2+….+13C13) for estimating E. coli loads. The Exhaustive Feature Selection is a technique of selecting a subset of relevant features for building robust models. The brute-force feature selection algorithm is applied to exhaustively evaluate all possible combinations of the input features, and then the best subset is chosen. The exhaustive search’s computational cost is prohibitively high, with a considerable danger of overfitting [Moore and Lee, 94; Skalak, 94]. Hence, for avoiding the over-fitting, K-fold (three-fold) cross validation is used in selecting the best subset. The aim of the feature selection is to choose a subset of the set of input features (physical, chemical, and biological factors) so that the subset can predict the output Y (E. coli loads) with accuracy akin to the performance of the whole input set χ, and with a reduction of the computational cost. For conducting the exhaustive feature selection, the following steps are outlined:
Shuffle the dataset and split into a training set of 2/3rd of the data and a test set of the remaining 1/3rd of the data.
Choose all possible combinations of various input variables.
Select each subset, and run the BNN model with leave-one-out cross-validation.
Store the Nash-Sutcliffe Efficiency (NSE) Coefficients (see section 4.4) of each run.
Select the feature set which has minimum root mean square error of NSE three-fold validation.
4.4 Model Performance
We computed the Nash–Sutcliffe efficiency (NSE) and Normalized Mean Squared Error (NMSE) as measures of the model performance.
The Nash–Sutcliffe efficiency (NSE) coefficient is given as:
| (11) |
The Normalized Mean Squared Error (NMSE) is given as:
| (12) |
where is observed E. coli loads, and is simulated E. coli at time t. is mean observed E. coli loads. denotes the variance of all the observed E. coli loads. NSE can range from −∞ to 1. An efficiency of 1 (NSE = 1) corresponds to a perfect match of simulated values to the observed data. An efficiency of 0 (NSE = 0) demonstrates that the model predictions are as accurate as the mean of the observed data. In essence, closer the efficiency of the model is to 1, the more accurate is the model. The NMSE of 0 indicates that the model predictions are perfect. The lower the NMSE, the better is the model performance.
The Exhaustive Feature Selection technique in conjunction with the BNN model rendered the best set of factors. In order to assess the relative importance of these factors, principal component analysis (PCA) is done.
4.5 Principal Component Analysis
Principal component analysis (PCA) is a multivariate statistical technique. The transformed features have a descriptive power that is more ordered than the original features. PCA has been applied in describing various aspects of streamflow regimes [Olden and Poff, 2003], understanding the spatial and temporal changes in water quality [Bengraïne, and Marhaba, 2003], determination of dominant biogeochemical processes in a contaminated aquifer [Cazull and McGuire, 2008]. In this study, PCA is used to identify major factors among water quality data that can explain most of the variation of E. coli loads.
PCA is an orthogonal linear transformation of the data (e.g., water quality data) to a new coordinate system such that the greatest variance by any projection of the data comes to lie on the first quadrant. The principal axis method is used to extract the components, followed by a varimax (orthogonal) rotation with Kaiser Normalization. A detailed description, of how the principal components are calculated, is provided elsewhere [Jolliffe, 2002].
4.6 Uncertainty Analysis
Monte Carlo based statistical techniques – resampling with replacement (“bootstrapping”) [Robert and Casella, 1999] is implemented to estimate the statistical uncertainty in predictions by the BNN and LOADEST models. To explore the uncertainty in the BNN predictions, 10000 realizations of E. coli loads are investigated. Bayesian networks are probabilistic models that combine prior distributions of uncertainty with data to yield an updated (posterior) set of distributions [Helton and Oberkampf, 2004]. Therefore, inputs are integrated over the weight space of the posterior probability distribution for finding the outputs (i.e., E. coli loads) of the networks.
The probability distribution of each output as a random variable is plotted utilizing the kernel density (Parzen window) estimation, which is a non-parametric method [Silverman, 1986]. If x1, x2… xN are samples drawn from the density function of a random variable, then the kernel density approximation of its probability density function is given as:
| (13) |
where K is some kernel and h is a smoothing parameter called the bandwidth. Here, a Gaussian kernel is chosen with mean zero and unit variance:
| (14) |
5. Results and Discussion
The results presented here provide insight into the different physical, chemical, and biological factors that are critical in the estimation of E. coli loads in surface streams. The exhaustive feature selection in conjunction with the BNN model (Fig. 1) identified the best combination of input variables for estimating E. coli loads. Out of the thirteen water quality factors, exhaustive feature selection identified six key variables: SS, phosphate, temperature, DO, ammonia, and chlorophyll. In the following section, we will focus on these key variables and their relative importance. Subsequently, we utilize these six key variables using the BNN model for estimating E. coli loads in Plum Creek.
