Abstract
Tethering a single lysozyme molecule to a carbon nanotube field effect transistor (FET) produced a stable, high-bandwidth transducer for protein motion. Electronic monitoring during 10-minute periods extended well beyond the limitations of fluorescence techniques to uncover dynamic disorder within a single molecule and establish lysozyme as a processive enzyme. On average, 100 chemical bonds are processively hydrolyzed, at 15-Hertz rates, before lysozyme returns to its nonproductive, 330-Hertz hinge motion. Statistical analysis differentiated single-step hinge closure from enzyme opening, which requires two steps. Seven independent time scales governing lysozyme’s activity we re observed. The pH dependence of lysozyme activity arises not from changes to its processive kinetics but rather from increasing time spent in either nonproductive rapid motions or an inactive, closed conformation.
Numerous experimental techniques have been developed to probe the dynamics of single molecules (1) and overcome the averaging effects of ensemble measurements. The most common experimental techniques employ fluorescence, encoding molecular motions and/or chemical reactions into a highly amplified photon flux that can be monitored and analyzed (2, 3). We show that single molecule dynamics can be monitored by attaching the molecule of interest to a FET device (Fig. 1A). Signal amplification was achieved by allowing the charged functionalities on the surface of the protein to electrostatically gate the underlying FET. By exploiting the low dimensionality and extreme local gate sensitivity of single-walled carbon nanotube (SWNT) FETs, molecular motions create changes in electrostatic potentials that can be converted into dynamically-changing electron fluxes. Such signals are similar to the photon fluxes in fluorescence experiments, but with advantageous bandwidth and shot noise limitations.
Figure 1.
(A) Schematic diagram of the single lysozyme being interrogated by a carbon nanocircuit. The partial PMMA coating is depicted in gr a y. (B) AFM topography of a SWNT FET before (inset) and after coating with the pyrene linker, lysozyme incubation, and washing to reduce nonspecifi c binding. The c ircle highlights the point of lysozyme attachment. (C) Response of current in a lysozyme device to electrolytic gating. (D) I (t) measured in phosphate buffer, with peptidoglycan substrate (25 μg/ml) added to the solution at t = 0. The ins et with a magnified time axis indicates a rapid response of <50 ms (inset).
The general concept of electronic transduction by low dimensional FETs has been explored using SWNT (4-7), silicon nanowire (8-11), nanocluster (12, 13), and graphene (14-16) devices. In most cases, slow changes in DC conductance have been suggestive of single-molecule detection, but the absence of dynamic responses undermined the general premise of high bandwidth detection. Recently, high bandwidth dynamic transduction has been achieved in two single-molecule electronic architectures. In the first, DNA molecules threading through solid state pores have generated high fidelity electronic signals that offer opportunities for DNA sequencing (15, 16). Separately, a more traditional FET architecture has leveraged amplification at SWNT point defects (17) to demonstrate kinetic binding and unbinding, both for molecules interacting directly with the defect site (18) and indirectly through an attached biomolecule (19, 20).
Here, we accomplished single molecule transduction very similar to the work of Sorgenfrei et al. (19, 20), but using a noncovalent bioconjugation strategy that provides a high device-fabrication yield. Rather than introducing a defect, tailoring its chemistry, and then conjugating that site to the target molecule, our method used noncovalent immobilization based on pyrene linkers (21). The pyrenes adhered to SWNTs through pi-pi stacking, and could provide dilute anchor points for further derivatization of the surface (22-24). In our particular implementation, a thiol from a single cysteine variant of T4 lysozyme (S90C) was covalently conjugated to a pyrene-maleimide anchor site [see Figs. S1 and S2 for characterization details (25)]. Similar site-specific conjugation of lysozyme to a solid surface has previously been shown to have no impact on the catalytic activity of lysozyme (26) used here as a template for introducing the S90C substitution; the pseudo-wild-type mutant of lysozyme (C54T, C97A) has identical activity and stability to wild-type lysozyme (27). To minimize potential perturbations to the enzyme, lysozyme was not conjugated to a His6 epitope, and was instead purified to >95% homogeneity by cation exchange followed by size exclusion chromatography. Standard biochemical conjugation protocols, followed by a strict rinsing protocol to minimize nonspecific binding, readily produced lysozyme attachments with a mean separation of 0.5 μm along the SWNT. Matching this empirical spacing to the SWNT channel length is a surprisingly simple method that produced active single molecule devices in 8 out of 10 fabrication attempts.
