Abstract
The cells present in amniotic fluid (AF) are currently used for prenatal diagnosis of fetal anomalies but are also a potential source of cells for cells therapy. To better characterize putative progenitor cell populations present in AF, we used culture conditions that support self-renewal to determine if these promoted generation of stable cell lines from AF-derived cells.
Cells isolated from E11.5 mouse were cultured on irradiated STO fibroblast feeder layers in human embryonic germ cell-derivation conditions. The cultures grew multicellular epithelial colonies that could be re-propagated from single cells. RT-QPCR analysis of established cell lines showed they belonged to the extraembryonic endoderm expressing high levels of Gata6, Gata4, Sox17, Foxa2 and Sox7 mRNA. Hierarchical clustering based on whole transcriptome expression profile of the AF-derived cell lines (AFCL) shows significant correlation between transcription profiles of AFCL and blastocyst-derived XEN. In vitro differentiation of AFCL results in generation of cells expressing Albumin and Alpha-fetoprotein (AFP), while intramuscular injection of AFCL into immunodeficient mice produced AFP+ tumors with primitive endodermal appearance.
Hence, E11.5 mouse AF contains cells that efficiently produce XEN lines. These AF derived XEN lines do not spontaneously differentiate into embryonic-type cells but are phenotypically stable and have the capacity for extensive expansion. The lack of requirement for reprogramming factors to turn AF-derived progenitor cells into stable cell lines capable of massive expansion together with the known ability of ExEn to contribute to embryonic tissues suggests that this cell type may be a candidate for banking for cell therapies.
Keywords: Primitive Endoderm, Amniotic Fluid, Self Renewal, Fetal
Introduction
In amniotes, the fetus develops inside an amniotic sac that is filled with amniotic fluid (AF) that both contributes to a stable environment and allows movement of the developing fetus. The fluid also contains several cell types of fetal and perhaps amniotic membrane origin. These cells were previously classified by differences in morphology [Gosden, 1983], surface phenotype, and differentiation potential [De Coppi et al., 2007; Klein and Fauza, 2011] and are also considered to be of potential clinical significance for cell therapy of several types of congenital tissue defects [Kunisaki et al., 2007; Steigman et al., 2009; Gray et al., 2012]
To characterize the progenitor potential of cell populations from AF it is necessary to achieve ex vivo expansion of relatively homogenous cell populations. Previously, this was accomplished using several methodologies including protocols for culture of Mesenchymal Stem Cells (MSC) [Kaviani et al., 2003; Klein and Fauza, 2011], expansion protocols used for clinical cytogenetics [De Coppi et al., 2007], or a combination of MSC derivation protocol followed by culture in serum-free media for human Embryonic Stem Cells (ESC) and induced Pluripotent Stem Cells (iPSC) [Moschidou et al., 2013]. These approaches establish multipotent populations of cells with limited self-renewal ability, further described as either MSC [Klein and Fauza, 2011] or c-KIT+ Amniotic Fluid Stem Cells (AFSC) with ability to initiate adipogenic, osteogenic, myogenic, endothelial, neuronal, and hepatic differentiation in vitro [De Coppi et al., 2007; Moschidou et al., 2012]. Using an alternative approach, we sought to establish stable c-KIT+ cell lines with self-renewal capacity by explanting mouse AF-derived cells in Embryonic Germ Cells (EGC) derivation conditions, previously used to establish stable cell lines from c-KIT+ primordial germ cells [Shamblott et al., 1998].
Explantation has been used to generate different types of self-renewing cell lines [Jaenisch and Young, 2008], including embryonic stem cells from different species [Evans and Kaufman, 1981; Martin, 1981; Thomson et al., 1995; Thomson et al., 1996; Thomson et al., 1998], mouse epiblast stem cells [Brons et al., 2007; Tesar et al., 2007], and mouse [Matsui et al., 1992; Resnick et al., 1992] and human embryonic germ cells [Shamblott et al., 1998] and it is also an important step in the culture of iPSC [Takahashi et al., 2007]. During explantation, primary progenitor cells are cultured in conditions that support and stimulate self renewal, typically through the addition of growth factors such as Leukemia Inhibitory Factor (LIF) and/or Human Recombinant Basic Fibroblast Growth Factor (FGF-2), mitotically inactivated mouse embryonic fibroblasts, and specially screened lots of fetal bovine serum or commercial serum replacer until successful generation of stable cell lines is achieved. In addition to its usefulness in generation of pluripotent stem cell lines, explantation can also be used to derive lineage committed permanent cell lines such as Extraembryonic Endoderm Cell Lines (XEN) [Kunath et al., 2005; Brown et al., 2010] and trophoblast cell lines [Tanaka et al., 1998].
In this report we describe the successful derivation of self-renewing cell lines from E11.5 mouse amniotic fluid using EGC-type explantation [Shamblott et al., 1998]. In addition, we show that these cell lines have the phenotypic and gene-expression profiles most similar to blastocyst-derived XEN cells, and we demonstrate their in vitro and in vivo Primitive Endoderm (PrE) lineage differentiation potential.
Material and Methods
AF cell line generation and culture
Cell lines were derived from mouse strain 129X1/SvJ (The Jackson Laboratory). Mouse amniotic fluid was obtained from dissected intact E11.5 amniotic sacs through a micropuncture. The collected cells were filtered using a 40 μm cell strainer (BD Bioscience) followed by a single wash step in High Glucose DMEM (Hyclone) with 10% fetal bovine serum (Sigma). Cells isolated from five amniotic sacs were plated into a single well of a tissue culture treated 12-well plate containing irradiated STO feeders (56-X, ATCC) at a density of 110,000 cells per cm2. The plating media consisted of Knockout DMEM/F12 (Gibco) with 15% of ESC-screened Fetal Bovine Serum (FBS) (Sigma), 0.1 mM nonessential amino acids, 2 mM glutamine, 1 mM Sodium Pyruvate (Gibco), 1X EmbryoMax nucleosides (Millipore), 0.14 mM 2-mercaptoethanol (Sigma), 1000u/mL ESGRO (Millipore), 2 ng/mL FGF-2 (Invitrogen), 10 μM Forskolin (Sigma) and 25 ng/mL Mouse Recombinant Stem Cell Factor (SCF) (R&D Systems). During the first four passages culture splitting was performed every 8-9 days using 0.25% Trypsin EDTA solution followed by vigorous pipetting to obtain a single cell suspension. Upon the appearance of the first colonies (~4 weeks), the culture of AF-derived cell lines (AFCL) was continued using mitomycin C treated mouse embryo fibroblast feeder cells, strain CF-1 (Millipore), in the absence of forskolin or SCF. During routine culture established cell lines were grown to subconfluence and passaged every 3-4 days using 0.05% Trypsin EDTA or TrypLE Express solution (Invitrogen). We cryopreserved cells in freezing media containing 10% DMSO (Acros Organics). Mouse ESC (CCE line) were cultured using standard methods (ATCC). XEN10 cell line, a kind gift of Drs. A.C. Foley and A.K. Hadjantonakis, was cultured as described in [Brown et al., 2010].
Doubling time analysis
For this analysis cells of each of AFCL were initially plated at 15,625 cells/cm2 in gelatinized 6-well plate pre-seeded with 20,000 cells/cm2 of irradiated CF-1 fibroblasts, in a regular AFCL growth medium. Upon reaching 80-90% confluence, each AFCL was re-plated in triplicate at 15,625 cells/cm2 in gelatinized well of a 6-well plate pre-seeded with above described number of irradiated CF-1 fibroblasts. After 46 hours, cells were treated with TrypLE Express (Invitrogen) and live cells were counted. The counts were normalized for the numbers of remaining irradiated CF-1 fibroblasts. The doubling time was calculated as previously described [Moschidou et al., 2012]: DT=t/(Log2(y/m)), t=time in culture, y=number of cells at end of culture, m=number of cells at beginning of culture.
Semi-Quantitative PCR (QPCR)
Total RNA extraction was performed using RNeasy Plus Mini Kit (Qiagen) followed by reverse transcription (RT) using QuantiTect Reverse Transcription Kit (Qiagen) using 500 ng of total RNA in each reaction. Real time Polymerase Chain Reaction (PCR) was performed on 1/20 of the RT reaction volume as a template, using Power SYBR Green PCR Master Mix (Applied Biosystems). The primers for the Gata6, Gata4, Pou5f1, Sox2, Nanog, Sox7, Sox17, Sall4, Lin28a and Alb were synthesized by Sigma-Aldrich based on sequences suggested by the Primer Express 3.0 software (Applied Biosystems). Primers for mouse 18s RNA, Afp, Foxa2 and Utf1 were manufactured by Qiagen. Samples were analyzed using Applied Biosystems StepOne Real-Time PCR instrument. We analyzed results using one-way ANOVA followed by Tukey’s Multiple Comparison Test functions by GraphPad Prism software. For the list of primer sequences and cycling conditions, see Table S1 in the supplementary information.