5.1 Identification of the key factors responsible for the E. coli Loads in Plum Creek
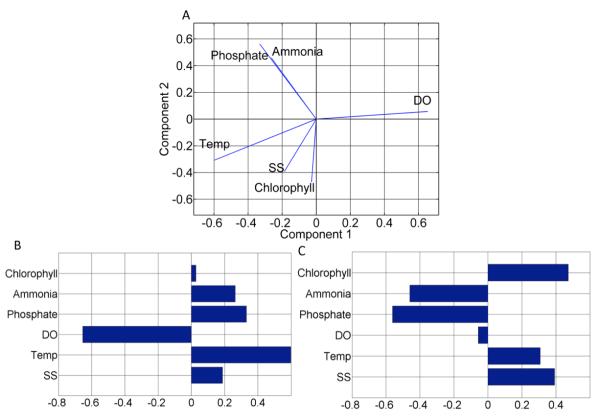
The exhaustive feature selection identified six factors in estimating E. coli loads in Plum Creek namely SS, phosphate, temperature, DO, ammonia, and chlorophyll. To investigate the relative importance of the key factors for E. coli loads in streams, a principal component analysis (PCA) was performed as shown in Figs. 3 and 4. The PCA explored the relationship among water quality factors such as SS, phosphate, temperature, DO, ammonia, and chlorophyll. The first two components explain 60.0% of the variance; component 1 and component 2 account for 35.6% and 24.4% of the variance, respectively (Fig. 4). The first principal component (PC) captures the variance of DO and temperature. The second PC captures the variance of SS, phosphate, ammonia, and chlorophyll. The PCA biplot (Fig. 4) illustrates a visual interpretation of the factor loadings that result from a bi-cluster system of variables projected onto the first and second PC axes. The biplot tells about the relative positions of the factors, and the angles between the factors give approximate estimates of the correlation among factors; small angles between projected axes imply a high correlation. The direction of axes gives the sign of correlation among factors displayed on the biplot [Jolliffe, 2002].
Figure 3.
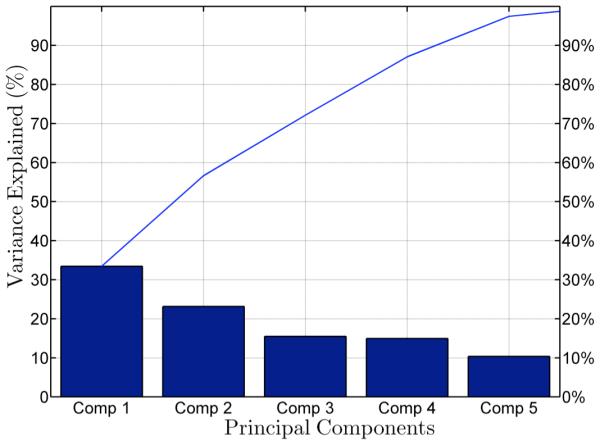
Pareto diagram of principal components shows the percentage explained by each component. The first three components explain 70% of variation in the data set.
Figure 4.
Biplot (A) of PCA is plotted by projecting the first principal component against the second principal component. Factor loadings on the first (B) and second (C) principal components reflect the relative importance of each factor.
Turning now to the interpretation of the PCs in the present work, the six factors can be divided into three groups. The group 1 includes temperature and DO (physical factors); the group 2 includes phosphate and ammonia (chemical factors); the group 3 includes the SS and the chlorophyll (biological factors). The central idea of this classification is based on the fact that groups of variables often move together, and more than one factor measures the same driving force. The first PC clearly measured physical factors, as DO and temperature have the maximum loadings (Fig. (4B), moreover, they also have a high negative correlation with each other (almost 180° separated in biplot (Fig. 4A)). Therefore, DO and temperature were classified as the group 1. The second PC accounted for the chemical and biological factors (Fig. 4C). Since, phosphate and ammonia are the dominant factors on the second PC, and phosphate and ammonia also have a high positive correlation with each other (almost overlapping in biplot (Fig. 4A)). For this reason, they were classified as group 2. Similarly, the third PC also accounted for biological factors, as SS and chlorophyll have a medium positive correlation with each other (a small angle between them in biplot Fig. 4A). Hence, they were grouped together.
The biological tolerance of E. coli to different physical, chemical, and biological factors has been well studied, albeit mostly in the laboratory. It has been observed that E. coli are sensitive to changes in temperature [Maeda et al., 1976; Berg, 2004]. The rate of die-off depends on temperature [Flint et al., 1987]. Moreover, E. coli are anaerobic bacteria, and thus E. coli also responds to oxygen gradient. The majority of E. coli cannot live in oxygen rich environment [Berg, 2004]. This clearly explains the selection of DO and temperature as important physical factors in estimating E. coli loads in our study and their negative correlation. Temperature affects positively, whereas DO affects negatively in estimating E. coli loads by the BNN model. In the biplot, approximately 180° separation of temperature and DO corroborates this behavior of E. coli (Fig. 4A).