Atomic force microscopy before functionalization and after completion of the measurements (Fig. 1B) confirmed the presence of a single attached lysozyme. Lysozyme is approximately 7 nm in size, which made it easily distin guishable when attached to a 1 to 2 nm diameter SWNT [see Fig. S3 for additional examples and height profiles (25)]. In addition to microscopy, electrical characterization was performed at each fabrication stage. Figure 1C shows the gate-dependence of the source-drain current I(Vg) in a completed lysozyme device, measured with aqueous electrolyte (phosphate buffered saline) in direct contact with the SWNT sidewall (28). The shape of this curve reflects SWNT band structure, contact resistance effects, and the role of scattering induced by the pyrenes and attached lysozyme; these issues are discussed in greater detail in the SOM (25). In general, the coating technique added 1 to 2 megohms of series resistance to the SWNT conductor and shifts its I(Vg) curve, in accord with previous reports (4, 29, 30). Finally, Fig. 1D displays the typical device response I(t; Vg =0) upon introduction of the lysozyme substrate, peptidoglycan (Sigma-Aldrich). A polysaccharide found in bacterial cell walls, peptidoglycan consists of N-acetylmuramate (NAM)-N-acetylglucosamine (NAG) repeating units, and lysozyme catalyzes the hydrolysis of its glycosidic bonds (31). Chemoresistive responses of this nature are widely reported for nanodevices, and SWNT conductance can be quite sensitive to slight environmental changes, even when they are not decorated with proteins (32-34).
The time-averaged, DC response depicted in Fig. 1D underlies the classification of such devices as chemical or biological “sensors.” However, analysis of the dynamic response, not the DC level, can provide insights into conformational changes of the attached single protein. The magnitude of I(t) fluctuations increased immediately when peptidoglycan substrate was added, and, after one or more seconds of equilibration, these fluctuations developed into a two-level, random telegraph signal (RTS) that can be statistically analyzed. Control experiments probing bare SWNTs [Fig. S4 (25)] and lysozyme-free, pyrene-coated SWNTs [Fig. S5 (25)] revealed no RTS response to peptidoglycan substrate; further controls included 12 devices fabricated with either of two, inactive variants of lysozyme [Fig. S6 (25)], none of which exhibited RTS signals. In every experiment, the presence or absence of RTS fluctuations was a reliable predictor of the simultaneous presence of the peptidoglycan substrate and a SWNT-bound, catalytically functional variant of lysozyme.
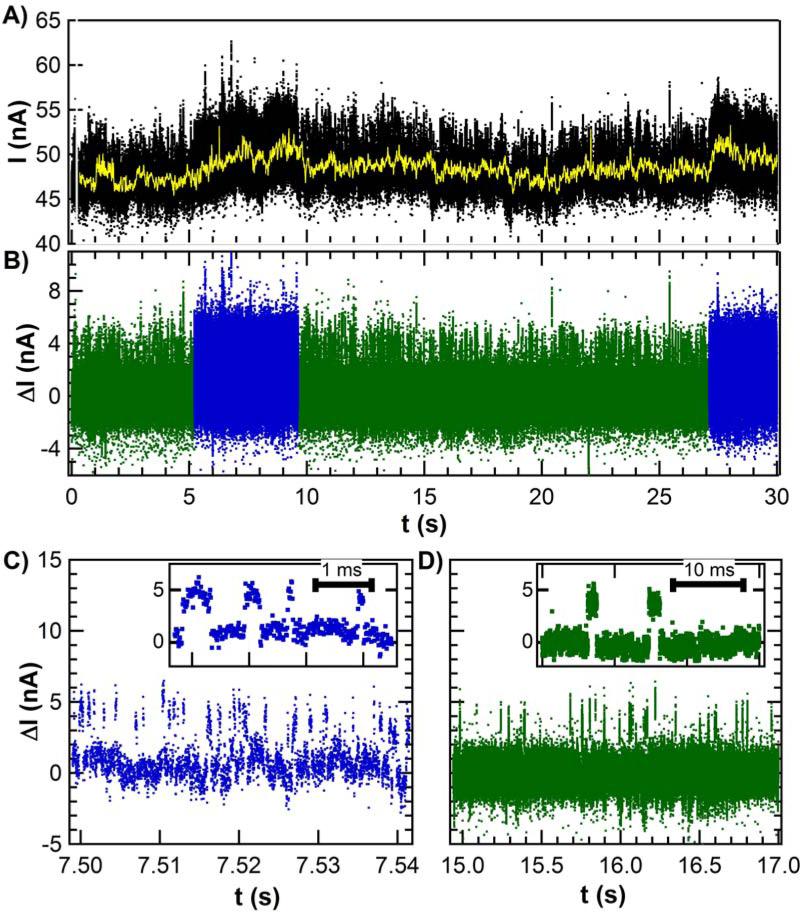
Fig. 2A shows 30 s of raw data collected from a single lysozyme device, along with the time-varying mean computed with a 10 Hz digital filter. The lowest frequency fluctuations had a 1/f spectral dependence and were indistinguishable from the noise incurred by a pristine SWNT in solution. Removing this slowly changing component, as shown in Fig. 2B, greatly simplified further analysis. The higher frequency components were revealed to be a two-level RTS with a constant amplitude distribution but two distinct RTS fluctuation rates. During some time periods, the RTS oscillated with a “fast” mean frequency of 316 Hz (Fig. 2C). At other times, the RTS oscillated with a “slow” mean frequency of 15.4 Hz (Fig. 2D). The data in Figure 2C is colored to differentiate the two types of RTS response and to emphasize their typical durations. Both the fast and the slow RTS segments lasted many seconds, suggestive of a long-term, two-state “memory” that was independent of the RTS itself. Accurate determination of the mean duration of this memory effect <τmem> required that measurements be conducted for at least 600 s.