Immunostaining of cultured cells
We stained cells cultured in a 4-well CultureSlide (BD Falcon), coated with 0.1% Gelatin solution (Millipore) for 1 hr at RT, using SALL4, UTF1, LIN28, GATA6 and FOXA2 antibodies using the following dilutions: SALL4 1:200 ab29112 Abcam, UTF1 1:300 ab24273 Abcam, LIN28 1:300 ab63740 Abcam, GATA6 1:200 ab22600 Abcam, FOXA2 1:300 sc-6554 Santa Cruz Biotech. Cells cultured on Collagen I coated 4-well CultureSlide (BD Falcon) were stained for Alpha-fetoprotein (AFP) and Albumin (ALB) as previously described [Gouon-Evans et al., 2006]. For all other antibodies, cells were fixed with 4% methanol-free formaldehyde (EMS) for 10 min at room temperature and then washed three times with Phosphate Buffered Saline (PBS) followed by permeabilization with 0.2% Triton X-100 in PBS for 5 min (ALB and AFP) or 10 min (SALL4, UTF1, GATA6 and FOXA2). No additional permeabilization was performed for LIN28 staining. After 3 washes in PBS, the cells were blocked for 2 hours at room temperature in PBS containing 5% Milk, 10% normal goat serum (Jackson ImmunoResearch) and 0.1% Tween 20 (Sigma-Aldrich). Subsequently the cells were stained over night at 4°C with primary antibody or rabbit IgG (Santa Cruz Biotech) diluted in blocking buffer. The next day, cells were washed 3 × with PBS and stained for 1 hr at room temperature with Alexa488-conjugated secondary antibody (1:200, Invitrogen). At the end of the staining period, the cells were washed in PBS three times and counterstained in DAPI (Invitrogen) for 30 min. The staining was visualized using either Zeiss LSM 510 confocal system (UTF1, LIN28, FOXA2) or DeltaVision Deconvolution Microscope (SALL4, ALB and AFP) or Nikon A1 confocal microscope (GATA6). We used ImageJ software (NIH) for all image processing.
Tumor formation assay and tissue immunohistochemistry
The in vivo lineage potency of generated cell lines was assessed by intramuscular injection of 1×106 AFCL or 0.5×106 ESC (CCE, positive control) into the upper hindlimb of NOD-SCID IL2Rgammanull immunodeficient mice (Jackson labs) and teratoma/tumor formation monitored for 3 months. Collected tumors were fixed in 25% Formalin and processed by the Pathology Core of Dana-Farber/Harvard Cancer Center. Paraffin embedded tissue was processed for AFP staining as described (R&D Systems) except that the antigen retrieval step was performed for 45 min at 95°C using pH9 antigen retrieval solution (DAKO). Primary antibody against AFP (Ab-2, Neomarkers) was used at 1:100 dilution. Chromogenic detection of bound antibody was performed using anti-rabbit HRP-DAB cell and tissue staining kit (R&D Systems).
In vitro differentiation
AFCL were grown to near confluence in DMEM/F12 (Invitrogen) media containing 15% FBS and 5 ng/mL FGF-2 (R&D Systems). Culture was performed on 0.1% Gelatin coated tissue culture plates containing mitotically inactivated CF-1 feeders at 35,000/cm2. The differentiation assay was initiated on day 0 by plating ~1000 cells/cm2 on Collagen I coated 6-well plates or 4 well-CultureSlide (BD Bioscience) in stage I serum-free media containing DMEM/F12, 1 × N2 and B27 supplements (Invitrogen), 20 ng/mL human Activin A, 20 ng/mL Mouse Recombinant Bone Morphogenic Protein 4 (BMP4), 10 ng/mL FGF-2 (all from R&D Systems), 0.45 μM monothioglycerol (Sigma) and 50 μM ascorbic-acid-2-phosphate (Sigma). On day 2 Stage II of differentiation was initiated by changing the media to serum-free basic media, as described above, except that the growth factor combination used was mouse BMP4, 20 ng/mL, Mouse Recombinant Epidermal Growth Factor (EGF) 20 ng/mL (R&D Systems), FGF-2 10 ng/mL. After 14 days, cells were either submitted to immunofluorescent analysis of AFP and ALB expression or to RNA extraction followed by QPCR analysis, as described above.
Microarray analysis
For microarray analysis each AFCL was cultured to subconfluence. Subsequently, cells were trypsinized with TrypLE Express solution and submitted to high speed sorting using DAKO-Cytomation MoFlo high speed sorter; the c-KIT+ Platelet-Derived Growth Factor Receptor-α (PDGFRA+) DAPI- population of cells was sorted from each cell line in triplicate. Mouse ESC, CCE line, were submitted to the Mouse Embryo Fibroblast (MEF) removal step by allowing suspensions of trypsinized cells to attach to gelatinized tissue culture dish for 45 min. Subsequently non-attached cells were gently removed and submitted to RNA extraction. Total RNA from each sample was isolated using RNeasy Plus Mini Kit (Qiagen), and subsequently analyzed by the Microarray Core at the University of Texas Health Science Center at Houston. Briefly, 300 ng of total RNA was amplified and purified using Illumina TotalPrep RNA Amplification Kit (Ambion). An aliquot of 1.5 μg of amplified products was loaded onto Illumina Sentrix Beadchip Array mouse WG6.V2 arrays, hybridized at 58°C in an Illumina Hybridization Oven (Illumina) for 17 hours, washed and incubated with streptavidin-Cy3 to detect biotin-labeled cRNA on the arrays. Arrays were dried and scanned with BeadArray Reader (Illumina). Microarray data was further analyzed as follows: The data was processed using the loess normalization algorithm. After normalization, additional processing was performed using a batch effect removal algorithm [Johnson et al., 2007] based on empirical Bayes methods. All arrays were adjusted so that they have similar distribution. To improve the outcome of the batch effect removal algorithm, all samples processed together with Xen10, Pys2, End2 and ESC samples were used as input. The intensities of all 45281 probes were included in the calculation of R2. Hierarchical clustering was performed using the “complete” algorithm and 1-R2 as a metric. Heatmap tables were generated using the detection p value of 0.01 as a threshold. In case of multiple probes for the same gene, the probe with the highest intensity was reported. The Venn diagram analysis was based on genes whose probes were detected in all biological replicates. A detection p-value of 0.01 was used.
Flow cytometric analysis of surface marker expression and cell cycle
Monolayer cultures of AFCL were dissociated using TrypLE express (Invitrogen) (5 min). Subsequently, isolated cells were stained with directly conjugated antibodies to c-KIT (Cat 561074, BD Bioscience) and PDGFRA (Cat. 562776, BD Bioscience) or c-KIT (Cat. 561075, BD Bioscience) and C-X-C Chemokine Receptor Type 4 (CXCR4) (Cat. 558644, BD Bioscience) in staining buffer (HBSS+2% FBS) for 30 minutes followed by a single wash step. DAPI nuclear staining was used to exclude dead cells. Negative gates were established using isotype controls (Cat. 551139 and 555844, BD Biosciences).
Cell cycle analysis was performed as previously described [Mullen, 2004]. Stained cells were analyzed using BD FACSCalibur, LSRII and FlowJo software (Tree Star).
Single cell colony-formation assay
AFCL and ESC (CCE) were both cultured in the DMEM/F12 based media containing 15% ESC-screened FBS with 1000u/mL ESGRO, until 60-70% confluency. To avoid sorting of feeder cells, AFCL were stained for flow cytometry using fluorophore conjugated antibodies against c-KIT and PDGFRA (see Cat# above, BD Bioscience) (1:100) and CCE were stained with primary fluorescently labeled antibodies for SSEA-1 (Cat. 560142, BD Bioscience) and EPCAM (Cat. 17-5791; eBioscience) (1:100), utilizing above described flow cytometry protocol. Double positive live cells were sorted as single cell/well into 96-well gelatinized flat bottom tissue culture treated plates (BD Falcon) containing previously seeded irradiated CF-1 fibroblasts (R&D Systems) using DAKO-Cytomation MoFlo high speed sorter. Three plates were used for each cell line tested. For secondary sorting experiment three individual wells from 96-well plates, containing colonies, were expanded separately as sub cloned cell lines in the original culture conditions for 7 days. Subsequently, the cells from each sub clone were single-cell sorted into three 96-well plates each. All secondary plates were examined for colony formation after 7 days of culture. Statistical analysis and data plotting was performed using one-way ANOVA followed by Tukey’s Multiple Comparison Test in GraphPad Prism software.
Electron microscopy
The sample preparation and imaging was performed by MD Anderson Cancer Center Electron Microscopy Core facility as follows: samples were treated with a fixative containing 2% glutaraldehyde in 0.1 M PBS, pH 7.3 at 4°C overnight and then washed with 0.1 M PBS buffer, pH 7.3, 3 × 10 min. The samples were post fixed with 1% PBS buffered osmium tetroxide for 1 hour, washed with 0.1M PBS buffer, 3 × 10 min., followed by rinsing with distilled water, 2 × 10 min. Dehydration was performed with increasing concentrations of ethanol for 5 min each followed by treatment with hexamethyldisilazane (HMDS) and air drying overnight. Samples were mounted on to double-stick carbon tabs (Ted Pella. Inc., Redding, CA) followed by coating under vacuum. Subsequently, platinum coating was performed under vacuum for a thickness of 25 nm, followed by carbon coating. Imaging was performed using JSM-5910 scanning electron microscope (JEOL Inc., Peabody, MA) at an accelerating voltage of 5 kV.
Results
Irradiated STO cells are required for generation of c-KIT+ AF-derived cell lines
To initiate the EGC-type explantation protocol, we plated AF-derived cells on irradiated STO feeder cells in serum containing media in the presence of LIF, FGF-2, SCF and Forskolin. After three to four weeks in culture we noted the first appearance of epithelial colonies (Fig. 1A). Using these conditions, we reproducibly generated three stem cell lines from 129X1/SvJ mice. These cell lines were termed K82, K83 and K84, in chronological order. Importantly, when AF cells were cultured on mitotically inactivated CF-1 fibroblast feeder cells plated at densities varying from 25,000 to 45,000/cm2, using the above described seeding and splitting protocol and media combination (including LIF, FGF-2, SCF and Forskolin), no observable colony formation occurred after 6 weeks of culture.
