Abstract
Liposarcoma is the most common soft tissue sarcoma, but little is known about the genomic basis of this disease. Given the low cell content of this tumor type, we utilized flow cytometry to isolate the diploid normal and aneuploid tumor populations from a well-differentiated liposarcoma prior to array comparative genomic hybridization and whole genome sequencing. This work revealed massive highly focal amplifications throughout the aneuploid tumor genome including MDM2, a gene that has previously been found to be amplified in well-differentiated liposarcoma. Structural analysis revealed massive rearrangement of chromosome 12 and 11 gene fusions, some of which may be part of double minute chromosomes commonly present in well-differentiated liposarcoma. We identified a hotspot of genomic instability localized to a region of chromosome 12 that includes a highly conserved, putative L1 retrotransposon element, LOC100507498 which resides within a gene cluster (NAV3, SYT1, PAWR) where 6 of the 11 fusion events occurred. Interestingly, a potential gene fusion was also identified in amplified DDR2, which is a potential therapeutic target of kinase inhibitors such as dastinib, that are not routinely used in the treatment of patients with liposarcoma. Furthermore, 7 somatic, damaging single nucleotide variants have also been identified, including D125N in the PTPRQ protein. In conclusion, this work is the first to report the entire genome of a well-differentiated liposarcoma with novel chromosomal rearrangements associated with amplification of therapeutically targetable genes such as MDM2 and DDR2.
Introduction
Adipocytic tumors are classified by the World Health Organization (WHO) International Agency for Research on Cancer (IARC) into benign, intermediate and malignant classes [1]. Intermediate tumors include atypical lipomatous tumors/well-differentiated liposarcoma (WDLS) that constitute locally aggressive mature adipocytes [1]–[3]. Among those diagnosed with liposarcoma, 40–45% will have WDLS [4], [5]. Surgical removal of the tumor is the primary treatment modality for WDLS as generally WDLS do not respond to chemotherapy and therapeutic options are limited for those with metastatic disease [4], [6]. WDLS occur commonly in the retroperitoneum and in the extremities, but can also occur in the mediastinum and paratesticular region [7]. WDLS tumors in the retroperitoneum or mediastinum are more likely to recur than tumors at other sites with this frequent recurrence resulting in death from local effects of the disease [2], [4]. While WDLS does not typically metastasize, it can dedifferentiate and progress to a more aggressive and potentially metastatic tumor [2], [4].
A number of cytogenetic abnormalities have been associated with WDLS. Supernumerary rings and giant marker chromosomes are the most frequent cytogenetic abnormalities associated with WDLS [8]–[10] that often contain amplifications of chromosome 12, specifically in the 12q13-q15 region [3], [11]. Interestingly, benign lipomas also contain chromosomal rearrangements in the 12q14-q15 region [3], [12]. A number of genes have been identified in these amplified regions including those with a potential oncogenic role including: MDM2, CDK4, HMGA2, and TSPAN31 [9], [10], [13]–[15]. While amplification of MDM2 and CDK4 frequently occur together [9], [10], [13], [15], the amplicons for MDM2 and CDK4 have been identified as being separate [13]. Patients with amplification of MDM2 but no amplification of CDK4 have a more favorable prognostic outlook than patients with amplifications in both genes [16]. MDM2 amplification has also been found to occur together with amplification of a neighboring gene, CPM [17].
Amplifications have also been found in genes flanking CDK4 (STAT6, B4GALNT1, OS9, CENTG1, TSPAN31, METTL1 and XRCC6BP1) and MDM2 (FRS2, CCT2, LRRC10, and BEST3) [15]. Additional genes potentially of interest located in the 12q13-q15 region include: amplified genes HMGIC and GLI, as well as a non-amplified gene, CHOP (also known as DDIT3) that is part of mixoid liposarcoma translocations [9], [13], [17], [18]. Amplifications have also been identified in regions 12q21-q22 and associated with overexpression of CCDC131, GLIPR1, BBS10, ZDHHC17, KITLG and WDR51B [15].
While the above studies have led to a greater understanding of the genetics underlying WDLS, they have not significantly advanced the standard of care for WDLS patients. In order to better understand the genetic basis of disease in liposarcoma, and to identify potential therapeutic targets, we sought to complete whole genome sequencing (WGS) in a WDLS patient. One challenge of studying the liposarcoma genome is the low DNA content in the tissue due to the majority of the cell volume consisting of lipid. As a result, DNA yields are low utilizing standard tumor purification and DNA extraction methods. In order to improve tumor purity and to extract DNA from highly purified tumor cells, we utilized flow cytometry to isolate the diploid and aneuploid populations from the tumor sample prior to array comparative genomic hybridization (aCGH) and whole genome sequencing (WGS) of a WDLS patient. This work revealed 7 damaging single nucleotide variants in 7 genes, massive amplification across multiple chromosomes, massive rearrangement on chromosome 12, the presence of a putative retrotransposon and 11 fusions between genes.