Figure 2.
(A) Long duration I (t) sequences exhibit dynamic noise on top of low frequency fluctuations (yellow line) having a 1/f distribution. (B) Subtracting the meandering mean produces a filtered data set that clarifies the fluctuations as two-level, simplifies further analysis, and reveals that the two-level switching rates vary over 5 – 15 second periods. (C) The faster RTS oscillates about 300 times per second, whereas (D) the slower RTS oscillates 15 times per second. The insets show individual switching events for each case.
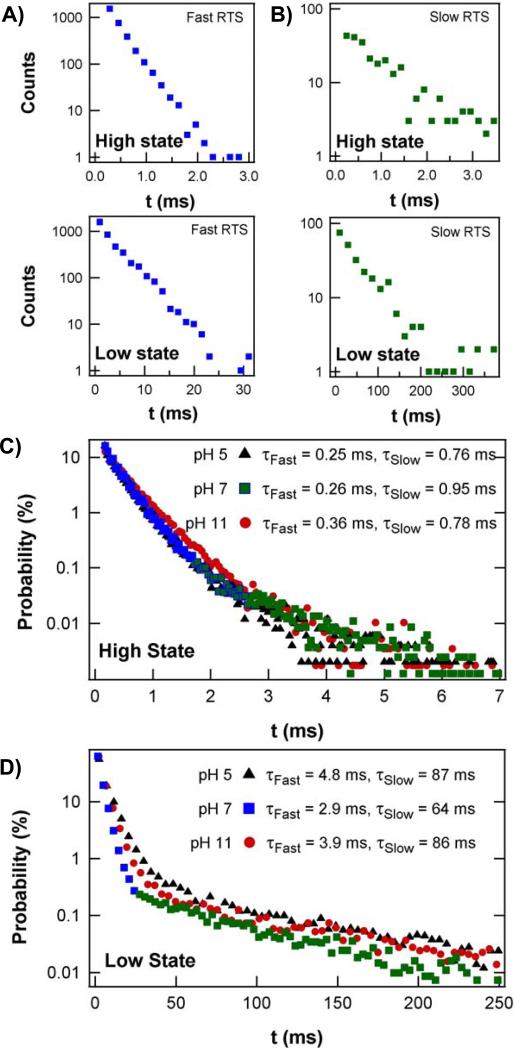
The sequences of fast and slow RTS oscillations could be separated for independent analysis. Each type of oscillation had a high and low current state characterized by durations τhi and τlo, respectively. Probability distributions for τhi and τlo are shown for a sequence of fast RTS fluctuations in Figure 3A and for slow RTS fluctuations in Figure 3B; the color scheme corresponds to the data in Fig. 2, B to D. All four distributions were well fit by single exponential time constants for periods of analysis shorter than <τmem>. Analysis of much longer time periods resulted in bi-exponential distributions (Fig. 3, C and D), and reflected the presence in data sets extending over hundreds of seconds of many sequences of both fast and slow RTS. However, the fast and slow rates were sufficiently different that they appeared as two distinguishable slopes in Fig. 3, C and D. As a guide, blue and green colors have been applied to portions of the distribution that correspond to the fast and slow fluctuations, respectively.
Figure 3.
(A, B) Direct comparison of the probability distributions of the high state (τhi) and low (τlo) state durations during (A) fast and (B) slow RTS switching at pH 7. Both pairs of distributions derive from uninterrupted, 15 second data sets, though many fewer counts occur during the slower activity represented by (B). (C, D) Probability distributions of continuous, 300 second data sets include as many as 105 transitions extending over many conversions between fast and slow RTS. The inclusion of both fast and slow RTS produces distinct, double-exponential distributions. Color has been applied to portions of the distributions in (A-D) to highlight correspondences with Figure 2. Measurements at three different pH values show that the high current state has almost no pH dependence (C). On the other hand, the low current state is at least 25% faster at pH 7 than at pH 5 or 11 (D).