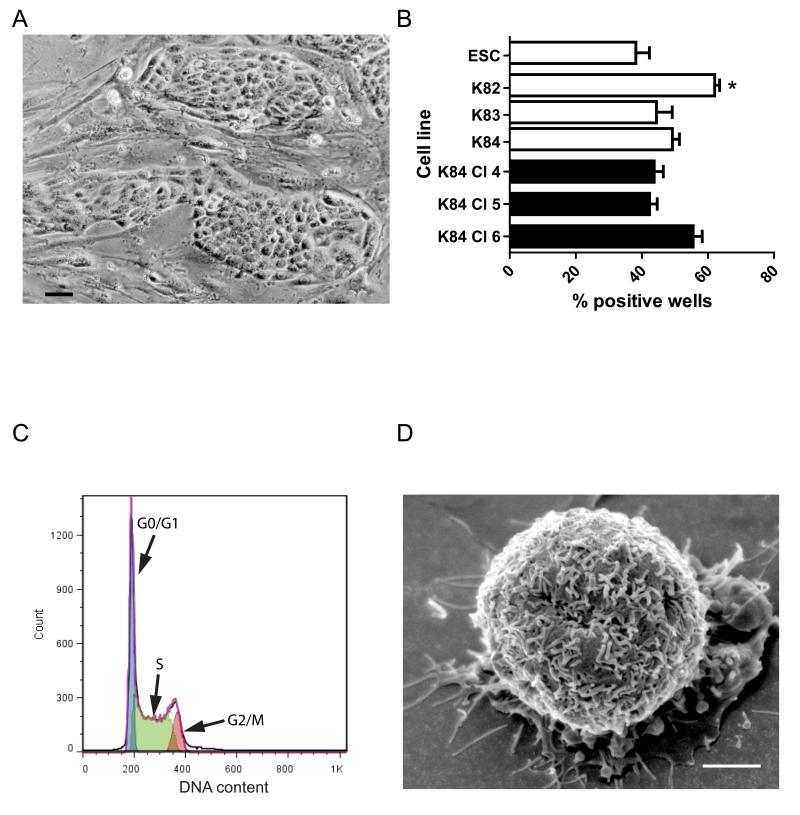
Figure 1.
Morphology, cell cycle analysis and single cell cloning of AFC.
A) Image of colonies of AFC nested among mitotically inactivated CF-1 fibroblast feeders (K82). 50 μm scale bar.
B) Results of primary single cell sorting experiment are depicted as open bars. The secondary single cell sorting experiment is depicted as black bars. Error bars denote standard error of the mean. *P<0.05 for K82 vs. ESC and K82 vs. K83
C) Cell cycle analysis: representative graph of K83 cell line is shown.
D) Scanning electron microscopy of K82 cell line. Rounded cell with numerous microvilli. 2 μm scale bar
To assess the phenotypic stability of the above cell lines, we maintained each in culture for 20 passages over 10 weeks. All three cell lines had stable morphology, viability (data not shown), growth characteristics (Fig. S1) and surface phenotype (Figs. S2 and S3).
A hallmark of self-renewal is the potential of cells to regenerate colonies from a single cell, so we used a high speed cell-sorter to deposit individual double positive (CD117 and CD140a for AFCL or SSEA-1 and CD326 for ESC) live cells in each well of a 96-well plate, previously seeded with fibroblast feeder cells. The results suggest that all AFCL cell lines have the capacity for clonal growth (Fig. 1B) Importantly, the efficiency of single cell sorting, calculated as the percent positive wells, from each of the AFCL lines is at least similar or higher (K82) compared to ESC. The maintenance of the self-renewal phenotype is further confirmed through retention of a single cell cloning efficiency of cells from each of the 3 previously sorted individual clonal lines of K84 parent cell line (Fig. 1B, dark bars)
We determined the ploidy of cultured AFCL by flow cytometric cell cycle analysis, which showed that 25-32% of the cells are diploid in G1 phase, 49- 51% are in S phase and 13-19% are in G2 phase of cell cycle; about 5% of cells had intermediate DNA content. The representative propidium iodide staining of K83 cells is shown in Fig. 1C. In addition, analysis of growth kinetics of exponentially growing AFCL suggest a similar growth rate of all three cell lines with an average doubling time of 12.8±0.5 hours (Fig. S1)
Morphology and surface phenotype of AFCL lines is similar to blastocyst-derived extraembryonic endoderm stem cell lines
The initial appearance of AFCL was characterized by epithelial colony formation. These colonies are nested in between mouse embryo fibroblast feeders (Fig. 1A). When the AFCL are grown on a gelatin-coated surface in the absence of feeder cells, however, they assume several different morphologies including rounded cells and epithelial sheets, similar to the previously described morphologies of cultured XEN [Kunath et al., 2005] (Fig. S4). Scanning electron microscopic examination of AFCL grown on gelatin-coated surfaces showed the presence of rounded cells rich in microvilli, also similar to previously described XEN cells [Kunath et al., 2005](Fig. 1D).
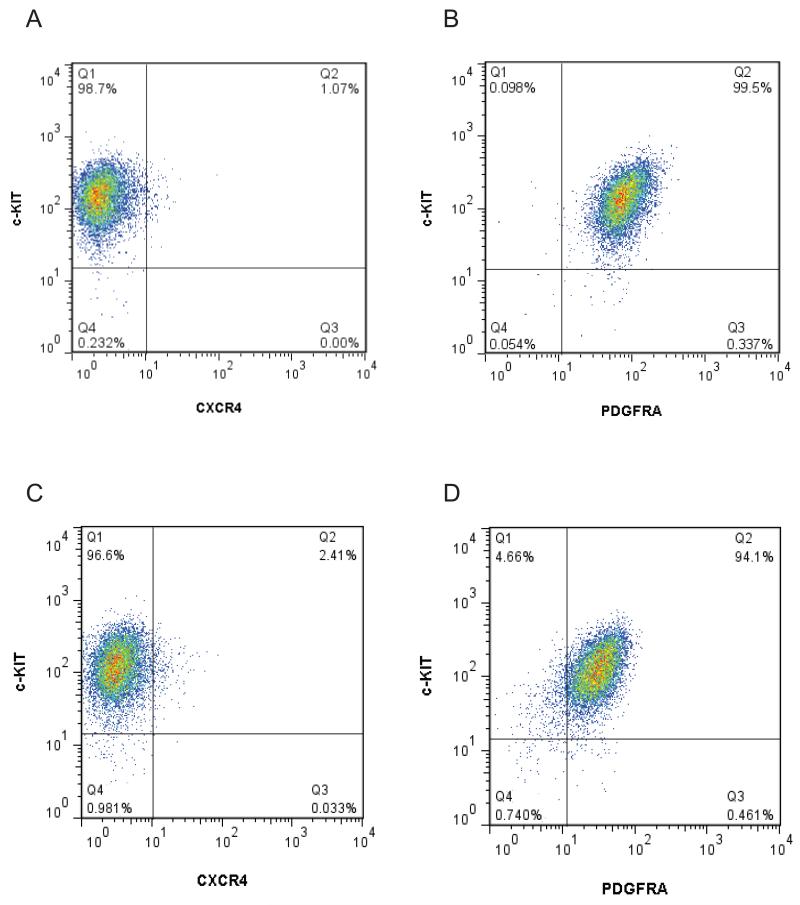
To evaluate phenotypic similarities between AFCL and XEN lineage, we performed flow cytometric analysis of the surface phenotype of AFCL and XEN10 using a panel of antibodies against surface markers, previously shown to be expressed on either AF-derived progenitors (c-KIT) [De Coppi et al., 2007; Moschidou et al., 2012], or definitive endoderm-derived cells: c-KIT [Orr-Urtreger et al., 1990] and CXCR4 [Yasunaga et al., 2005; Gouon-Evans et al., 2006], or XEN, PrE, Parietal Endoderm (PE) and extraembryonic Visceral Endoderm (VE) (PDGFRA) [Takakura et al., 1997; Plusa et al., 2008; Artus et al., 2010]. As shown in Figure 2, both AFCL (Fig. 2B) and XEN (Fig. 2D) are c-KIT and PDGFRA double positive. At the same time, both cell types are mostly negative for CXCR4 expression (Fig. 2A and 2C), consistent with assignment of E11.5-derived AFCL to the XEN lineage. In addition, Fluorescence Activated Cell Sorting (FACS) examination of above mentioned marker expression in early (Fig. S2) and late passage AFCL (Fig. S3) confirms their phenotypic stability over multiple passages.
Figure 2.
Flow cytometric analysis of surface expression of CD117 (c-KIT), CD140a (PDGFRA) and CD184 (CXCR4) on K82 (A and B) and XEN (C and D). Approximately 9,000 DAPI negative events are shown on each graph.