Materials and Methods
Samples
Samples were acquired after written informed consent was obtained in compliance with, and approval by, the Mayo Clinic Institutional Review Board. Peripheral blood was acquired for sequencing of the constitutional genome. DNA was isolated from peripheral blood with the Puregene kit (Qiagen) following the manufacturer's protocol. The tumor was acquired from an abdominal mass debulking and flash frozen. The tissue was then minced in DAPI (4, 6-diamindine-2phenylindole dihydrochloride) stock solution at 10 μg/mL, passed through a 40 μM Nylon Cell Strainer filter (BD Biosciences) to disaggregate nuclei and prepare a single particle suspension. Minced and disaggregated nuclei were sorted based on DNA content with the BD Influx™ flow cytometer (BD Biosciences) equipped with UV excitation at 358 nm and emission at 460 nm. This resulted in >95% purity of tumor cells in sorted samples (Figure S1). A minimum of 10,000 events (after exclusion of doublets) were collected for the MultiCycle analysis and a total of 953,000 events collected in three fractions for DNA extraction. Samples were analyzed at rates below 1000 cells/second in order to yield a good signal of discrimination between singlets and doublets. In order to identify the position of the nuclei with the normal diploid amount of DNA, reference cells obtained from normal fibroblast of healthy volunteers were included. DAPI binds stoichiometrically to the DNA. The stained material has incorporated an amount of dye proportional to the amount of DNA. DNA content analysis included determination of the mean channel fluorescence and the coefficient of variation (CV) of the diploid and aneuploid G0/G1 and G2/M peaks. DNA content and cell cycle were analyzed using the software program MultiCycle (Phoenix Flow System). The ploidy of the aneuploid population was 2.3N and included a large (14%) G2/M (4.6N) fraction. DNA extraction was performed separately for each of the sorted aneuploid and diploid populations with the QIAGEN QIAamp DNA Micro Kit according to the manufacturer's protocol. Samples were eluted twice from each column with 100ul of water for a final volume of 200ul. In order to capture residual nuclei and maximize the final volume of genomic DNA, the original microcentrifuge tubes were rinsed with water, pooled and extracted using the protocol above. The products of this second “rescue” extraction were then added to the initial pooled, extracted samples. Following ethanol precipitation the samples were resuspended in water.
Array Comparative Genomic Hybridization
Array CGH (aCGH) was conducted as described previously [19]. Briefly, prior to hybridization 100 ng of genomic DNA from each sorted fraction and a commercial 46, XX reference (Promega) were amplified with GenomiPhi amplification kit (GE Healthcare). Tumor and control DNA samples were digested with DNAseI and fragmented DNA was labeled using the BioPrime Array CGH Genomic Labeling kit (Invitrogen). Labeled DNA from each sorted fraction was pooled with the labeled DNA from the reference then hybridized to the Agilent 400K Human Genome CGH Microarray according to the manufacturer's protocol.
Paired End Library Preparation
Paired-end libraries were prepared using NEBNext DNA sample preparation kit following the manufacturer's protocol (New England Biolab). Briefly, DNA was fragmented using the Covaris E210 sonicator to generate double-stranded DNA fragments with a fragment size of 400–500 bp. The ends were repaired, phosphorylated, followed by 3′ end adenylation. Paired end DNA adaptors were ligated and the resulting constructs size selected for ∼500 bp fragments. The excised gel band was purified following manufacturer's protocol utilizing Qiagen Gel Extraction Kits. These fragments were enriched with 12 cycles of PCR. The concentration and size distribution of the libraries was determined on an Agilent Bioanalyzer DNA 1000 chip.
Libraries were loaded onto paired end flow cells and sequenced as 101 by 2 paired end indexed reads on Illumina HiSeq 2000 and base-calling performed using Illumina's RTA version 1.7.45.0.
Whole Genome Sequencing Analysis
Alignment and variant detection of the WGS reads were performed utilizing TREAT (Targeted RE-sequencing Annotation Tool) [20]. TREAT is an analytical tool that utilizes open source tools in a pipeline that aligns, identifies and annotates variants. Raw sequence reads were aligned to hg18 with Burrows-Wheeler Aligner (BWA). Post-alignment processing included local realignment with Genome Analysis Toolkit (GATK) [21].