Figures 3C and D further provide a comparison of the probability distributions acquired at different pH values. The stability of the lysozyme devices allowed statistics to be accumulated for many minutes at each pH, all from the same attached enzyme. Figure 3C shows that τhi was nearly independent over a pH range from 5 to 11; outside this pH range, lysozyme was no longer catalytically active (35-37). Figure 3D, however shows that τlo is much faster at pH 7 than at pH 5 or 11. The pH dependence of τlo in the long-duration, slow-switching region of the histogram, being much longer than the other three time constants, dominates the time-averaged properties of the lysozyme molecule.
Two important physical parameters could be calculated from the mean values of <τlo> and <τhi>. Their sum represents one complete oscillation, and defines a mean turnover rate for the activity, k = (<τlo> + <τhi>)−1. The calculation of enzymatic reaction rates from single molecule data has been reviewed by Xie (38). The ratio of <τlo> and <τhi> also determines an energy separation, ΔE, between the two physical states responsible for the high and low I(t) values. Boltzman statistics provides the relative thermodynamic probability of being in one state versus the other as ΔE = KBT ln(<τhi> / <τlo>). In total, we identified five independent parameters <τhi>, <τlo>, <τmem>, k, and ΔE, all of which change when the lysozyme switches from its fast RTS state to its slow RTS state. Table 1 summarizes these parameters and their pH dependence for a single lysozyme molecule.
Table 1.
Lysozyme activity rates.
| Parameter | pH 5 | pH 7 | pH 11 | ||||
|---|---|---|---|---|---|---|---|
| Processing (slow) | <τhi>, ms | 0.76 | ± 0.05 | 0.95 | ± 0.08 | 0.78 | ± 0.09 |
| <τlo>, ms | 87 | ± 3.0 | 64 | ± 2.0 | 86 | ± 3.0 | |
| ΔE, kcal/mol | 2.84 | 2.53 | 2.82 | ||||
| k, Hz | 11.4 | 15.4 | 11.5 | ||||
| <τmem>, s | 9.3 | ± 5.1 | 8.0 | ± 3.0 | 12.0 | ± 4.4 | |
| % time in state | 16.3% | 41.1% | 21.1% | ||||
| time-averaged catalytic rate, Hz | 1.8 | 6.3 | 2.4 | ||||
| Nonproductive (fast) | <τhi>, ms | 0.25 | ± 0.01 | 0.26 | ± 0.01 | 0.36 | ± 0.01 |
| <τlo>, ms | 4.80 | ± 0.35 | 2.90 | ± 0.10 | 3.90 | ± 0.17 | |
| ΔE, kcal/mol | 1.77 | 1.45 | 1.43 | ||||
| k, Hz | 198 | 316 | 235 | ||||
| <τmem>, s | 6.2 | ± 4.0 | 7.9 | ± 2.3 | 5.4 | ± 1.8 | |
| % time in state | 72.4% | 52.1% | 63.8% | ||||
| Inactive | <τmem>, s | 0.83 | ± 0.63 | 0.72 | ± 0.25 | 0.96 | ± 0.53 |
| % time in state | 11.3% | 6.8% | 15.0% | ||||
Table 1 further lists the overall percentage of time spent in the fast or slow RTS states. This percentage is yet another independent parameter, being a nontrivial combination of the duration <τmem> and the regularity with which each behavior is observed. At pH 7, <τmem> was nearly equal for the fast and the slow RTS state, and the time in either state approached 50%. At non-neutral pH values, however, multiple changes skewed this balance. First, the number of inactive periods in which no switching was observed nearly doubled. This doubling, amplified by a modest increase in <τmem> of the inactive duration, resulted in a rapidly growing proportion of total inactive time. More importantly, we observed that inactive periods always interrupted sequences of fast RTS oscillations and doubled the number of fast RTS intervals observed while only modestly decreasing their duration <τmem>. The increased time spent in inactive or fast RTS intervals both came at a cost to the percentage of time spent in the slow-switching RTS state, which was reduced to 19.7% at pH 11 and 15.7% at pH 5. Interestingly, this decrease occurred despite a substantial increase in <τmem>, which considered in isolation would indicate an improving stability of the slow RTS state. We further note that the fast and slow RTS oscillation rates k both decreased by 25 to 35% away from pH 7; because they both decreased proportionally, the k values only minimally contributed to the pH dependence of the time spent in the fast or slow RTS states.