Phosphate and ammonia are also found to be important factors in the estimation of E. coli loads by the BNN model. This is because phosphate and ammonia act as nutrients or substrates, and the presence of nutrients increases E. coli concentrations in streams [van der Steen et al., 2000]. These nutrients have significant positive correlation and therefore signify the importance of chemical factors on E. coli loads.
In our study, SS and chlorophyll are also important factors in the estimation of E. coli loads by the BNN model. In literature, there is evidence to suggest that high concentrations of chlorophyll and suspended sediments are associated with high E. coli concentrations [Nevers and Whitman, 2005]. However, Money et al. (2009) examined the relationship between turbidity and E. coli and found a significant correlation between both the parameters. Turbidity indicates high volumes of suspended sediments. SS and chlorophyll correspond to the biological factors, as they are sources of organic carbon [de Jonge et al., 1980]. These biological factors were measured by the second PC.
It should be noted that the sign of any PC is completely arbitrary. If every coefficient in a PC has its sign reversed, the variance is unchanged, and so is the orthogonality [Jolliffe, 2002]. Therefore, the biplot and loadings only show the relative importance of the factors, they do not demonstrate if a factor is positively or negatively affecting the E. coli loads. However, the biplot exhibits how each factor can affect the E. coli loads. For example, it is evident from Fig. 4A that all the factors on the left side of the plot (phosphate, ammonia, temperature, SS, and chlorophyll) are positively associated with E. coli loads, whereas the only factor on the right hand side of the plot is DO, and it is negatively associated with E. coli loads. This graphic examination further substantiates our findings.
5.2 Estimation of E. coli Loads
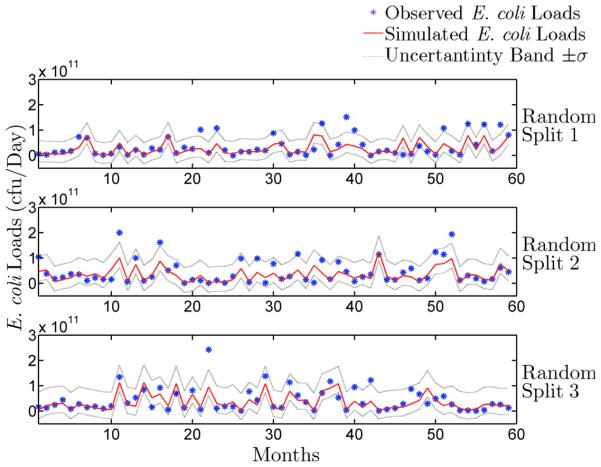
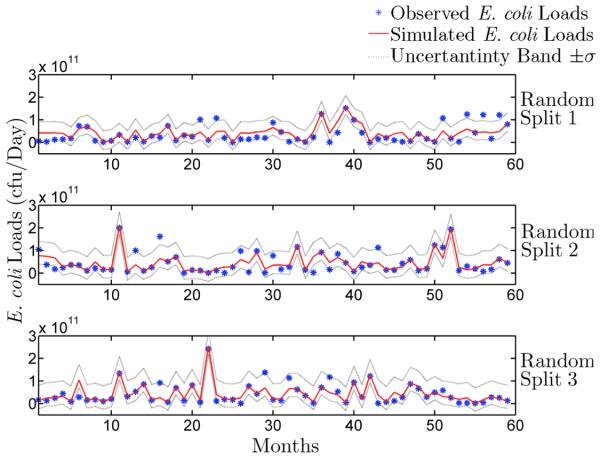
In this section, we discuss the discrepancy between simulated and observed E. coli loads using the BNN model (using the six key variables) and compare its performance to the LOADEST model. Figs. 5 and 6 show measured and simulated E. coli loads in Plum creek using the LOADEST and BNN models respectively for three random splits. Table 1 shows the measures of the models’ performance. A three-fold cross validation results show that both modeling approaches (BNN and LOADEST) reproduce observed E. coli loads reasonably well, with all NSE values greater than or equal to 0.39 and all NMSE values smaller than or equal to 0.59 (Table 1). However, the BNN is able to estimate E. coli loads better in all the three random splits (Table 1). The uncertainty bands (Figs. 5 and 6) show that the BNN is also able to capture higher E. coli loads more accurately than the LOADEST model. This is expected because the BNN model provides more flexible choices for the functional dependence in estimating E. coli loads based on physical, chemical, and biological factors (e.g., SS, phosphate, temperature, DO, ammonia, and chlorophyll), whereas the LOADEST model uses only the E. coli and flow data.
Figure 5.