Single nucleotide variants (SNV) and insertions/deletions (indel) were detected utilizing GATK [21] and SNVMix [22]. Identified variants were then placed in the custom annotation pipeline and SNV and indel reports created. SNVMix filtered (probability ≥0.8) variant calls from TREAT were used to extract tumor only variants. Annotation of these files utilized SeattleSeq (http://gvs.gs.washington.edu/SeattleSeqAnnotation/) for variant classification, as well as Sorting Intolerant from Tolerant (SIFT) [23] and Polyphen-2 [24] (http://genetics.bwh.harvard.edu/pph2/) for functional impact prediction of the variants. Variants were then visually validated in the Integrative Genomics Viewer (IGV) [25] and any reads with the variant allele present in the normal were removed. Candidate SNV were then selected for validation by capillary sequencing if they were predicted to result in a damaging mutation by SIFT/Polyphen2.
Detection of Structural Variants
Potential gene fusions were detected with two methods: an in-house computational tool and visual confirmation of CGH breakpoints in the WGS data. Breakpoints for the amplifications observed in the aCGH data were visually confirmed with IGV in the WGS data to determine potential breakpoints and gene fusions. In addition, bioinformatics identified anomalous reads using a sliding window type approach quantifying the number of anomalous reads pointing to a distinct window elsewhere in the genome. Window sizes were based on the insert size. Regions where the reference or germline genome aligns with either a high number of anomalous reads or a high number of poorly mapped reads were ignored. All anomalous read pairs mapping to coding regions were identified as potential fusion genes and were visually confirmed in IGV. Due to the likely presence of double minute chromosomes in this patient, only potential fusion genes that presented with at least two different potential fusion partners were considered for validation.
Validation Of Structural Variants
Potential fusion genes were then subjected to PCR followed by capillary sequencing to validate the presence of the fusion (Table S1). Fluorescent in-situ hybridization (FISH) was performed as previously described [26] to validate amplification of key genes of interest including MDM2, SYT1 and DDR2. Bacterial artificial chromosomes mapping to the appropriate region for each gene were identified in the UCSC Genome browser [27] (http://genome.ucsc.edu/.) (Table S2).
Identification and characterization of LOC100507498
A genome-wide search across species for sequence elements related to LOC100507498 was done using a megablast search against the nonredundant nucleotide database [28]. A selection of unique hits that were highly similar to the LOC100507498 sequence (>90% identity) were translated in all 6 reading frames using transeq [29]. Translated sequences were used to query the Repbase database of repeat element sequences using the repeatmasker algorithm [30]. Nucleotide sequences were also used to identify closely related transposon HMM profiles using dfam_scan.pl [31]. Sequence alignments of LOC100507498 with known L1 elements [32], [33] was done with clustalw to characterize regions of high conservation [34].
RNA Sequencing
Frozen tumor tissue was cryofractured with the Cryoprep Impactor (Covaris), and lysed in RLT buffer containing 1% beta-mercaptoethanol. Lysate was passed through a Qiashredder column for homogenization followed by the addition of Qiazol lysis buffer to homogenate. Chloroform was added to the homogenate and mixed in Phaselock tubes (5 Prime, Gaithersburg, MD). The tubes were centrifuged at 16,000 g. The aqueous layer was transferred to a new tube, and 70% ethanol added. The sample was transferred to RNeasy spin columns. The columns were washed, and RNA eluted with nuclease-free water.
RNA-Sequencing data was analyzed utilizing the MAP-RSeq pipeline, developed at the Mayo Clinic. Detailed quality control data is generated with RSeQC software [35]. Paired-end reads were aligned by TopHat 2.0.6 [36] against the hg19 genome build using the bowtie1 aligner option [37]. Gene counts were generated using HTseq software (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) and gene annotation files were obtained from Illumina (http://cufflinks.cbcb.umd.edu/igenomes.html). Fusions were predicted with the TopHat-Fusion algorithm [38] and analyzed using custom scripts.
Pathway analysis
Pathway analysis of genes containing amplifications by aCGH, and SNV or fusion genes by WGS was conducted utilizing IPA Core Analysis build 172788 (Ingenuity® Systems, www.ingenuity.com) and MetaCore Pathway Analysis (GeneGo v6.11.41105, www.genego.com). IPA Core analysis was completed utilizing default parameters. MetaCore Pathway Analysis was completed with threshold = 0 and P-value = 0.1. P-values in MetaCore were determined utilizing hypergeometric intersection.