The conductance signal of a single lysozyme device allowed us to directly determine multiple independent parameters, including seven independent time constants and their pH dependence, all without foreknowledge of the properties of either the enzyme or the SWNT. Out of 50 single-molecule devices, the presence of fast and slow RTS components was very reproducible, although there were variations in the numerical rates observed. For example, among seven lysozyme devices with high quality signal-to-noise, the rates of the slow RTS state at pH 7 varied from 10 to 50 Hz, with a mean k = 24 ± 15 Hz. The rate of the fast RTS state varied from 127 to 461 Hz, with a mean k = 284 ± 127 Hz.
Next, we applied these empirical observations to examine the mechanism and catalytic activity of lysozyme, through detailed analysis of the electronic device signal from lysozyme-tethered nanocircuits. As noted above and demonstrated in Figures S4, S5, and S6 (25), the RTS can be ascribed to the presence of protein-substrate interactions. By comparing the I(t) signal to lysozyme dynamics known from ensemble and single molecule FRET experiments (39-42), we can draw several parallels (e.g., lysozyme remains static in the absence of substrate). During substrate processing, lysozyme undergoes an 8 Å, hinge-like mechanical motion with two domains closing around the substrate (41-45). FRET observations reveal that this motion occurs at two different rates: a slow hinge oscillation of 20-90 Hz corresponding to enzymatic turnover events, or else a more rapid, nonproductive movement at 200-400 Hz (37, 41, 42).
These FRET rates are in excellent agreement with our fast RTS and slow RTS oscillations, and the interconversion rate matches our <τmem> values. Thus, we conclude that the two-level electronic signal is caused by the lysozyme hinge motion, with slow RTS oscillations resulting from the transduction of catalytic turnover events and fast RTS oscillations corresponding to lysozyme’s nonproductive binding events. These kinetic rates using surface-bound lysozyme might differ from bulk rates, but FRET measurements with freely diffusing lysozyme and surface-bound peptidoglycan (40) yield the same range of rates, suggesting that the consequences of tethering the lysozyme to a surface are minor.
The agreement demonstrates the equivalence of the lysozyme device data with FRET measurements, but in other ways the SWNT-lysozyme device data is more informative. Fluorophore bleaching and quenching limit the duration of FRET measurements on a single molecule, and constrain the ability to observe slow conformational interconversions by FRET (37). The I(t) measurement duration is not similarly limited, and the <τmem> values in Table 1 represent hundreds of such events by the same single molecule. Using such long time-scales, we directly observed the same molecule changing from its productive conformation to its unproductive one. We also obtained the average percentage time spent in the slow RTS state, which when multiplied by k gave a time-averaged, effective catalytic rate for the single molecule. By collecting a true average over many conformational changes, this single molecule rate approached the kinetics of an ensemble (Table 1). Furthermore, our I(t) records extend this single-molecule insight to different pH conditions.
Furthermore, our measurements of a long duration <τmem> support a processive catalytic mechanism in which each lysozyme hydrolyzes on average 100 glysodic bonds before dissociation and reassociation of the substrate (40). During the slow RTS state, no breaks in activity were observed, and so no product dissociation and substrate rebinding occurred. Furthermore, the substrate remained bound during both nonproductive fast RTS and the substrate-bound inactive state before returning to processing.
Our long duration data sets also enable analysis of the statistical variance of the τlo and τhi values. Any single-step Poisson process has a statistical variance σ2 = <τ>2, and the normalized variance
is a powerful tool in single molecule studies for distinguishing hidden intermediate steps along a reaction coordinate (46-48). As shown in Table 2, analysis of individual τlo durations concludes that r ≈ 1, indicating that the physical processes underlying the transition from Ilo to Ihi is in fact governed by a simple, single step Poisson process. This conclusion remains true for both fast and slow RTS data at the different pH values tested, even though τlo differs by a factor of 20. In contrast, the physical process underlying the Ihi to Ilo transition has values of r < 1, indicating to more complex processes. For example, n identical Poisson processes in succession will produce a distribution of durations τ that have a variance r = 1/n (47, 49). Thus, lysozyme forms a closed conformation in a single step, but the transition back to its open configuration requires at least two steps. Although this finding is the same for both fast and slow RTS oscillations, the extra intermediate step required for opening is not necessarily the same in both cases. During processive sequences, the intermediate step could play a role in the catalysis and substrate turnover (50). During fast sequences of nonproductive binding, it might be involved in repositioning the substrate within the active site in an attempt to find a reactive bond for hydrolysis. Notably, the inactive state of lysozyme occurs when the enzyme closes around the substrate.