Measured and simulated loads of E. coli by the LOADEST model in Plum Creek are shown above. E. coli loads are presented here for the three random splits. These random splits were used for three-fold cross validation of the BNN model.
Figure 6.
Measured and simulated loads of E. coli by the BNN model in Plum Creek are shown above. The simulations were tested by three-fold cross validation.
Table 1.
Nash Sutcliffe Efficiency (NSE) and Normalized Mean Squared Error (NMSE) of estimated E. coli loads by BNN and LOADEST models and observed E. coli loads in Plum Creek
| Random Splits | Models | NSE | NMSE |
|---|---|---|---|
| Random Split 1 | BNN | 0.48 | 0.51 |
| LOADEST | 0.39 | 0.59 | |
| Random Split 2 | BNN | 0.69 | 0.30 |
| LOADEST | 0.55 | 0.44 | |
| Random Split 3 | BNN | 0.75 | 0.23 |
| LOADEST | 0.52 | 0.46 |
Fig. 7 shows the cumulative distribution functions (CDFs) of the observed, BNN simulated (all the three random splits), and LOADEST simulated (all the three random splits) E. coli loads in Plum Creek. The region A signifies the smaller E. coli loads (smaller than 0.5×1011 cfu/Day), which are better estimated by the LOADEST model. The BNN model is underestimating the E. coli loads in this region. The region C encompasses the higher E. coli loads (greater than 1.5×1011 cfu/Day), which are better estimated by the BNN model. For best management practices, it is essential to be able to estimate higher E. coli loads, and the BNN model is able to estimate values with greater accuracy in this range. The region B (0.5×1011 cfu/Day to 1.5×1011 cfu/Day) constitutes the region with medium loads between regions A and C, and where both the LOADEST and BNN models are overestimating the E. coli loads. However, the BNN model is closer to the observed values than the LOADEST model in this region.
Figure 7.
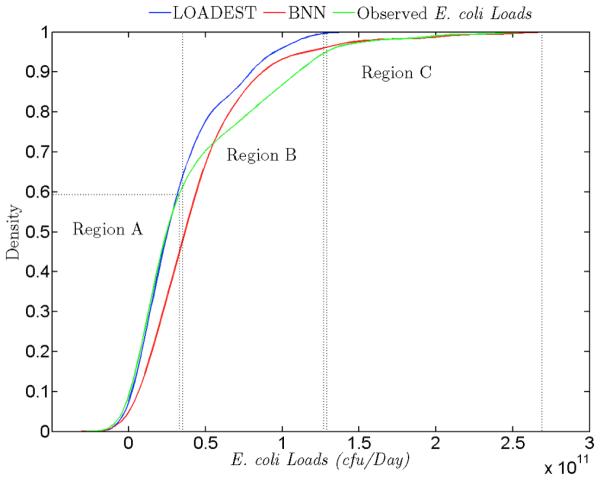
The cumulative density functions of the observed E. coli loads, estimated E. coli loads by the BNN, and estimated E. coli loads by the LOADEST are presented above. There are three regions in the figure. The region A signifies that the LOADEST model is able to estimate E. coli loads better. In the region B and C, the BNN model predicts better than the LOADEST model.
5.3 Sensitivity Analysis
In this study, the objective of the sensitivity analysis is to demonstrate the relative response (E. coli loads) of the BNN model for each physical (temperature and DO), chemical (phosphate and ammonia), and biological (SS and chlorophyll) factor. One-factor-at-a-time (OFAT) approach has been deemed appropriate for evaluating the sensitivity of different explanatory variables in the neural network models [for example Xie et al. (2007) and Delen et al. (2006)]. The OFAT approach involves perturbing each factor individually within a reasonable interval (±20%) and keeping the rest of the factors constant at their baseline values. The effect of perturbation of a single factor is quantified by recording the corresponding variation in the BNN output using NMSE and percentage change in the E. coli loads. NMSE values are calculated by using residuals between E. coli loads estimated by original variables and E. coli loads estimated by perturbing each factor. A higher NMSE value implies a higher sensitivity to the factor under consideration. Correspondingly, a higher percentage change in the E. coli loads means a higher sensitivity to that factor. Table 2 lists the relative sensitivity of each factor for all the three random splits. This ranking suggests that each factor shows comparable sensitivity; however, DO and SS show higher sensitivity as compared to other factors (temperature, phosphate, ammonia, and chlorophyll) in estimating E. coli loads.
Table 2.