Analysis of the gene expression profile of AFCL confirms that they belong to the family of extra-embryonic endoderm stem cells
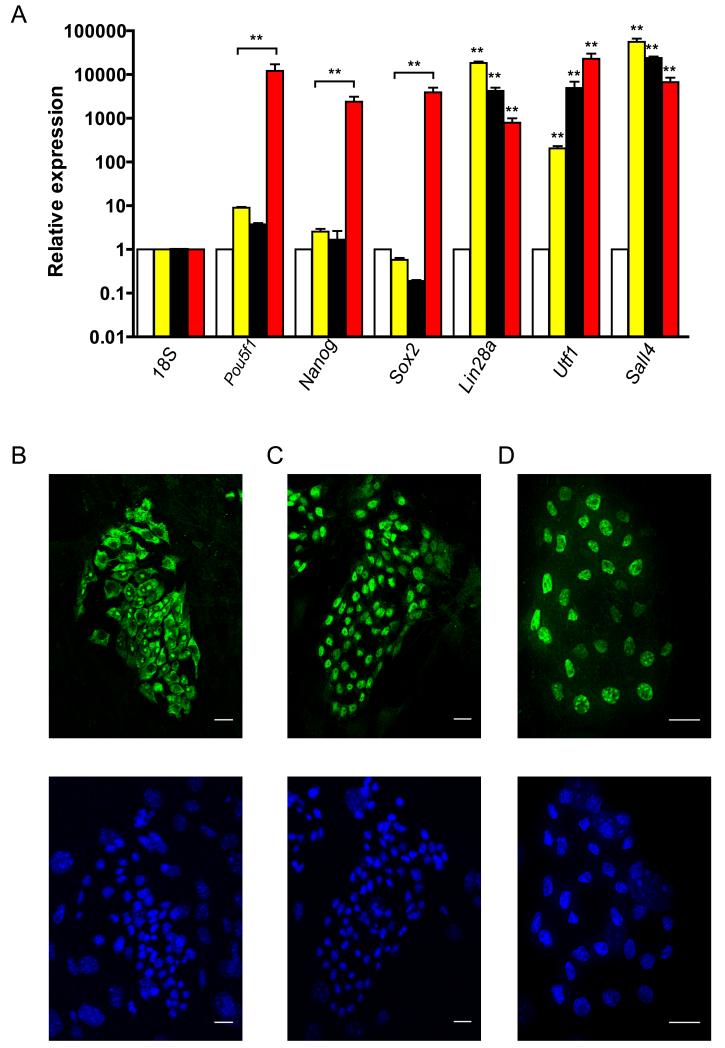
To analyze the gene expression pattern potentially associated with self-renewal of AFCL in culture, we initially examined the mRNA expression of six genes that are notably involved in self-renewal of embryonic stem cells, including: Pou5f1, Nanog, Sox2, Lin28a, Utf1, and Sall4. Our results showed that both AFCL and XEN10 express levels of Lin28a, Utf1, and Sall4 mRNA comparable to those expressed in ESC (Fig. 3A). To verify the corresponding protein expression, we used immunofluorescence analysis of cultured AFCL-K83 stained with LIN28, UTF1, and SALL4 antibodies, and showed that K83 cells express significant levels of the three proteins (Fig. 3B-D). Because self-renewal of XEN cells is dependent on Sall4 expression [Lim et al., 2008], we hypothesize that Sall4 expression has a similar importance in AFCL self-renewal, while the specific roles of Lin28 and Utf1 remain to be tested. Importantly, neither AFCL nor XEN express significant levels of the three key pluripotency-related genes, Pou5f1, Sox2, and Nanog, suggesting that AFCL, like XEN, exist in a lineage-committed self-renewal state.
Figure 3.
Analysis of mRNA and protein expression of self-renewal related genes.
A) QPCR analysis of mRNA expression in MEF (open bars), K83 (black bars), XEN10 (yellow bars) and mouse ESC (red bars). The data is normalized to mRNA expression level of each gene in CF-1 mouse embryo fibroblasts. Error bars denote standard error of the mean. ** P<0.01.
B) Immunofluorescent analysis of LIN28 expression in K83 (top image). DAPI nuclear staining, bottom image, 20 μm scale bar.
C) Immunofluorescent analysis of UTF1 expression in K83 (top image). DAPI nuclear staining, bottom image, 20 μm scale bar.
D) Immunofluorescent analysis of SALL4 expression in K83 (top image). DAPI nuclear staining, bottom image, 20 μm scale bar.
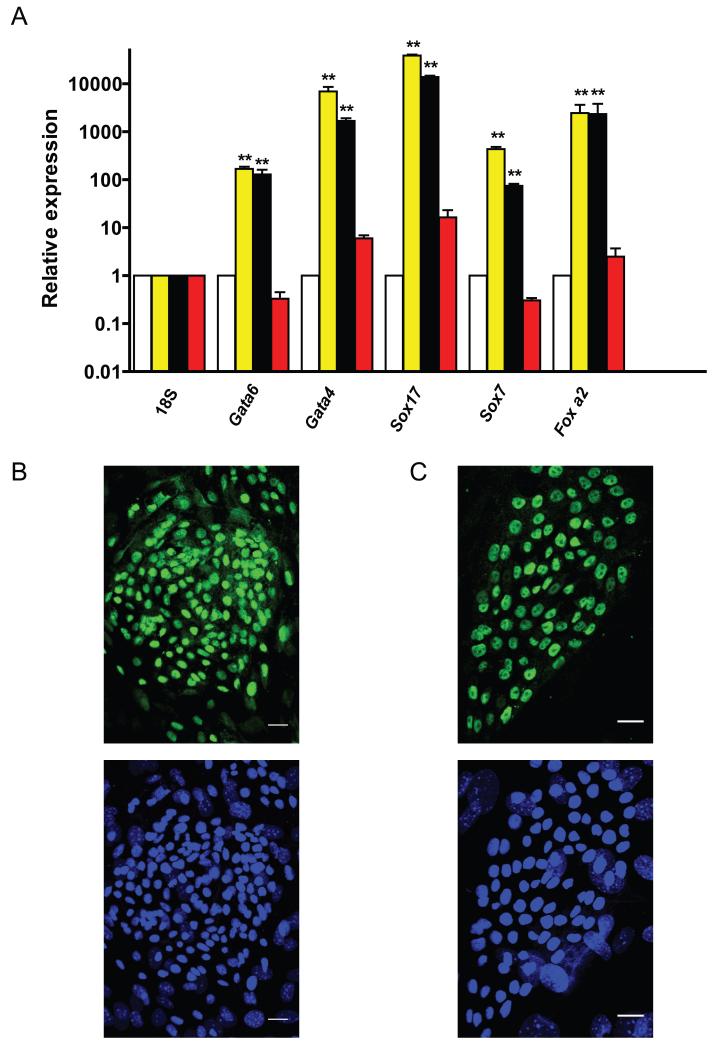
While both PrE and definitive endoderm share a significant overlap in gene expression patterns including Gata4, Gata6, Sox17, Foxa2, the cells of primitive endoderm also express Sox7 [Seguin et al., 2008]. We therefore compared the expression of Gata4, Gata6, Foxa2, Sox17, and Sox7 mRNA in ESC, XEN, and AFCL. As shown in Fig. 4A, AFCL-K83 express similar levels of primitive endoderm lineage genes as XEN cells. In addition, comparative QPCR analysis of Gata6, Gata4, Sox7 and Sox17 in all three AFCL suggests similar expression levels of the four PrE-related genes (Fig. S5A), except that in this experiment K82 appears to have somewhat higher expression of Gata6 compared with K83 and K84. Protein expression and localization of GATA6 and FOXA2 transcription factors in K83 cells were also confirmed using immunofluorescent analysis of cells grown on culture slides (Fig. 4B and 4C).
Figure 4.
Analysis of mRNA and protein expression of endoderm lineage genes
A) QPCR analysis of mRNA expression in MEF (open bars), K83 cells (black bars), XEN10 (yellow bars) and mouse ESC (red bars). The data is normalized to mRNA expression level of each gene in CF-1 mouse embryo fibroblasts. Error bars denote standard error of the mean. ** P<0.01
B) Immunofluorescent analysis of GATA6 expression in K83 cells (top image). DAPI nuclear staining, bottom image, 20 μm scale bar.
C) Immunofluorescent analysis of FOXA2 expression in K83 cells (top image). DAPI nuclear staining, bottom image, 20 μm scale bar.