Results
We depict the case of a 72 year-old Caucasian male with a 23-year history of WDLS of the abdomen. The clinical course was characterized by multiple surgical debulking procedures over this period requiring left adrenal gland resection, partial pancreatectomy, large and small bowel resections, abdominal/pelvic lymphadenectomy, left nephrectomy and splenectomy. Post-operative adjuvant radiotherapy was received after two of the eleven surgical resections during this period. No systemic therapy was administered for his extensive disease. Pathological evaluation of the debulked tumor that was used for WGS demonstrated a well-differentiated liposarcoma consistent with prior evaluations. There was evidence of low mitotic activity (<1 mitosis/10 high power field), high adipose content, FNCLCC grade of 1, >90% tumor cellularity but with very low tumor cell/fat nuclear ratio (<1%). At the time of the debulking associated with WGS analysis, it was felt that surgical options would become extremely limited over time due to the decreasing ability to obtain significant debulking and increasing post surgical recovery times. Along with aCGH and WGS performed on the flow sorted nuclei, a commercially available molecular profiling assay (TargetNow®, Caris Life Sciences) was obtained. The TargetNow® assay demonstrated increased levels of topoisomerase I by immunohistochemistry (2+ in 95% of cells analyzed) and no detection of thymidylate synthetase by immunohistochemistry (in 100% of cells analyzed). The patient was treated with leucovorin calcium, 5-fluorouracil, and irinotecan hydrocloride (FOLFIRI) systemic therapy but obtained no clinical benefit and had disease progression. Unfortunately, the patient's clinical condition deteriorated over the ensuing six-month period with protracted debilitation related to post-surgical recovery and comorbidity from unresected tumor that led to his eventual demise.
Whole genome sequencing was conducted on germline and tumor DNA (SRA accession numbers SAMN02214846, and SAMN02214847). An average of 1.7×109 reads/germline sample and 1.4×109 reads/tumor sample were obtained with 1.5×109 (90%) and 1.3×109 (94%) of these reads respectively aligning to the hg18 reference (Table 1). After filtering for reads with a quality score ≥20, coverage was 43X for the germline and 38X for the tumor. Over 2000 somatic single nucleotide variants (SNV) were identified (Table S3) of which 12 were predicted as potentially deleterious by SIFT/Polyphen-2 and seven were validated by capillary sequencing (Table 2). Eight somatic small insertions and deletions (indels) were identified, but none validated by capillary sequencing.
Table 1. Metrics and summary statistics.
| Normal | Tumor | |
| Total reads | 1676273096 | 1363626456 |
| Aligned reads | 1508120220 | 1276332896 |
| Percent aligned | 90.0% | 93.6% |
| Coverage depth (raw) | 50 | 43 |
| Coverage depth (quality ≥20) | 43 | 38 |
| Total SNV | 3865355 | 3830574 |
| Somatic SNV | NA | 2334 |
| Somatic small insertions/deletions | NA | 8 |
Table 2. Validated functionally damaging SNV.
| Chr | Position (NCBI36) | Allele change | Amino acid change | Gene |
| 2 | 210491597 | G > A | G1704R | UNC80 |
| 6 | 33025040 | T > G | K256T | HLA-DMA |
| 10 | 5129651 | A > T | E93V | AKR1C3 |
| 11 | 57183888 | C > G | P122A | CLP1 |
| 12 | 79591146 | G > A | D125N | PTPRQ |
| 16 | 69268358 | C > T | D253N | MTSS1L |