Table 2.
Normalized variances of lysozyme rates.
| Parameter | pH 5 | pH 7 | pH 11 | ||||
|---|---|---|---|---|---|---|---|
| Processing (slow) | rhi | 0.68 | ± 0.15 | 0.74 | ± 0.12 | 0.60 | ± 0.15 |
| rio | 1.00 | ± 0.18 | 1.06 | ± 0.15 | 1.11 | ± 0.23 | |
| Nonproductive (fast) | rhi | 0.48 | ± 0.10 | 0.43 | ± 0.06 | 0.61 | ± 0.08 |
| rio | 0.97 | ± 0.13 | 0.99 | ± 0.09 | 1.00 | ± 0.10 | |
Finally, we conclude with a brief discussion of the device’s transduction mechanism. The current step that occurs with each opening and closing of the lysozyme is surely not additional carriers flowing into the SWNT from the protein or the electrolyte. The entire current I(t) flows from source to drain electrode, with variability introduced by lysozyme’s hinge-bending motion. The change from open to closed conformations moves lysozyme’s charge residues (51). The surrounding electrolyte, which has a Debye length of 0.8 nm, screens most of the residues but not two, positively charged residues, Lys83 and Arg119, that are located on the protein surface within 1.0 – 1.2 nm of the SWNT attachment site. Both of these residues lie close enough to the SWNT that their movement can electrostatically gate the channel conductance (52-54), according to the mechanism most often attributed to similar sensing experiments (6, 29, 55).
To test the applicability of this mechanism, we compare the transduction by different SWNT devices. Using the average slope dI/dVg indicated in Fig. 1C, the RTS magnitude ΔI of a fluctuation can be converted into an effective swing in the applied gate, ΔVg = ΔI(dI/dVg)−1. For the device in Fig. 2, for example, ΔI = 5 nA and ΔVg = 0.20 V. Table S2 summarizes device properties of five semiconducting SWNT devices and five metallic SWNT ones, each having a different contact resistance and mean current (25). For these ten devices, the switching magnitudes ΔI vary from 3 to 300%, but the calculated values ΔVg are all narrowly clustered around ΔVg = 0.19 ± 0.02 V. This reproducibility in ΔVg demonstrates that the signal transduction mechanism is indeed electrostatic, with every protein-pyrene attachment modulating its underlying SWNT channel with the same effective field. Metallic SWNTs display the smallest ΔI signals, but only because of their modest sensitivity to gating.
It would be a mistake, however, to infer that the lysozyme is gating the entire SWNT, or to convert ΔVg into a carrier concentration. T he pyrene-protein attachment site is a highly localized scattering center that, because the SWNT is quasi-one dimensional, can modulate the entire channel conductance. The resistance increase that occurs upon conjugation (Table S2) is believed to be concentrated primarily at the attachment site, similar to the case of a defect being introduced (17-19). Scanning probe measurements prove these local scattering sites to be strongly gate-dependent conduction barriers (56). During substrate binding and release, the charged sidechains of Lys83 and Arg119 move by 1 - 2 Å relative to the SWNT (41-44, 57). This motion is sufficient to modulate the local chemical potential in the SWNT and account for our observed ΔI (56).
This nanocircuit architecture is complement ary to more traditional fluorescence techniques, but with the advantages that fluorescent labels are not required, the transduction mechanism does not bleach, and electronic bandwidths extend temporal resolution into the single microsecond regime. There advantages provide a framework for exploring dynamics of other molecules with charged functionalities.
Supplementary Material
Acknowledgements
This work was sponsored by the NCI of the NIH (R01 CA133592-01) and NSF (DM R-0801271, ECCS-0802077, and the Center for C hemical Innovation on Chemistry at the Space-Time Limit CHE-0802 913).