Normalized Mean Squared Error (NMSE) and percentage change of estimated E. coli loads due to perturbation of each factor (±20%) one-at-a-time and estimated E. coli loads while keeping other factors constant at their baseline values by the BNN model
| Factors | Variation | Random Split-1 |
Random Split-2 |
Random Split-3 |
% change in E. coli
loads (range) |
|---|---|---|---|---|---|
| Temperature | Lower Upper |
0.22 0.27 |
0.31 0.34 |
0.32 0.27 |
−5 to 5 |
| DO | Lower Upper |
0.43 0.35 |
0.45 0.37 |
0.56 0.32 |
−25 to 25 |
| Phosphate | Lower Upper |
0.18 0.21 |
0.10 0.23 |
0.21 0.22 |
−2 to 12 |
| Ammonia | Lower Upper |
0.15 0.14 |
0.21 0.11 |
0.22 0.12 |
−2 to 12 |
| SS | Lower Upper |
0.39 0.51 |
0.37 0.43 |
0.43 0.47 |
−20 to 20 |
| Chlorophyll | Lower Upper |
0.23 0.22 |
0.14 0.24 |
0.17 0.18 |
−2 to 5 |
5.4 Uncertainty Analysis
Uncertainty analysis is conducted to further compare the performance of BNN and LOADEST models in estimating E. coli loads in Plum Creek. The uncertainty bands (±σ with 95% confidence) computed using bootstrap samples show that there is more uncertainty for larger loads than smaller loads (Figs. 5 and 6). There is evidence that uncertainties of discrete E. coli samples are greater than 30% while the uncertainty in storm water flow measurements are greater than 97% [McCarthy et al., 2008]. Therefore, E. coli loads will have more uncertainty due to storm events. As high E. coli loads are often associated with storm events, the upper limit of the uncertainty band is also wider for higher loads. These uncertainties in the inputs propagate into larger uncertainties in the output.
Fig. 8 shows six E. coli loads estimated by the BNN model (low and high E. coli loads for each random split), and the probability distribution functions (PDFs) of 10,000 realizations for each E. coli loads were plotted. As stated previously, Bayesian Neural Networks use a range of weight sets instead of a single set. Each weight gives a realization of E. coli loads. The final predicted E. coli loads were generated from the average of 10,000 such realizations. The center of mass of a PDF shows the mean of the prediction and spread around the mean shows the uncertainty. It is clear from the Fig. 8 that E. coli loads, estimated by the BNN model, were closer to the centers of the PDFs with high density values (4 of them are >0.4). It should be noted that the BNN model estimates lower E. coli loads with a small bias (observed E. coli loads falling close to the center of mass of the pdfs) and higher E. coli loads with a large bias (observed E. coli loads falling on the tails of the pdfs); however, the performance of the BNN model is better than the LOADEST model for estimating higher E. coli loads. The large variance in the PDFs is due to various uncertainties, which stem mainly from (1) the uncertainties in input data (e.g., flow rate and water quality data); (2) uncertainties in data used for calibration, (e.g., E. coli loads). Input data (flow rate and water quality data) and E. coli loads have large inherent uncertainties, and these uncertainties cannot be removed from the model predictions in the existing data. However, the advent of newer technologies and careful data collection may help in minimizing these uncertainties in the future. The other source of uncertainties is from model parameters (weights and biases). These uncertainties are related to the fact that a small bias in the estimation, using a neural network with a training set of fixed size, can only be achieved with a large variance [Geman et al., 1992; Haykin, 1996]. This dilemma can be avoided if the training set is made very large, but the total amount of data is limited in our case. However, a possibility of making training sets larger can be plausible in the future.
Figure 8.
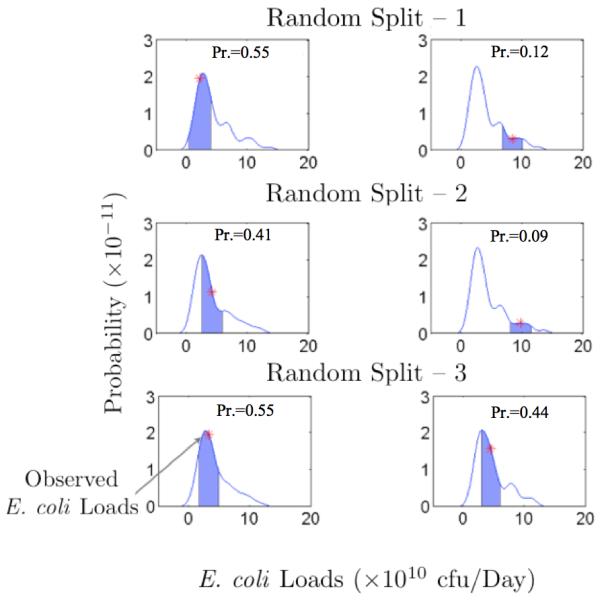
Probability distributions of low (left panels) and high (right panels) E. coli loads of each random split by the BNN model. PDFs show that higher loads are associated with multi-modality. The blue shaded area (Pr.) represents the probability of observed E. coli loads with in ± 10% uncertainty bands.