| 20 | 49059955 | C > T | V110I | KCNG1 |
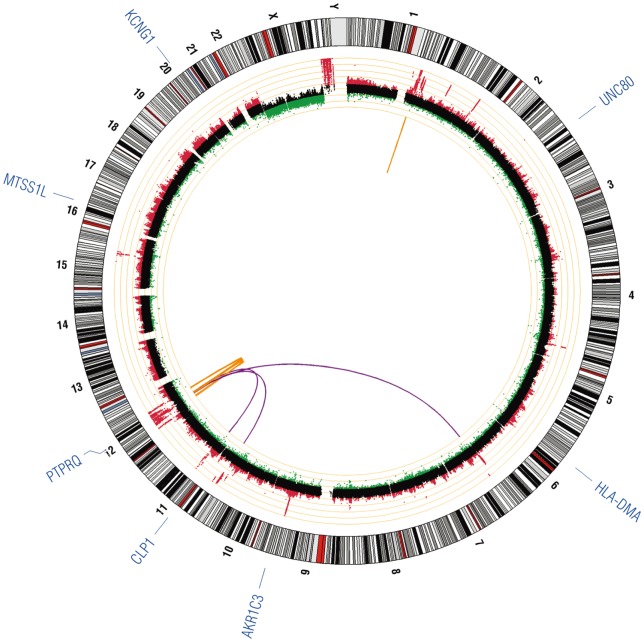
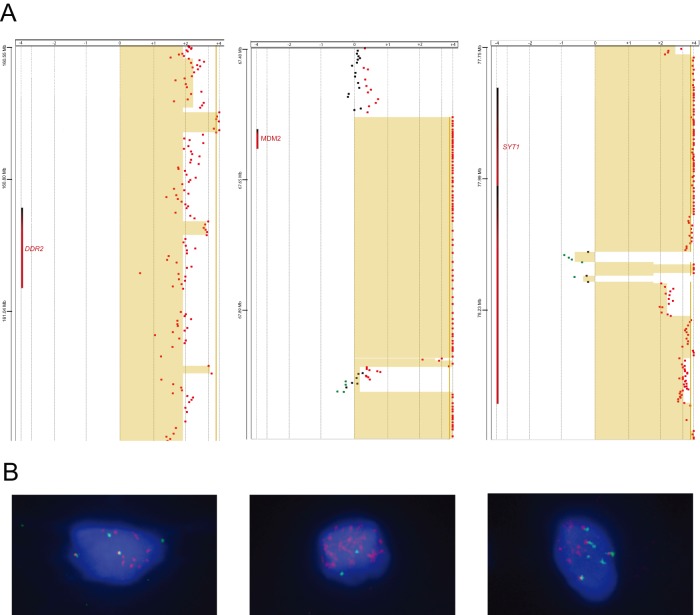
Array CGH (GEO accession number GSE47701) revealed massive amplification involving numerous chromosomes. Chromosome 12, which underwent massive rearrangement, was the most significantly affected (Figure 1, middle ring). Breakpoint analysis of these amplified regions in the WGS data revealed 40 potential fusions (Table S4) between genes of which 22 were selected for validation with PCR. Gene fusions validated in DNA by PCR are presented in Table 3. All the genes identified as potential fusion partners were checked in the COSMIC database [39]. None of the genes had reported fusions and all had reported substitution mutations with the exception of UHMK1. Amplification of MDM2 was identified by array comparative genomic hybridization (aCGH) (Figure 2A) and confirmed by FISH (Figure 2B) although we were unable to confirm a potential fusion partner with MDM2 suggesting the possible presence of MDM2 on double minute chromosomes. Overexpression of MDM2 is associated with reduced expression of the key tumor suppressor TP53 [40]. Interestingly, WGS revealed SYT1 presenting with three structural rearrangements that include potential fusions with 3 different genes (FGD6, C12orf26 and NELL1) of which 2 are likely part of a ring chromosome due to multiple rearrangements between these genes. RNA Sequencing revealed 17 putative fusion transcripts including a SYT1-C12orf63 fusion (Table S5).
Figure 1. Circos plot of validated genetic variation in a well-differentiated liposarcoma.
Inner-most circle contains validated structural rearrangements of fusion genes with translocations indicated in purple, and intra-chromosomal rearrangements indicated in orange. The middle ring contains the aCGH plot with copy number loss indicated in green and copy number gain in red; each orange ring corresponds to a log2 value of 1. The outer-most ring indicates validated, damaging single nucleotide variants.
Table 3. DNA validated fusion genes.
| Chr | Breakpoint 1 | Gene 1 | Chr | Breakpoint 2 | Gene 2 | Reads |
| Intrachromosomal rearrangements | ||||||
| 1 | 160738159 | UHMK1 | 1 | 160878664 | DDR2 | Facing |
| 12 | 70553166 | TBC1D15 | 12 | 81370954 | C12orf26 | Same direction – reverse |
| 12 | 77820664 | SYT1 | 12 | 94015001 | FGD6 | Same direction – forward |
| 12 | 78116536 | SYT1 | 12 | 81379417 | C12orf26 | Opposing |
| 12 | 78600788 | PAWR | 12 | 94875477 | AMDHD1 | Facing |
| 12 | 81369919 | C12orf26 | 12 | 94014762 | FGD6 | Opposing |
| 12 | 81371116 | C12orf26 | 12 | 94875230 | AMDHD1 | Opposing |
| Translocations | ||||||
| 6 | 157853571 | ZDHHC14 | 12 | 78591928 | PAWR | Same direction – forward |
| 11 | 21368102 | NELL1 | 12 | 77114108 | NAV3 | Same direction – reverse |
| 11 | 21368605 | NELL1 | 12 | 78114565 | SYT1 | Opposing |
| 11 | 71472153 | LRTOMT | 12 | 81346827 | C12orf26 | Opposing |
Figure 2. aCGH and Fluorescent in-situ hybridization of DDR2, MDM2, and SYT1.
(A) aCGH dot plots of the chromosomal region of each gene. Tan colored shading indicates regions of significant copy number change. Red dots indicate copy number gain and green dots indicate copy number loss. (B) FISH images of cells probed for the indicated genes with SpectrumOrange probe and the chromosome control indicated with the SpectrumGreen probe.