References
- 1.Claridge SA, Schwartz JJ, Weiss PS. ACS Nano. 2011;5:693. doi: 10.1021/nn103298x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Min W, et al. Accounts of Chemical Research. 2005;38:923. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- 3.Roy R, Hohng S, Ha T. Nat. Methods. 2008;5:507. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Star A, Gabriel JCP, Bradley K, Gruner G. Nano Lett. 2003;3:459. [Google Scholar]
- 5.Star A, et al. Organic Letters. 2004;6:2089. doi: 10.1021/ol0495826. [DOI] [PubMed] [Google Scholar]
- 6.Besteman K, Lee JO, Wiertz FGM, Heering HA, Dekker C. Nano Lett. 2003;3:727. [Google Scholar]
- 7.Gruner G. Analytical and Bioanalytical Chemistry. 2006;384:322. doi: 10.1007/s00216-005-3400-4. [DOI] [PubMed] [Google Scholar]
- 8.Cui Y, Wei QQ, Park HK, Lieber CM. Science. 2001;293:1289. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 9.Patolsky F, et al. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14017. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patolsky F, et al. Science. 2006;313:1100. doi: 10.1126/science.1128640. [DOI] [PubMed] [Google Scholar]
- 11.Patolsky F, Zheng GF, Lieber CM. Analytical Chemistry. 2006;78:4260. doi: 10.1021/ac069419j. [DOI] [PubMed] [Google Scholar]
- 12.Park SJ, Taton TA, Mirkin CA. Science. 2002;295:1503. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 13.Xiao Y, Patolsky F, Katz E, Hainfeld JF, Willner I. Science. 2003;299:1877. doi: 10.1126/science.1080664. [DOI] [PubMed] [Google Scholar]
- 14.Schedin F, et al. Nat. Mat. 2007;6:652. doi: 10.1038/nmat1967. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, et al. Nat Nano. 2010;5:868. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsutsui M, et al. Sci. Rep. 1. 2011 doi: 10.1038/srep00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldsmith BR, et al. Science. 2007;315:77. doi: 10.1126/science.1135303. [DOI] [PubMed] [Google Scholar]
- 18.Goldsmith BR, Coroneus JG, Kane AA, Weiss GA, Collins PG. Nano Lett. 2008;8:189. doi: 10.1021/nl0724079. [DOI] [PubMed] [Google Scholar]
- 19.Sorgenfrei S, et al. Nat Nano. 2011;6:126. doi: 10.1038/nnano.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorgenfrei S, Chiu C.-y., Johnston M, Nuckolls C, Shepard KL. Nano Lett. 2011;11:3739. doi: 10.1021/nl201781q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen RJ, Zhan YG, Wang DW, Dai HJ. J. Am. Chem. Soc. 2001;123:3838. doi: 10.1021/ja010172b. [DOI] [PubMed] [Google Scholar]
- 22.Artyukhin AB, Bakajin O, Stroeve P, Noy A. Langmuir. 2004;20:1442. doi: 10.1021/la035699b. [DOI] [PubMed] [Google Scholar]
- 23.Zhao YL, Hu LB, Stoddart JF, Gruner G. Adv. Mater. 2008;20:1910. [Google Scholar]
- 24.Perello DJ, et al. J. App. Phys. 2009;124309:5. [Google Scholar]
- 25.Materials, methods, and additional control data are available as supporting material on Science Online., pp.
- 26.Heredia KL, et al. J. Am. Chem. Soc. 2005;127:16955. doi: 10.1021/ja054482w. [DOI] [PubMed] [Google Scholar]
- 27.Matsumura M, Matthews BW. Science. 1989;243:792. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- 28.Rosenblatt S, et al. Nano Lett. 2002;2:869. [Google Scholar]
- 29.Chen RJ, et al. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4984. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen RJ, et al. J. Am. Chem. Soc. 2004;126:1563. doi: 10.1021/ja038702m. [DOI] [PubMed] [Google Scholar]
- 31.Meroueh SO, et al. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4404. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong J, et al. Science. 2000;287:622. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- 33.Collins PG, Bradley K, Ishigami M, Zettl A. Science. 2000;287:1801. doi: 10.1126/science.287.5459.1801. [DOI] [PubMed] [Google Scholar]
- 34.Zhang T, Mubeen S, Myung NV, Deshusses MA. Nanotechnology 19. 2008 doi: 10.1088/0957-4484/19/33/332001. [DOI] [PubMed] [Google Scholar]
- 35.Tsugita A, Inouye M, Terzaghi E, Streisin G. Journal of Biological Chemistry. 1968;243:391. [PubMed] [Google Scholar]
- 36.Jensen HB, Kleppe K. European Journal of Biochemistry. 1972;28:116. doi: 10.1111/j.1432-1033.1972.tb01891.x. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Hu D, Vorpagel ER, Lu HP. J. Phys. Chem. B. 2003;107:7947. [Google Scholar]