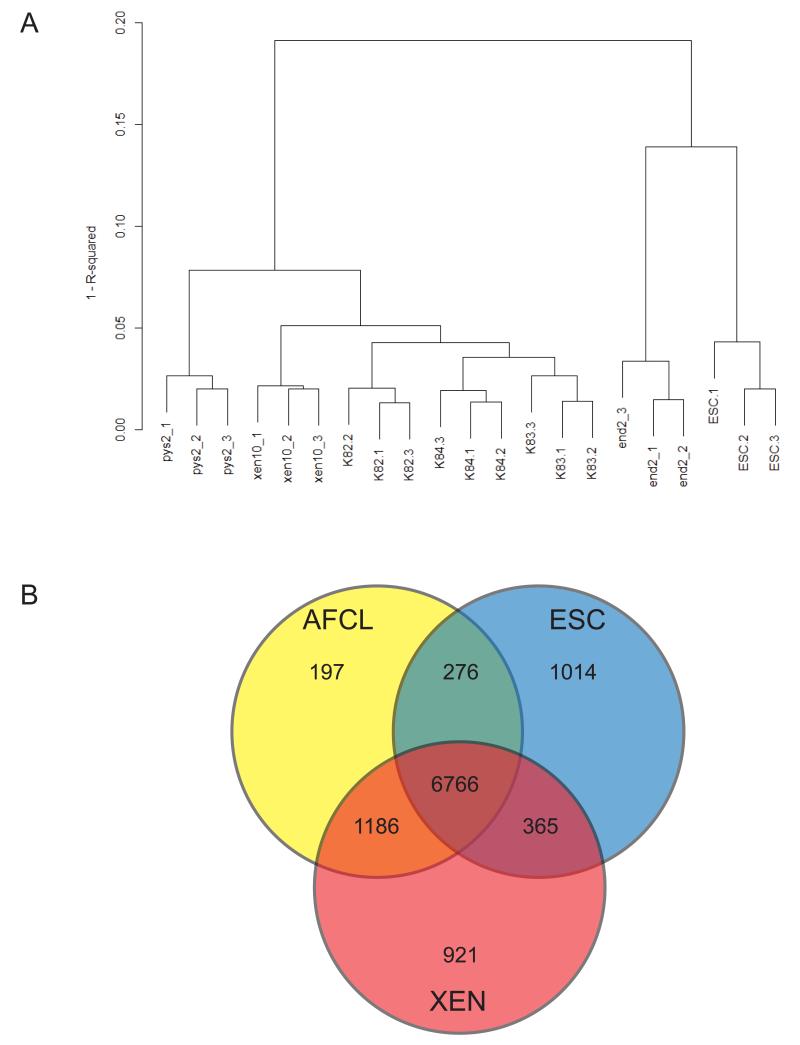
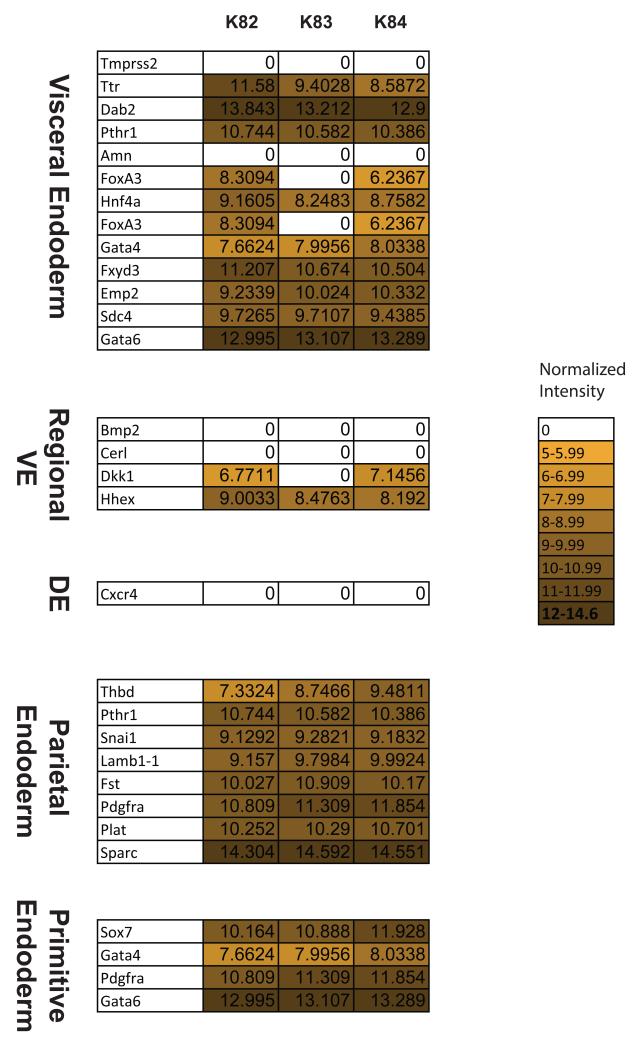
Because previous studies support a XEN lineage potential for AFCL, we performed an Illumina mouse whole genome 6.0 BeadChip-based microarray gene expression analysis on AFCL and compared the obtained data with the gene expression profile of mouse ESC and a previously published microarray data set of three different Extraembryonic Endoderm (ExEn)-like cell lines [Brown et al., 2010]. Hierarchical clustering of AFCLs, ESC and ExEn-like cell lines reveals that the whole transcriptome profile of AFCLs is most closely related to blastocyst-derived XEN cells, followed by Pys2 line, while it is well separated from the expression profiles of End2 line and ESC (Fig. 5A). To examine additional similarities and differences between AFCL, XEN and ESC we plotted Venn diagrams depicting number of common and unique genes between the three types of cell lines and we observed that the number of common genes between AFCL and XEN is 7952, compared with the number of commonly expressed genes between AFCL and ESC of 7042 and between XEN and ESC of 7131 confirming a closer relationship between AFCL and XEN than between AFCL and ESC (Fig. 5B), as suggested by dendrogram. To more precisely characterize endodermal potential of AFCL we examined expression of genes related to different endodermal subtypes by making a comparative expression heatmap of genes previously shown to be characteristic for various endodermal subtypes, including VE, regional VE, Definitive Endoderm (DE), PE and PrE [Yasunaga et al., 2005; Brown et al., 2010] (Fig. 6). These results suggest that all three cell lines express a number of markers for PrE, PE and VE. More specifically, there is a moderate expression of Thbd and Gata4 in K82 compared with K84 cells, and an intermediate level of these two genes in K83. All three cell lines also express most of listed markers of VE and regional VE (excluding Tmprss2 and Amn) with slightly lower levels of Ttr, Hnf4a, Foxa3, Fxyd3 and Hhex in K84 compared with K82, while K83 cell line does not express Foxa3 and Dkk1. However, the significance of these minor variations in endodermal lineage gene expression is not clear at this time. AFCL also have a notable absence of Cxcr4, a previously described DE marker [Yasunaga et al., 2005], which correlates well with FACS analysis of comparative surface marker expression depicted in Figs. 2, S2 and S3. In summary, AFCL display significant levels of expression of PrE-related genes and more specifically genes related to both PE and VE with some variation in expression levels of individual genes, suggesting that AFCL belong to ExEn lineage and more closely to XEN cells.
Figure 5.
Hierarchical clustering of AFCL. A) Dendrogram for AFCL (K82, K83 and K84), ESC and XEN10, Pys2 and End2 cell lines. The legend represents 1-R2. Hierarchical clustering included all 45K probes and was performed using the “complete” algorithm and the 1-R2 metric. Samples were grouped according to their condition B) Venn Diagram depiction of genes whose expression is similar or distinct between three cell types
Figure 6.
Heat map of genes characteristic for different endodermal subtypes. Normalized data from Illumina microarray for genes previously described as specifically expressed in each endodermal subtype. Intensity value was depicted as zero if detection p value was >0.01.
In vitro differentiation of AFCs results in the generation of AFP and ALB producing cells
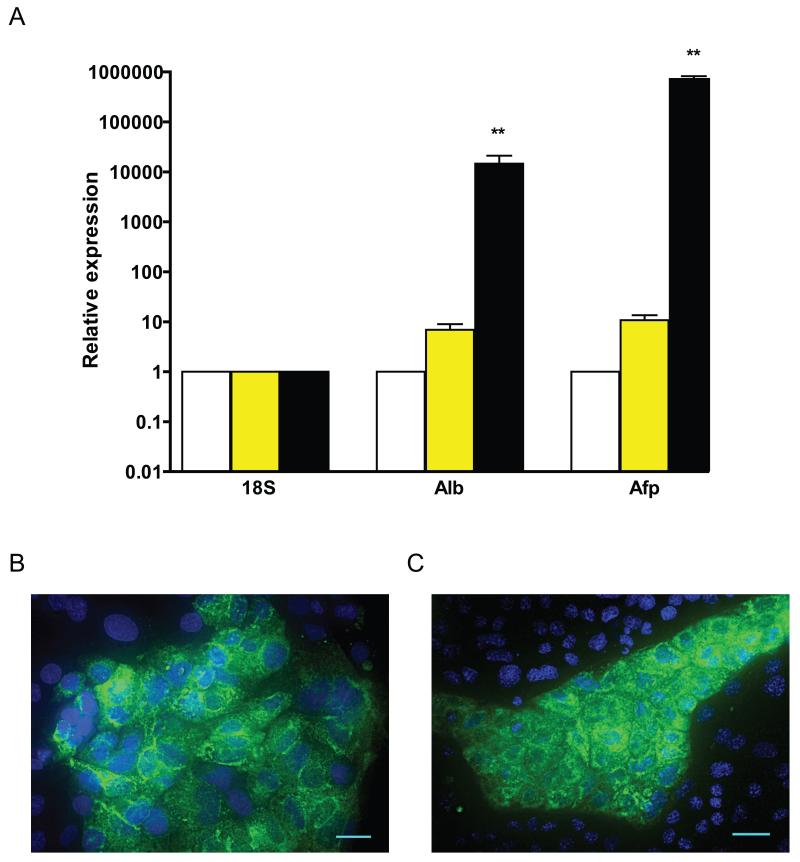
To examine the potential for in vitro endodermal lineage commitment of AFCL, we treated these cells with several different cytokines, including BMP4, Activin A, EGF, HGF, FGF-2, and FGF-4, individually and in combination, followed by measurement of mRNA and protein expression of AFP and ALB as markers of maturing embryonic and extraembryonic endodermal lineages. The most efficient differentiation protocol was based on the previously published study [Yasunaga et al., 2005] and it required the presence of BMP4 during both stages of differentiation (data not shown). The differentiation resulted in significant upregulation of Afp and Alb mRNA (Fig. 7A) and protein (Fig. 7B), consistent with the known role of BMP4 in differentiation of blastocyst-derived XEN into AFP expressing VE [Artus et al., 2012; Paca et al., 2012]. Comparative QPCR analysis of Afp and Alb expression before and after differentiation in all three AFCL is shown in Fig. S5B. These results confirm that all three AFCL are capable of in vitro differentiation in the presence of BMP4. In addition, it can also be noted that K82 has a slightly higher efficiency of differentiation (suggested by the difference in Afp and Alb levels between undifferentiated and differentiated state) compared with the other two cell lines, which did not reach level of statistical significance (Fig. S5B).
Figure 7.
Analysis of mRNA and protein expression of ALB and AFP after in vitro differentiation of K82 cells.
A) QPCR analysis of mRNA expression in MEF (open bars), K82 cells before differentiation (yellow bars) and K82 cells after differentiation (black bars). The data is normalized to mRNA expression level of Afp and Alb in CF-1 mouse embryo fibroblasts. Error bars denote standard error of the mean. ** P<0.01
B) Immunofluorescent analysis of ALB expression (left) and AFP (right) in differentiated K82 cells. DAPI nuclear stain is shown in blue. 20 μm scale bar.