6. Conclusions
This study provides a Bayesian Neural Network (BNN) model for E. coli prediction in streams. A significant contribution of this paper is in identifying six key variables from a selection of physical, chemical, and biological factors that influence E. coli loads in surface streams. An exhaustive feature selection technique used in conjunction with BNN and the principal component analysis (PCA) indicated the importance and correlation among these six variables. Physical factors included temperature and DO; chemical factors included phosphate and ammonia; biological factors included SS and chlorophyll. The sensitivity analysis was conducted on these factors which demonstrated all the six factors to be sensitive and DO and SS to be the most sensitive with respect to estimating E. coli loads.
The BNN model was then run using these six factors and a comparison with a traditional model (LOADEST) developed by the USGS was also conducted. The inherent differences between the models are the calibration procedures using statistical (LOADEST) versus probabilistic (BNN) framework. The models were compared for estimation of E. coli loads based on available water quality data using Nash–Sutcliffe efficiency (NSE) and normalized mean squared error (NMSE) in three-fold cross validation. All the efficiency measures suggest that estimation of E. coli loads by the BNN model was better than the LOADEST model on all the occasions during three-fold cross validation. The results also highlight that the LOADEST model estimates E. coli loads better in the smaller ranges, whereas the BNN model estimates E. coli loads better in the higher ranges, as well. Hence, the BNN model can be useful to decision maker and environmental managers to design targeted monitoring programs and establishing regulatory control such as TMDL programs. An uncertainty analysis is also used to compare the predictive powers of the two models. These results suggest that more uncertainty is associated with larger E. coli loads, and signify that the major source of uncertainty comes from storm events associated with E. coli loads.
7. References
- Andrieu C, de Freitas N, Doucet A. Robust full Bayesian learning for radial basis networks. Neural Computation. 2001;13:2359–2407. doi: 10.1162/089976601750541831. [DOI] [PubMed] [Google Scholar]
- Arnold JG, Fohrer N. SWAT2000: current capabilities and research opportunities in applied watershed modelling. Hydrol. Processes. 2005;19(3):563–572. [Google Scholar]
- Auer MT, Niehaus SL. Modeling fecal coliform bacteria: 1.Field and laboratory determination of loss kinetics. Water Res. 1993;27:693–701. [Google Scholar]
- Babbar-Sebens M, Karthikeyan R. Consideration of Sample Size For Estimating Contaminant Load Reductions Using Load Duration Curves. J. of Hydrol. 2009;372(1-4):118–123. [Google Scholar]
- Baez-Cazull SE, McGuire JT, Cozzarelli IM, Voytek MA. Determination of dominant biogeochemical processes in a contaminated aquifer-wetland system using multivariate statistical analyses. Journal of Environmental Quality. 2008;37(1):30–46. doi: 10.2134/jeq2007.0169. [DOI] [PubMed] [Google Scholar]
- Bates BC, Campbell EP. A Markov Chain Monte Carlo scheme for parameter estimation and inference in conceptual rainfall–runoff modeling. Water Resour. Res. 2001;37(4):937–947. [Google Scholar]
- Bengraïne K, Marhaba TF. Using principal component analysis to monitor spatial and temporal changes in water quality. Journal of Hazardous Materials B. 2003;100:179–195. doi: 10.1016/s0304-3894(03)00104-3. [DOI] [PubMed] [Google Scholar]
- Benham BL, et al. Modeling bacteria fate and transport in watersheds to support TMDLs. Transactions of the ASABE. 2006;49:987–1002. [Google Scholar]
- Berg HC. E. coli in Motion. Springer Verlag. 2004:10–30. [Google Scholar]
- Cilek EC, Yilmazer BZ. Effects of hydrodynamic parameters on entrainment and flotation performance. Miner. Eng. 2003;16:745–756. [Google Scholar]
- Ciocoiu IB. Hybrid feed forward neural networks for solving classification problems. Neural Process Lett. 2002;16:81–91. [Google Scholar]
- Cohn TA. Estimating contaminant loads in rivers: An application of adjusted maximum likelihood to type 1 censored data. Water Resour. Res. 2005;41:W07003. doi:10.1029/2004WR003833. [Google Scholar]