- 38.Xie SN. Single Molecules. 2001;2:229. [Google Scholar]
- 39.Lu HP, Xun LY, Xie XS. Science. 1998;282:1877. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 40.Hu D, Lu HP. Biophysical Journal. 2004;87:656. doi: 10.1529/biophysj.104.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu HP. Current Pharmaceutical Biotechnology. 2004;5:261. doi: 10.2174/1389201043376887. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Lu HP. J. Phys. Chem. B. 2010;114:6669. doi: 10.1021/jp1004506. [DOI] [PubMed] [Google Scholar]
- 43.Arnold GE, Manchester JI, Townsend BD, Ornstein RL. Journal of Biomolecular Structure & Dynamics. 1994;12:457. doi: 10.1080/07391102.1994.10508751. [DOI] [PubMed] [Google Scholar]
- 44.de Groot BL, Hayward S, van Aalten DMF, Amadei A, Berendsen HJC. Proteins-Structure Function and Genetics. 1998;31:116. doi: 10.1002/(sici)1097-0134(19980501)31:2<116::aid-prot2>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 45.Faber HR, Matthews BW. Nature. 1990;348:263. doi: 10.1038/348263a0. [DOI] [PubMed] [Google Scholar]
- 46.Svoboda K, Mitra PP, Block SM. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11782. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnitzer MJ, Block SM. Cold Spring Harbor Symposia on Quantitative Biology. 1995;60:793. doi: 10.1101/sqb.1995.060.01.085. [DOI] [PubMed] [Google Scholar]
- 48.Xu WL, Kong JS, Chen P. Journal of Physical Chemistry C. 2009;113:2393. [Google Scholar]
- 49.Schnitzer MJ, Block SM. Nature. 1997;388:386. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 50.Bhabha G, et al. Science. 2011;332:234. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mchaourab HS, Oh KJ, Fang CJ, Hubbell WL. Biochemistry. 1997;36:307. doi: 10.1021/bi962114m. [DOI] [PubMed] [Google Scholar]
- 52.Stern E, et al. Nano Lett. 2007;7:3405. doi: 10.1021/nl071792z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heller I, et al. Nano Lett. 2008;8:591. doi: 10.1021/nl072996i. [DOI] [PubMed] [Google Scholar]
- 54.Prisbrey L, Schneider G, Minot E. J. Phys. Chem. B. 2010;114:3330. doi: 10.1021/jp910946v. [DOI] [PubMed] [Google Scholar]
- 55.Li C, et al. J. Am. Chem. Soc. 2005;127:12484. doi: 10.1021/ja053761g. [DOI] [PubMed] [Google Scholar]
- 56.Hunt SR, Wan D, Khalap VR, Corso BL, Collins PG. Nano Lett. 2011;11:1055. doi: 10.1021/nl103935r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brooks B, Karplus M. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:4995. doi: 10.1073/pnas.82.15.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shiu Y-J, et al. Biophysical Journal. 2008;94:4828. doi: 10.1529/biophysj.107.124214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou RQ, Chen SG, Recsei P. Anal. Biochem. 1988;171:141. doi: 10.1016/0003-2697(88)90134-0. [DOI] [PubMed] [Google Scholar]
- 60.Müller A, et al. Angew. Chem.-Int. Edit. 2000;39:3414. [Google Scholar]
- 61.An L, Owens JM, McNeil LE, Liu J. J. Am. Chem. Soc. 2002;124:13688. doi: 10.1021/ja0274958. [DOI] [PubMed] [Google Scholar]
- 62.Yu WJ, et al. New Journal of Physics 10. 2008 [Google Scholar]
- 63.Shim M, Kam NWS, Chen RJ, Li YM, Dai HJ. Nano Lett. 2002;2:285. [Google Scholar]
- 64.Collins PG, Fuhrer MS, Zettl A. Appl. Phys. Lett. 2000;76:894. [Google Scholar]
- 65.Kingrey D, Khatib O, Collins PG. Nano Lett. 2006;6:1564. doi: 10.1021/nl060058x. [DOI] [PubMed] [Google Scholar]
- 66.Mann D, Javey A, Kong J, Wang Q, Dai HJ. Nano Lett. 2003;3:1541. [Google Scholar]
- 67.Kim W, et al. Appl. Phys. Lett. 2005;87:173101. [Google Scholar]
- 68.Chen Z, Appenzeller J, Knoch J, Lim Y, Avouris P. Nano Lett. 2005;5:1497. doi: 10.1021/nl0508624. [DOI] [PubMed] [Google Scholar]
- 69.Sun XY, Sun YG. Journal of Materials Science & Technology. 2008;24:569. [Google Scholar]
- 70.Coroneus JG, et al. ChemPhysChem. 2008;9:1053. doi: 10.1002/cphc.200700863. [DOI] [PubMed] [Google Scholar]
- 71.Grabarek Z, Gergely J. Anal. Biochem. 1990;185:131. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- 72.Goldsmith BR., Ph.D. University of California at Irvine; 2008. [Google Scholar]
- 73.Coroneus JG., Ph.D. University of California at Irvine; 2008. [Google Scholar]
- 74.Zechel DL, Withers SG. Accounts of Chemical Research. 2000;33:11. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.