AFCL generate primitive endoderm tumors in vivo
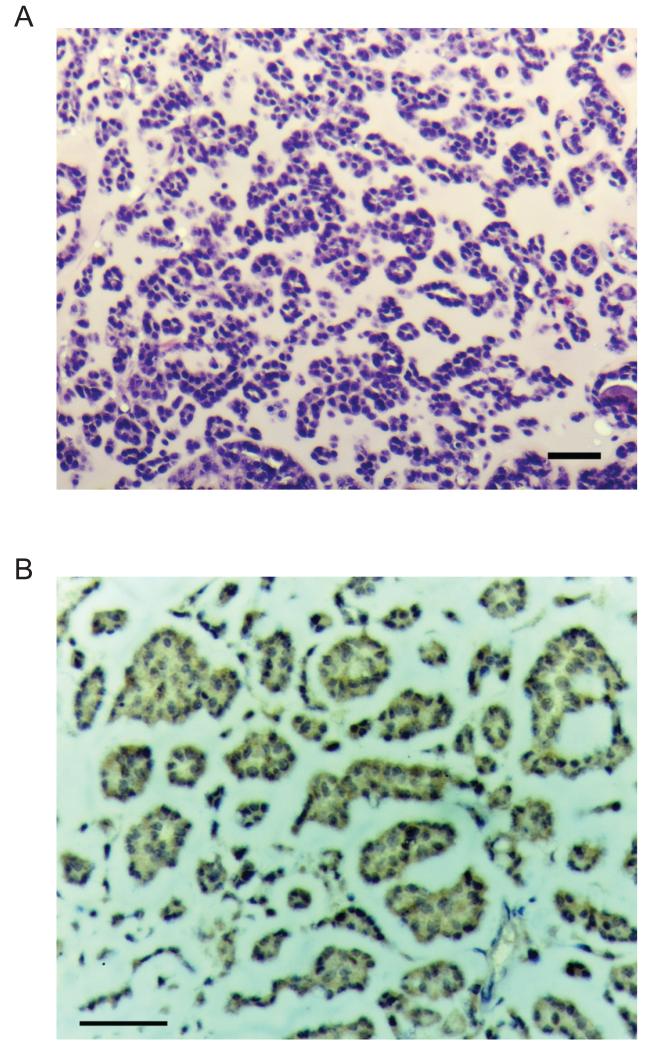
The teratoma formation assay is a standard assay for the multi-lineage potential of pluripotent cells in vivo. We therefore injected cells from either AFCL or ESC (positive control) intramuscularly into immunodeficient mice and observed the animals for teratoma formation. ESC-derived tumors appeared after approximately 3 weeks, but AFCL-derived tumors required 6-8 weeks to appear. Histological analysis confirmed the multilineage properties of the control ESC-derived teratoma (data not shown). AFCL-derived tumors, however, had a significantly different appearance from the ESC-derived tumors, with a glandular appearance (Fig. 8), and a significant amount of interglandular amorphous matrix, suggestive of an immature endodermal lineage origin. AFCL tumor cells also expressed significant levels of AFP (Fig. 8B). Detailed histological examination of AFCL tumors did not yield any evidence of definitive ectodermal, mesodermal or endodermal differentiation. Hence, AFCL cells likely represent primitive endoderm progenitors with limited potential for in vivo primitive endodermal differentiation during heterotopic transplantation.
Figure 8.
Histological and immunohistochemical analysis of tumors formed by K82 cells
A) Hematoxylin and eosin staining of a tumor section. 50 μm scale bar.
B) Immunoperoxidase staining of anti-AFP antibody labeled tumor section counterstained with hematoxylin. Antigen positive areas are labeled with brown color. 50 μm scale bar.
Discussion
Development of cell replacement therapy for degenerative diseases is one of the most eagerly-sought goals of stem cell research. A requirement for this goal is generation of pure populations of progenitor cells with extensive ability for expansion. While differentiation of ESC into lineage committed cells could fulfill the criteria for clinical application, the requirement for embryo destruction to generate new ESC lines raises substantial ethical and religious barriers to implementation. Induced pluripotent stem cells are an alternative source of pluripotent cells for lineage differentiation [Takahashi et al., 2007]. Although these cells avoid many of the ethical and availability drawbacks associated with the use of ESC, concerns exist about their true therapeutic value, due to the presence of oncogenic viruses and proteins used to immortalize the cells, the low efficiency of iPSC cell line generation [Sun et al., 2010], and their limited and unpredictable differentiation potential [Feng et al., 2010]. We therefore determined whether AF could be an alternative source of committed progenitor cells able to give rise to stable self-renewing cell lines that could be used in subsequent cell therapies.
AF contains several types of stem cells including MSCs and c-KIT+ AFSC. Human AF-derived MSCs have been consistently isolated from AF and have been previously well characterized as positive for surface expression of CD73, CD105, CD44, CD29, CD90, CD13, CD10, CD71 and several other markers [Klein and Fauza, 2011]. These cells are capable of chondrogenic [Kunisaki et al., 2006] and osteogenic [Turner et al., 2012] differentiation. However, wider application of MSCs in regenerative medicine applications may be limited by their finite lifespan and eventual loss of their stem cell properties during prolonged culture [Liu et al., 2009]. Alternatively, we considered c-KIT+ cells an appropriate candidate for our studies since c-KIT+ AF cells have been previously described as capable of limited self-renewal and multilineage differentiation [De Coppi et al., 2007; Moschidou et al., 2012]. As generation of a stable cell line from progenitor cells likely involves overcoming of a significant epigenetic barrier [Surani et al., 2007], we hypothesized that the culture conditions previously shown to convert c-KIT+ primordial germ cells to self-renewing c-KIT+ embryonic germ cell lines may also facilitate such conversion of AF cells [Matsui et al., 1992; Resnick et al., 1992; Shamblott et al., 1998].
An important aspect of generation of stem cell lines in vitro is maintenance of cultured cells in an in vitro “niche” environment [Davey and Zandstra, 2006]. To establish the key components of the initial AFCL culture that stimulated cell line formation, we varied several culture parameters including growth factor composition and feeder cell type. The resulting conclusion is that utilization of irradiated STO cells is critical for achieving reproducible success in cell line generation. Application of commonly used CF-1 feeder cells was unsuccessful, resulted in a gradual decrease in the total cell number in culture and subsequent failure to establish stable cell lines. The exact mechanism of benefit from STO cells is not clear, but it is likely related to differences in the amount and the type of cytokines and/or extracellular signaling molecules, including Activin A (high in CF-1), hepatocyte growth factor (high in STO), IL-6 (high in CF-1), SCF (high in STO) and others [Talbot et al., 2012]. Since AFCL are consistently c-KIT+ and fail to be generated on CF-1 cells, it is possible that c-KIT-SCF interaction is critical for AFCL generation. However, because we used an identical derivation medium (containing exogenous soluble SCF) with either STO or CF-1 feeders, we hypothesize that it is the transmembrane SCF that may play an important role in derivation of AFCL, based on previously shown differential effects of soluble and transmembrane SCF in other cell types [Ashman, 1999]. Importantly, comparison of derivation conditions for blastocyst-derived XEN cells [Kunath et al., 2005] and our results with AFCL suggests that the more advanced developmental stage of AFCL require more elaborate culture conditions for cell line generation, as blastocyst-derived XEN can be successfully generated even on mitotically inactivated primary fibroblast feeders.
Self-renewal is one of the two key properties of stemness. The most detailed studies of stem cell self-renewal mechanisms have been performed in ESC system where a core self-renewal machinery was described as consisting of Pou5f1, Sox2 and Nanog genes [Boyer et al., 2005; Mallanna and Rizzino, 2012]. The proteins coded by the three genes maintain self-renewal through regulation of a large transcriptional network that includes targets such as Sall4 [Lim et al., 2008], Lin28 [Shyh-Chang and Daley, 2013], Utf1[Laskowski and Knoepfler, 2012] and others. To examine the genes potentially involved in self-renewal of AFCL we analyzed the expression of above mentioned genes and discovered that the self-renewal of AFCL, like XEN, can be maintained in the absence of gene expression of core self-renewal network of ESC. Further analysis of AFCL gene expression profile suggests that AFCL, like XEN, have a high expression of Sall4, Lin28 and Utf1, which may be able to participate in an XEN type self-renewal gene network. Importantly, previous studies described a critical role for Sall4 in XEN self-renewal [Lim et al., 2008], giving support to the above hypothesis. However, additional studies will be required to test the dependence of AFCL self-renewal on Sall4, Utf1 and/or Lin28.