Furthermore, WGS revealed a gene fusion between UHMK1 and DDR2 that was confirmed in DNA, but we were unable to confirm in RNA. Both of these genes are located in a region of copy number gain in this tumor. Copy number gains in 1q23.3 where UHMK1 and DDR2 are located, have been reported previously in WDLS [15]. Mutations and increased expression of DDR2 have been reported in Hodgkin's Lymphoma and anaplastic large cell lymphoma [41], lung squamous cell carcinoma [42], nasopharyngeal carcinoma [43], sarcoma [44], hepatocellular carcinoma [45], aneuploid papillary thyroid cancer [46] and non-small cell lung cancer [47]. DDR2 is of interest both mechanistically and therapeutically. It plays key roles in multiple cellular activities, including proliferation, migration, adhesion, and extracellular matrix remodeling [48], [49]. The kinase domain of DDR2 is predicted to remain intact and the presence of copy number gain is significant because DDR2 kinase activity has been inhibited with the kinase inhibitors imatinib, nilotinib and dasatinib [50]. Interestingly, the gene fusion event between UHMK1 and DDR2, located in a region of copy number gain, may disrupt the normal function of the UAP1 gene, encoding a UDP-N-acetylglucosamine pyrophosphorylase. The UAP1 gene is located between UHMK1 and DDR2, thus structural rearrangement between UHMK1 and DDR2 is likely disruptive to the normal function of UAP1. The UAP1 protein is predicted to interact with glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase (GNE) [51], [52], a kinase regulating the biosynthesis of sialic acids which play a critical role in cell adhesion, signal transduction and tumor metastasis [53]. We find that GNE also resides in an amplified region.
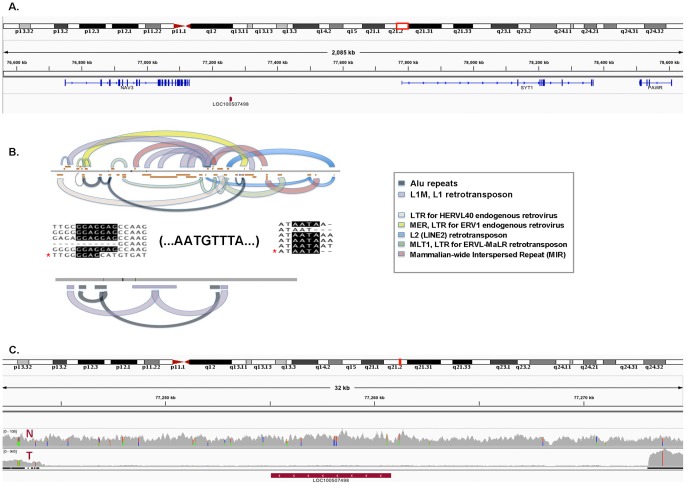
In depth analysis of a region of genomic instability hotspot on chromosome 12 revealed the presence of a highly conserved, putative L1 retrotransposon element, LOC100507498 (Figure 3A). This noncoding ∼33 kb RNA nucleotide sequence resides on the minus strand within the NAV3-SYT1-PAWR gene cluster on chromosome 12, in which 6 of the 11 validated gene fusion events occur. Two pseudogenes with homology to a mitochondrial ribosomal protein L11 (MRPL11P3) and a RAS related leukemia viral oncogene (LOC642550) also reside within the NAV3-SYT1-PAWR gene cluster, proximal to the LOC100507498 transposon. Characterization of LOC100507498 and closely related nucleotide (Figure 3B, top) and translated sequences (Figure 3B, bottom) show the highest similarity to L1 retrotransposon and Alu elements. L1 retrotransposons are non-LTR (non-Long Terminal Repeat) elements that have greatly expanded the human genome by autonomous retrotransposition, as well as non-autonomous retrotransposition of other mobile elements (e.g. Alu) which do not have their own transposases [54]. Sub-sequences of the LOC100507498 element were highly conserved (>95% similarity) in other species including Pan Troglodytes, Pan Paniscus, Gorilla, Macaca mulatta, and Nomascus leucogenys. Sequence alignment comparisons of the LOC100507498 element with known L1 retrotransposons showed highest overall conservation with Class 3 L1's (Table 1, [32]) known to be associated with 3′ transduction. A genomic deletion present specifically in patient tumor samples was identified by sequence read alignments to the LOC100507498 locus and surrounding region, indicating that this locus is a hotspot of genomic instability (Figure 3C).
Figure 3. Depiction of genomic rearrangement hotspot on chromosome 12.