- de Jonge VN. Fluctuations in the organic carbon to chlorophyll a ratios for estuarine benthic diatom populations. Mar. Ecol. Prog. Ser. 1980;2:345–353. [Google Scholar]
- Delen D, Sharda R, Bessonov M. Identifying significant predictors of injury severity in traffic accidents using a series of artificial neural networks. Accid. Anal. Prev. 2006;38(3):434–444. doi: 10.1016/j.aap.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Dorner SM, Anderson WB, Slawson RM, Kouwen N, Huck PM. Hydrologic modeling of pathogen fate and transport. Environ. Sci. Technol. 2006;40:4746–4753. doi: 10.1021/es060426z. [DOI] [PubMed] [Google Scholar]
- Fincher LM, Parker CD, Chauret CP. Occurrence and Antibiotic Resistance of Escherichia coli O157:H7 in a Watershed in North-Central Indiana. J. Environ. Qual. 2009;38:997–1004. doi: 10.2134/jeq2008.0077. doi:10.2134/jeq2008.0077. [DOI] [PubMed] [Google Scholar]
- Flint KP. The Long-Term Survival of Escherichia-Coli in River Water. J. Appl. Bacteriol. 1987;63:261–270. doi: 10.1111/j.1365-2672.1987.tb04945.x. [DOI] [PubMed] [Google Scholar]
- Gelman A, Carlin BJ, Stern HS, Rubin DB. CRC Press; London, U. K.: 1995. Bayesian Data Analysis; pp. 3–14. [Google Scholar]
- Geman S, Bienenstock E, Doursat R. Neural Networks and the Bias Variance Dilemma. Neural Computation. 1992;4:1–58. [Google Scholar]
- Haykin S. Neural networks expand SP’s horizons. IEEE Signal Process Mag. 1996;13:24–49. [Google Scholar]
- Helton JC, Oberkampf WL. Alternative representations of epistemic uncertainty. Reliab.Eng. Syst. Saf. 2004;85:1–10. [Google Scholar]
- Hipsey MR, Antenucci JP, Brookes JD. A generic, process-based model of microbial pollution in aquatic systems. Water Resour. Res. 2008;44:W07408. doi:10.1029/2007WR006395. [Google Scholar]
- Holmes CC, Mallick BK. Bayesian radial basis functions of variable dimension. Neural Computation. 1998;10:1217–1233. [Google Scholar]
- Jin G, Englande AJ, Liu A. A preliminary study on coastal water quality monitoring and modeling. J. Environ. Sci. Health. 2003;A38:493–509. doi: 10.1081/ese-120016909. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT. Springer-Verlag; New York, N.Y.: 2002. Principal Component Analysis; pp. 63–130. [Google Scholar]
- Lanouette R, Thibault J, Valade JL. Process modeling with neural networks using small experimental data sets. Comput. Chem. Eng. 1999;23:1167–1176. [Google Scholar]
- Lessard EJ, Sieburth JM. Survival of Natural Sewage Populations of Enteric Bacteria in Diffusion and Batch Chambers in the Marine-Environment. Appl. Environ. Microbiol. 1983;45:950–959. doi: 10.1128/aem.45.3.950-959.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Imae Y, Shioi JI, Oosawa F. Effect of temperature on motility and chemotaxis of Escherichia coli. Journal of bacteriology. 1976;127:1039–1046. doi: 10.1128/jb.127.3.1039-1046.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DT, Deletic A, Mitchell VG, Diaper C. Uncertainties in storm water E. coli levels. Water Res. 2008;42(6-7):1812–1824. doi: 10.1016/j.watres.2007.11.009. [DOI] [PubMed] [Google Scholar]
- McCorquodale JA, Georgiou I, Carnelos S, Englande AJ. Modeling coliforms in storm water plumes. J. Environ. Eng. Sci. 2004;3:419–431. [Google Scholar]
- McKergow LA, Davies-Colley RJ. Stormflow dynamics and loads of Escherichia coli in a large mixed land use catchment. Hydrol. Processes. 2009;24(3):276–289. doi: 10.1002/hyp.7480. [Google Scholar]
- Mead PS, Griffin PM. Escherichia coli O157 : H7. Lancet. 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- Medema GJ, Schijven JF. Modelling the sewage discharge and dispersion of Cryptosporidium and Giardia in surface water. Water Res. 2001;35:4307–4316. doi: 10.1016/s0043-1354(01)00161-0. [DOI] [PubMed] [Google Scholar]
- Money ES, Carter GP, Serre ML. Modern Space/Time Geostatistics Using River Distances: Data Integration of Turbidity and E. coli Measurements to Assess Fecal Contamination Along the Raritan River in New Jersey. Environ. Sci. Technol. 2009;43:3736–3742. doi: 10.1021/es803236j. doi: 10.1021/es803236j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AW, Lee MS. In Proc. ML-94. Morgan Kaufmann; 1994. Efficient algorithms for minimizing cross validation error. [Google Scholar]