The majority of previously examined mouse XEN cell lines exhibit several characteristics of PE in culture [Kunath et al., 2005; Brown et al., 2010; Paca et al., 2012]. These include poor maintenance of epithelial appearance during extended culture and predominant contribution to PE after blastocyst injection, suggestive of their predominant PE differentiation potential. However, rat XEN cell lines appear to have a more balanced VE vs. PE differentiation potential resulting in significant contribution of these cells to extraembryonic visceral endoderm upon blastocyst injection [Debeb et al., 2009] and also in activation of PE program with Forskolin and VE program with a small molecule GSK3 inhibitor [Chuykin et al., 2013]. To examine the in vitro differentiation potency of AFCL we began with an observation that AFCL grown on gelatin surface have growth characteristics similar to XEN, including ineffective maintenance of epithelial properties, which is consistent with PE potential [Hogan and Tilly, 1981]. Furthermore, statistical analysis of whole transcriptome profile of AFCL suggested that it correlates most closely with the profile of blastocyst-derived XEN cells and ExEn-like Pys2 cells, and this analysis showed that AFCL express transcripts of both PE and VE, suggestive of their bipotency. However, when we examined the gene expression profile of K82 cells cultured for 2-3 weeks after reaching confluence, using a QPCR array, we observed significant induction of Afp expression (data not shown), consistent with VE type differentiation, as Afp is mainly expressed in VE and not PE [Dziadek and Adamson, 1978]. To confirm our hypothesis of VE differentiation potential of AFCL we tested several different cytokines and observed that BMP4 is critical for induction of AFP and ALB expression suggesting that BMP4 can induce extraembryonic VE [Yasunaga et al., 2005] in AFCL, which is consistent with published reports describing BMP4 mediated VE induction in blastocyst derived XEN [Artus et al., 2012; Paca et al., 2012]. Further analysis of in vitro Afp and Alb expression in all three AFCL, submitted to an abbreviated differentiation protocol, confirmed that they are capable of extraembryonic VE type differentiation with somewhat different efficiencies. The molecular basis for these differences is not clear but it may come from differential expression of a number of PE and/or VE specific genes, as evidenced from microarray analysis, and/or a difference in Gata6 expression (higher in K82) as observed during comparative QPCR analysis. Of note, the same experiment also shows that the basic AFCL maintenance medium can be used to propagate cells with a higher baseline expression level of Afp and Alb, suggesting that modest increase in expression of VE-type markers does not interfere with cell line propagation. The elevated levels of Afp and Alb are likely related to a different lot of ESC-tested FBS used for routine maintenance of AFCL, as previous studies demonstrated presence of BMP-type activity in FBS [Kodaira et al., 2006] as well as a significant lot to lot variability in bone morphogenic protein content of FBS [Herrera and Inman, 2009].To examine in vivo differentiation potential of AFCL we analyzed histological characteristics of AFCL-derived tumors and we observed endodermal sinus-type glandular clusters that also showed presence of AFP, which can suggest a potentially significant in vivo VE differentiation potential of AFCL. However, further studies would be required to confirm this hypothesis because Afp may be expressed in tissues other than VE [Yasunaga et al., 2005] In summary, AFCL appear to display a context related-VE or PE differentiation potential similar to previously studied blastocyst-derived XEN cell lines.
The origin of AF cells capable of generating primitive endoderm cell lines in vitro is currently unknown. Such cell lines have mostly been generated from blastocyst-stage embryos in the past [Kunath et al., 2005] However, a recent report also suggested that PrE cell lines can be generated from adult rat bone marrow [Roelandt et al., 2010]. AF may therefore receive PrE endoderm progenitors through a transient communication between amniotic and yolk sacs via a transient neurenteric canal [Ozek et al., 2008]. Alternatively, XEN cell lines may derive from PrE derived cells in fetal gut [Kwon et al., 2008] which are then released into AF through regular emptying of fetal gut contents, an activity observed in both early and mid-gestation human fetuses [Kimble et al., 1999]. A less likely possibility is transdifferentiation of definitive endodermal progenitors into XEN cell lines when maintained under the culture conditions described above
In this report we describe derivation of AFCL from AF of 129X1/SvJ mouse strain. This strain was purposefully chosen as the initial donor of AF because of previous reports of high permissiveness for ESC derivation of the same mouse strain [Brook and Gardner, 1997]. XEN were initially derived from several different mouse strains including PO, ICR and 129 [Kunath et al., 2005] as well as from several different rat strains [Debeb et al., 2009], without the use of small molecule inhibitors which were required for efficient derivation of ESC from various mouse and rat strains [Blair et al., 2011]. This suggests that AFCL could potentially be established from different mouse and rat strains with minimal protocol modification. Importantly, ExEn cell lines have not yet been established from human blastocysts. The information we have about human PrE lineage mostly comes from studies on PrE cells that arise during human ESC and iPSC differentiation [Takayama et al., 2011; Feng et al., 2012]. Interestingly, a recent report describes a population of c-KIT+ cells isolated from first trimester human AF that express Sox7[Moschidou et al., 2012], a key factor in development of parietal endoderm. Whether these human AF-derived cells represent human counterparts of mouse AFCL-generating progenitors remains to be studied.
In summary, we have demonstrated that culture of AF-derived progenitor cells in culture conditions that support self-renewal can generate phenotypically stable self-renewing cell lines of the PrE lineage. Our approach has an excellent reproducibility of cell line derivation and the phenotypic stability of AFCL. These findings together with the previously published results that suggest presence of lineage plasticity in cells of PrE, allowing it to contribute to embryonic tissues at different stages of development [Kwon et al., 2008; Grabarek et al., 2012], would make potential human counterparts of such cell lines a candidate for banking and subsequent manipulation to prepare cell replacement therapies for regenerative medicine.
Supplementary Material
Acknowledments
We thank Dr Malcolm K. Brenner for critically reviewing the manuscript and for helpful discussions; Dr Leslie Silberstein for helpful discussions; Chris Threeton, Tatiana Goltsova and Amos Gaikwad of Texas Children’s Hospital Flow Cytometry Core facility for help with cell sorting experiments; Tuan Tran of the UT Health Sciences Center at Houston for microarray analysis; Laura Liles of BCM microarray core for help with microarray data analysis; Lihong Bu of IDDRC Cellular Imaging Core for help with Confocal Microscopy; James Broughman of Integrated Microscopy Core Facility at BCM for help with deconvolution microscopy; Kemi Cui of Advanced Cellular and Tissue Microscopy Core Laboratory at TMHRI for help with confocal microscopy; Dita Mayerova for help with teratoma assay; Kenneth Dunner of UTMDACC High Resolution Electron Microscopy Facility (supported by Institutional Core Grant #CA16672) for sample prep and imaging. Charlotte Su and Keri Watson for technical assistance. This work was supported by NHLBI K08 HL088508 through Brigham and Women’s Hospital and The Methodist Hospital, and Joint Program in Transfusion Medicine at Brigham and Women’s Hospital, Boston Children’s Hospital and Dana-Farber Cancer Institute and The Methodist Hospital Research Institute institutional funding.
List of abbreviations
- AF
Amniotic Fluid
- AFCL
Amniotic Fluid-derived Cell Lines
- AFP
Alpha-fetoprotein
- AFSC
Amniotic Fluid Stem Cells
- ALB
Albumin
- BMP4
Mouse Recombinant Bone Morphogenic Protein 4
- CXCR4
C-X-C Chemokine Receptor Type 4
- DE
Definitive Endoderm
- EGC
Embryonic Germ Cells
- EGF
Mouse Recombinant Epidermal Growth Factor
- ESC
Embryonic Stem Cells
- ExEn
Extraembryonic Endoderm
- FBS
Fetal Bovine Serum
- FGF-2
Human Recombinant Basic Fibroblast Growth Factor
- FACS
Fluorescence Activated Cell Sorting
- iPSC
Induced Pluripotent Stem Cells
- LIF
Mouse Recombinant Leukemia Inhibitory Factor
- MEF
Mouse Embryo Fibroblast
- MSC
Mesenchymal Stem Cells
- PCR
Polymerase Chain Reaction
- PE
Parietal Endoderm
- PrE
Primitive Endoderm
- PDGFRA
Platelet-Derived Growth Factor Receptor-α
- QPCR
Semi-Quantitative PCR
- RT
Reverse Transcription
- SCF
Mouse Recombinant Stem Cell Factor
- VE
Visceral Endoderm
- XEN
Extraembryonic Endoderm Cell Line
References
- Artus J, Douvaras P, Piliszek A, Isern J, Baron MH, Hadjantonakis AK. BMP4 signaling directs primitive endoderm-derived XEN cells to an extraembryonic visceral endoderm identity. Developmental biology. 2012;361(2):245–262. doi: 10.1016/j.ydbio.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Panthier JJ, Hadjantonakis AK. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development. 2010;137(20):3361–3372. doi: 10.1242/dev.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashman LK. The biology of stem cell factor and its receptor C-kit. The international journal of biochemistry & cell biology. 1999;31(10):1037–1051. doi: 10.1016/s1357-2725(99)00076-x. [DOI] [PubMed] [Google Scholar]
- Blair K, Wray J, Smith A. The liberation of embryonic stem cells. PLoS genetics. 2011;7(4):e1002019. doi: 10.1371/journal.pgen.1002019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 122(6):2005, 947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(11):5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Legros S, Artus J, Doss MX, Khanin R, Hadjantonakis AK, Foley A. A comparative analysis of extra-embryonic endoderm cell lines. PloS one. 2010;5(8):e12016. doi: 10.1371/journal.pone.0012016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuykin I, Schulz H, Guan K, Bader M. Activation of the PTHRP/adenylate cyclase pathway promotes differentiation of rat XEN cells into parietal endoderm, whereas Wnt/beta-catenin signaling promotes differentiation into visceral endoderm. Journal of cell science. 2013;126(Pt 1):128–138. doi: 10.1242/jcs.110239. [DOI] [PubMed] [Google Scholar]