We identified and further characterized a putative transposable element (LOC100507498) located on the (-) strand, within the PAWR-SYT1-NAV3 gene cluster (3A). The LOC100507498 and closely related sequences were characterized by comparing both nucleotide (3B,top) and translated (3B,bottom) sequences to known families of repetitive elements (Methods). Highly conserved sequence domains/motifs are color coded by known families of repetitive elements (Legend). Overall, these sequences exhibited the highest similarity to the L1 retrotransposon and Alu repeat elements (domain hit counts and similarity score). Sequence alignments of LOC100507498 (*) with known L1 elements [32], [33] exhibited the highest overall homology to Class 3 L1 elements as described by Pickeral et al. (Table 1, [32]) and in addition to the 5′-GGAG and 3′-AATA signature motifs, LOC100507498 carries several ‘AATGTTTA’ motifs that suggest multiple rounds of L1-mediated transduction [33]. The LOC100507498 locus resides within a genomic region that is deleted in the Tumor (T) sample, but present in the Normal (N) genome (3C).
SNV analysis by SIFT/Polyphen-2 revealed 7 SNV in 7 genes with potentially damaging functional significance (Figure 1, Table 2): AKR1C3, CLP1, HLA-DMA, KCNG1, MTSS1L, PTPRQ, and UNC80. Interestingly, AKR1C3 is upregulated in castration resistant prostate cancer [55]. All but one of these genes (HLA-DMA) have reported somatic mutations in the COSMIC database [39], but none of our SNV are shared and none of these genes have reported mutations in sarcomas.
In order to determine molecular pathways shared by genes containing mutations or copy number gains, pathway analysis was conducted. This analysis revealed expected pathways involved in cell cycle regulation, proliferation, survival, and cellular assembly as well as DNA replication, recombination and repair (Tables 4 and 5). Interestingly, both IPA and MetaCore identified lipid metabolism in their top eight pathways.
Table 4. IPA pathway analysis of genes containing SNV, gene fusions or amplifications.
| ID | Pathways | Score | Number of focus molecules | Genes from dataset in network |
| 1 | Cell Cycle, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair | 49 | 27 | AGAP2, AGPS, AIM2, ANAPC10, CCT2, CDK4, CUL1, DYRK2, E2F7, EGLN1, GRIP1, HELB, HMGA2, IFI16, KIF5A, MARS, MDM2, OS9, PNKD, PRRC2C, PYHIN1, RFTN1, SDHC, UHMK1, USP21, USP44, USP9Y |
| 2 | Cell-To-Cell Signaling and Interaction, Molecular Transport, Small Molecule Biochemistry | 41 | 23 | AKR1C3, ARHGAP9, CRP, DUSP6, F11R, FCER1A, FCER1G, FCGR2A, FCGR2B, FCGR3B, FCRLA, FCRLB, KLF13, NTN4, PAWR, PPFIA2, PTPRB, PTPRD, PTPRH, PTPRR, RGS4, RGS5, STK24 |
| 3 | Cell-To-Cell Signaling and Interaction, Nervous System Development and Function, Post-Translational Modification | 38 | 22 | AP2A2, caspase, CLP1, CPSF6, CYP27B1, FRS2, GAS2, MLL2, NCOR2, NLGN1, NR1I3, NR2C1, NRXN1, NUMA1, PFDN2, SYT1, TAB2, TBL1Y, UBAC1, VAMP4, ZDHHC17, ZFPM1 |
| 4 | Cell Morphology, Cellular Assembly and Organization, Connective Tissue Disorders | 27 | 17 | APOA2, AVIL, CSRP2, DDIT3, DEDD, ESPN, GDA, HLA-DMA, LAMA5, LYZ, MYOC, NELL1, OSBPL8, RGMA, RRBP1, SACS, SGSM2, |
| 5 | Connective Tissue Disorders, Developmental Disorder, Skeletal and Muscular Disorders | 25 | 16 | ADAMTS4, AKAP6, BAIAP2, DCTN2, DDR2, DISC1, GLI3, HERC2, KCNC2, KDM5D, MPZ, MRC2, NIT1, RAB3IP, SHC2, ZIC3 |
| 6 | Cellular Compromise, Cellular Function and Maintenance, Small Molecule Biochemistry | 23 | 15 | CADM3, CAMSAP1, DDX3Y, DERL3, FGD6, KCNG1, METTL1, NACC2, NAV3, PPOX, RSF1, SCAPER, TOX2, TSFM, UFC1 |
| 7 | Lipid Metabolism, Molecular Transport, Ophthalmic Disease | 21 | 14 | ATP11A, BBS10, CHD2, DNM3, EHD2, FANCF, LCN1, NUF2, PIP4K2C, PTK7, SASH1, STXBP5, TBC1D15, TOMM40L |
| 8 | Cell-To-Cell Signaling and Interaction, Infectious Disease, Inflammatory Response | 21 | 14 | C11orf67, CCDC41, CCDC94, CPM, GALNTL4, INTS4, KCNT1, OR10J1, PRKRIR, SBNO2, SEPT9, USF1, XRCC6BP1, ZDHHC14 |
Table 5. GeneGo pathway analysis of genes containing SNV, gene fusions or amplifications.