- Nevers BM, Whitman RL. Nowcast modeling of Escherichia coli concentrations at multiple urban beaches of southern Lake Michigan. Water Res. 2005;39(20):5250–5260. doi: 10.1016/j.watres.2005.10.012. doi.org/10.1016/j.watres. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Nakajima H, Aono R. Effects of oxygen and nitrate on growth of Escherichia coli and Pseudomonas aeruginosa in the presence of organic solvents. Extremophiles. 1997;1:193–198. doi: 10.1007/s007920050033. [DOI] [PubMed] [Google Scholar]
- Olden JD, Poff NL. Redundancy and the choice of hydrologic indices for characterizing streamflow regimes. River Res. Appl. 2003;19:101–121. [Google Scholar]
- Pachepsky Ya. A., Sadeghi AM, Bradford SA, Shelton DR, Guber AK, Dao TH. Transport and Fate of Manure-Borne Pathogens: Modeling Perspective. Agric. Water Managmt. 2006;86:81–92. [Google Scholar]
- Reckhow KH. Water quality prediction and probability network models. Can. J. Fish. Aquat. Sci. 1999;56:1150–1158. [Google Scholar]
- Robakis N, Cenatiempo Y, Meza-Basso L, Brot N, Weissbach H. A coupled DNA-directed in vitro system to study gene expression based on di- and tripeptide formation. Methods Enzymol. 1983;101:690–706. doi: 10.1016/0076-6879(83)01048-4. [DOI] [PubMed] [Google Scholar]
- Robert CP, Casella G. Monte Carlo Statistical Methods. Springer; New York: 1999. pp. 32–35. [Google Scholar]
- Runkel R, Crawford CG, Cohn TA. U.S. Geol. Surv. Tech. Methods. Reston, Va: 2004. Load Estimator (LOADEST): A Fortran Program for Estimating Constituent Loads in Streams and Rivers. Book 4, chap. A5. [Google Scholar]
- Sims CA, Zha T. Bayesian methods for dynamic multivariate models. International Economic Review. 1998;39:949–968. [Google Scholar]
- Silverman BW. Density Estimation for Statistics and Data Analysis. Chapman and Hall; London, U.K.: 1986. [Google Scholar]
- Sjogren RE, Gibson MJ. Bacterial Survival in a Dilute Environment. Appl. Environ. Microbial. 1981;41:1331–1336. doi: 10.1128/aem.41.6.1331-1336.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalak DB. Prototype and feature selection by sampling and random mutation hill climbing algorithms; Proceedings of the 11th International Conference on Machine Learning, ML-94; New Brunswick, NJ. Morgan Kaufmann; 1994. [Google Scholar]
- Steets BM, Holden PA. A mechanistic model of runoff associated fecal coliform fate and transport through a coastal lagoon. Water Res. 2003;37:589–608. doi: 10.1016/S0043-1354(02)00312-3. [DOI] [PubMed] [Google Scholar]
- Teague A, Karthikeyan R, Babbar-Sebens M, Srinivasan R, Persyn RA. Spatially Explicit Load Enrichment Calculation Tool to Identify Potential E. coli Sources in Watersheds. Transactions of the ASABE. 2009;52(4):1109–1120. [Google Scholar]
- Tian YQ, Gonga P, Radkeb JD, Scarborough J. Spatial and temporal modeling of microbial contaminants on grazing farmland. J. Environ. Qual. 2002;31:860–869. doi: 10.2134/jeq2002.8600. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency 2006 Available at http://www.epa.gov/volunteer/stream/vms50.html [accessed 26 June 2009] and http://www.gbra.org/CRP/Sites [accessed 12May 2009] [PubMed]
- Whitman RL, Nevers MB, Korinek GC, Byappanahalli MN. Solar and temporal effects on Escherichia coli concentration at a lake Michigan swimming beach. Appl. Environ. Microbiol. 2004;70:4276–4285. doi: 10.1128/AEM.70.7.4276-4285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Steen P, Brenner A, Shabtai Y, Oron G. Improved fecal coliform decay in integrated duckweed and algal ponds. Water science and technology. 2000:363–370. [Google Scholar]
- Vidon P, Tedesco LP, Wilson J, Campbell MA, Casey LR, Gray M. Direct and Indirect Hydrological Controls on Concentration and Loading in Midwestern Streams. J Environ Qual. 2008;37(5):1761–8. doi: 10.2134/jeq2007.0311. doi: 10.2134/jeq2007.0311. [DOI] [PubMed] [Google Scholar]
- Walker FR, Stedinger JR. Fate and transport model of Cryptosporidium. J. Environ. Eng. 1999;125:325–333. [Google Scholar]
- Wilkinson J, Jenkins A, Wyer M, Kay D. Modelling faecal coliform dynamics in streams and rivers. Water Res. 1995;29:847–855. [Google Scholar]
- Xie Y, Lord D, Zhang Y. Predicting motor vehicle collisions using Bayesian neural network models: An empirical analysis. Accident Analysis and Prevention. 2006;2007;39:922–933. doi: 10.1016/j.aap.2006.12.014. [DOI] [PubMed] [Google Scholar]