- Davey RE, Zandstra PW. Spatial organization of embryonic stem cell responsiveness to autocrine gp130 ligands reveals an autoregulatory stem cell niche. Stem cells. 2006;24(11):2538–2548. doi: 10.1634/stemcells.2006-0216. [DOI] [PubMed] [Google Scholar]
- De Coppi P, Bartsch G, Jr., Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, Furth ME, Soker S, Atala A. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25(1):100–106. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- Debeb BG, Galat V, Epple-Farmer J, Iannaccone S, Woodward WA, Bader M, Iannaccone P, Binas B. Isolation of Oct4-expressing extraembryonic endoderm precursor cell lines. PloS one. 2009;4(9):e7216. doi: 10.1371/journal.pone.0007216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziadek M, Adamson E. Localization and synthesis of alphafoetoprotein in post-implantation mouse embryos. Journal of embryology and experimental morphology. 1978;43:289–313. [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim KS, Lanza R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem cells. 2010;28(4):704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- Feng X, Zhang J, Smuga-Otto K, Tian S, Yu J, Stewart R, Thomson JA. Protein kinase C mediated extraembryonic endoderm differentiation of human embryonic stem cells. Stem cells. 2012;30(3):461–470. doi: 10.1002/stem.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosden CM. Amniotic fluid cell types and culture. Br Med Bull. 1983;39(4):348–354. doi: 10.1093/oxfordjournals.bmb.a071847. [DOI] [PubMed] [Google Scholar]
- Gouon-Evans V, Boussemart L, Gadue P, Nierhoff D, Koehler CI, Kubo A, Shafritz DA, Keller G. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol. 2006;24(11):1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- Grabarek JB, Zyzynska K, Saiz N, Piliszek A, Frankenberg S, Nichols J, Hadjantonakis AK, Plusa B. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development. 2012;139(1):129–139. doi: 10.1242/dev.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray FL, Turner CG, Ahmed A, Calvert CE, Zurakowski D, Fauza DO. Prenatal tracheal reconstruction with a hybrid amniotic mesenchymal stem cells-engineered construct derived from decellularized airway. Journal of pediatric surgery. 2012;47(6):1072–1079. doi: 10.1016/j.jpedsurg.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Herrera B, Inman GJ. A rapid and sensitive bioassay for the simultaneous measurement of multiple bone morphogenetic proteins. Identification and quantification of BMP4, BMP6 and BMP9 in bovine and human serum. BMC cell biology. 2009;10:20. doi: 10.1186/1471-2121-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Tilly R. Cell interactions and endoderm differentiation in cultured mouse embryos. Journal of embryology and experimental morphology. 1981;62:379–394. [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132(4):567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kaviani A, Guleserian K, Perry TE, Jennings RW, Ziegler MM, Fauza DO. Fetal tissue engineering from amniotic fluid. Journal of the American College of Surgeons. 2003;196(4):592–597. doi: 10.1016/s1072-7515(02)01834-3. [DOI] [PubMed] [Google Scholar]
- Kimble RM, Trudenger B, Cass D. Fetal defaecation: is it a normal physiological process? J Paediatr Child Health. 1999;35(2):116–119. doi: 10.1046/j.1440-1754.1999.00314.x. [DOI] [PubMed] [Google Scholar]
- Klein JD, Fauza DO. Amniotic and placental mesenchymal stem cell isolation and culture. Methods Mol Biol. 2011;698:75–88. doi: 10.1007/978-1-60761-999-4_7. [DOI] [PubMed] [Google Scholar]
- Kodaira K, Imada M, Goto M, Tomoyasu A, Fukuda T, Kamijo R, Suda T, Higashio K, Katagiri T. Purification and identification of a BMP-like factor from bovine serum. Biochemical and biophysical research communications. 2006;345(3):1224–1231. doi: 10.1016/j.bbrc.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Kunath T, Arnaud D, Uy GD, Okamoto I, Chureau C, Yamanaka Y, Heard E, Gardner RL, Avner P, Rossant J. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132(7):1649–1661. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- Kunisaki SM, Armant M, Kao GS, Stevenson K, Kim H, Fauza DO. Tissue engineering from human mesenchymal amniocytes: a prelude to clinical trials. Journal of pediatric surgery. 2007;42(6):974–979. doi: 10.1016/j.jpedsurg.2007.01.031. discussion 979-980. [DOI] [PubMed] [Google Scholar]
- Kunisaki SM, Jennings RW, Fauza DO. Fetal cartilage engineering from amniotic mesenchymal progenitor cells. Stem cells and development. 2006;15(2):245–253. doi: 10.1089/scd.2006.15.245. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell. 2008;15(4):509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski AI, Knoepfler PS. Utf1: Goldilocks for ESC bivalency. Cell stem cell. 2012;11(6):732–734. doi: 10.1016/j.stem.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Tam WL, Zhang J, Ang HS, Jia H, Lipovich L, Ng HH, Wei CL, Sung WK, Robson P, Yang H, Lim B. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell stem cell. 2008;3(5):543–554. doi: 10.1016/j.stem.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Liu TM, Wu YN, Guo XM, Hui JH, Lee EH, Lim B. Effects of ectopic Nanog and Oct4 overexpression on mesenchymal stem cells. Stem cells and development. 2009;18(7):1013–1022. doi: 10.1089/scd.2008.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna SK, Rizzino A. Systems biology provides new insights into the molecular mechanisms that control the fate of embryonic stem cells. Journal of cellular physiology. 2012;227(1):27–34. doi: 10.1002/jcp.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70(5):841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Moschidou D, Drews K, Eddaoudi A, Adjaye J, De Coppi P, Guillot PV. Molecular Signature of Human amniotic Fluid Stem Cells During Fetal Development. Current stem cell research & therapy. 2013;8(1):73–81. doi: 10.2174/1574888x11308010009. [DOI] [PubMed] [Google Scholar]
- Moschidou D, Mukherjee S, Blundell MP, Drews K, Jones GN, Abdulrazzak H, Nowakowska B, Phoolchund A, Lay K, Ramasamy TS, Cananzi M, Nettersheim D, Sullivan M, Frost J, Moore G, Vermeesch JR, Fisk NM, Thrasher AJ, Atala A, Adjaye J, Schorle H, De Coppi P, Guillot PV. Valproic acid confers functional pluripotency to human amniotic fluid stem cells in a transgene-free approach. Mol Ther. 2012;20(10):1953–1967. doi: 10.1038/mt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen P. Flow cytometric DNA analysis of human cancer cell lines. Methods Mol Med. 2004;88:247–255. doi: 10.1385/1-59259-406-9:247. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Avivi A, Zimmer Y, Givol D, Yarden Y, Lonai P. Developmental expression of c-kit, a proto-oncogene encoded by the W locus. Development. 1990;109(4):911–923. doi: 10.1242/dev.109.4.911. [DOI] [PubMed] [Google Scholar]
- Ozek MM, Cinalli G, Maixner WJ. Spina Bifida: Management and Outcome. Springer-Verlag; Italia: 2008. [Google Scholar]
- Paca A, Seguin CA, Clements M, Ryczko M, Rossant J, Rodriguez TA, Kunath T. BMP signaling induces visceral endoderm differentiation of XEN cells and parietal endoderm. Developmental biology. 2012;361(1):90–102. doi: 10.1016/j.ydbio.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135(18):3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359(6395):550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- Roelandt P, Pauwelyn KA, Sancho-Bru P, Subramanian K, Bose B, Ordovas L, Vanuytsel K, Geraerts M, Firpo M, De Vos R, Fevery J, Nevens F, Hu WS, Verfaillie CM. Human embryonic and rat adult stem cells with primitive endoderm-like phenotype can be fated to definitive endoderm, and finally hepatocyte-like cells. PloS one. 2010;5(8):e12101. doi: 10.1371/journal.pone.0012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin CA, Draper JS, Nagy A, Rossant J. Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell stem cell. 2008;3(2):182–195. doi: 10.1016/j.stem.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefield JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(23):13726–13731. doi: 10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell stem cell. 2013;12(4):395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigman SA, Ahmed A, Shanti RM, Tuan RS, Valim C, Fauza DO. Sternal repair with bone grafts engineered from amniotic mesenchymal stem cells. Journal of pediatric surgery. 2009;44(6):1120–1126. doi: 10.1016/j.jpedsurg.2009.02.038. discussion 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Longaker MT, Wu JC. Human iPS cell-based therapy: considerations before clinical applications. Cell Cycle. 2010;9(5):880–885. doi: 10.4161/cc.9.5.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takakura N, Yoshida H, Ogura Y, Kataoka H, Nishikawa S, Nishikawa S. PDGFR alpha expression during mouse embryogenesis: immunolocalization analyzed by whole-mount immunohistostaining using the monoclonal anti-mouse PDGFR alpha antibody APA5. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 1997;45(6):883–893. doi: 10.1177/002215549704500613. [DOI] [PubMed] [Google Scholar]
- Takayama K, Inamura M, Kawabata K, Tashiro K, Katayama K, Sakurai F, Hayakawa T, Furue MK, Mizuguchi H. Efficient and directive generation of two distinct endoderm lineages from human ESCs and iPSCs by differentiation stage-specific SOX17 transduction. PloS one. 2011;6(7):e21780. doi: 10.1371/journal.pone.0021780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot NC, Sparks WO, Powell AM, Kahl S, Caperna TJ. Quantitative and semiquantitative immunoassay of growth factors and cytokines in the conditioned medium of STO and CF-1 mouse feeder cells. In Vitro Cell Dev Biol Anim. 2012;48(1):1–11. doi: 10.1007/s11626-011-9467-7. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282(5396):2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, J Itskovitz-Eldor,, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Becker RA, Hearn JP. Isolation of a primate embryonic stem cell line. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(17):7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Hearn JP. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biology of reproduction. 1996;55(2):254–259. doi: 10.1095/biolreprod55.2.254. [DOI] [PubMed] [Google Scholar]
- Turner CG, Klein JD, Gray FL, Ahmed A, Zurakowski D, Fauza DO. Craniofacial repair with fetal bone grafts engineered from amniotic mesenchymal stem cells. The Journal of surgical research. 2012;178(2):785–790. doi: 10.1016/j.jss.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, Jakt LM, Nishikawa S, Chiba T, Era T. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol. 2005;23(12):1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.