| Network # | Pathways | Total | In Data | pValue | Genes from Active Data |
| 1 | Development Hedgehog signaling | 46 | 4 | 2.991E-04 | Cullin 1, GLI-3R, DYRK2, GLI-3 |
| 2 | Cell cycle Influence of Ras and Rho proteins on G1/S Transition | 53 | 3 | 6.183E-03 | CDK4, MLCP (reg), MDM2 |
| 3 | Cytoskeleton remodeling Neurofilaments | 25 | 2 | 1.337E-02 | Kinesin heavy chain, DCTN2 |
| 4 | Cell cycle Role of SCF complex in cell cycle regulation | 29 | 2 | 1.777E-02 | Cullin 1, CDK4 |
| 5 | Regulation of lipid metabolism RXR-dependent regulation of lipid metabolism via PPAR, RAR and VDR | 30 | 2 | 1.896E-02 | CAR, NCOA2 (GRIP1/TIF2) |
| 6 | DNA damage ATM/ATR regulation of G1/S checkpoint | 32 | 2 | 2.142E-02 | CDK4, MDM2 |
| 7 | LRRK2 in neurons in Parkinson's disease | 33 | 2 | 2.270E-02 | AP-2 alpha subunits, AP2A2 |
| 8 | Cell cycle Spindle assembly and chromosome separation | 33 | 2 | 2.270E-02 | NUMA1, DCTN2 |
| 9 | Cell cycle ESR1 regulation of G1/S transition | 33 | 2 | 2.270E-02 | Cullin 1, CDK4 |
| 10 | Signal transduction Erk Interactions: Inhibition of Erk | 34 | 2 | 2.401E-02 | MKP-3, PTPRR |
Discussion
Previous studies in liposarcoma have contributed significantly to the understanding of the genetics underlying WDLS, but none have evaluated these in the context of the entire genome. This work reports the use of flow cytometry to isolate tumor cells from a WDLS prior to whole genome sequencing. Structural rearrangements potentially contributing to tumor development were detected in addition to identification of potential therapeutic targets of interest.
The presence of LOC100507498 with high similarity to L1 retrotransposon and Alu elements in the NAV3-SYT1-PAWR gene cluster that was prone to massive rearrangement has potentially significant functional consequences. First, although the majority of L1 and Alu elements are inactive sequence relics of ancient evolutionary events [54], many are still active during development and cancer [54], [56]. Second, in addition to mediating genomic rearrangements, the presence of L1 retrotransposons, which preferentially act in cis [57], can impact genomic stability and gene expression of neighboring genes via a number of different mechanisms [56]. The E2F7 transcription factor that plays an important role in cell cycle regulation [58], [59], is 5′ of the gene cluster, and is in cis with the L1 retrotransposon on the minus strand. Furthermore, the gene protein tyrosine phosphatase receptor type Q (PTPRQ) that has been shown to regulate adipogenesis in mesenchymal stem cells [60], resides just 3′ of the NAV3-LOC100507498-SYT1-PAWR gene cluster. Interestingly, a related protein tyrosine phosphatase, PTPRM, has been identified as an insertional mutagenesis target by L1 retrotransposons in colon tumors [56].
The role of transposons in cancer screening [61], [62] as well as gene therapy [63], [64] has expanded over recent years and applications continue to broaden as transposon-based techniques improve. Recent studies of a number of murine and human cancer cell lines have established the efficacy of both Non-Nucleoside and Nucleoside Reverse Transcription Inhibitors (NRTIs) in suppressing L1 retrotransposition, inducing significant cell growth and differentiation phenotypes [65], [66]. Collectively, these studies suggest that L1 retrotransposases may be highly effective as molecular targets in cancer therapy.
The identification of ∼600 bp of a RAS related leukemia viral oncogene (LOC642550) proximal to the L1 retrotransposon within the NAV3 -SYT1-PAWR gene cluster is also of interest given the known and shared role of rearrangement mediated FUS gene fusions between myxoid liposarcoma and leukemia [67]–[71]. Although the rearrangement hotspot and gene fusions identified in our studies are different, the presence of a pseudogene with homology to a RAS related leukemia viral oncogene within the identified hotspot is suggestive that perhaps gene fusions involving NAV3-SYT1-PAWR may also play a yet unidentified role in other cancers in addition to well-differentiated liposarcoma.
Structural variants resulting in potential fusion genes are often associated with amplified genes. This work identified amplification in two genes of particular interest. The first is SYT1, a calcium sensor involved in conducting neurotransmitter signals, which contained structural rearrangements with three different genes and is possibly part of a ring chromosome, commonly present in WDLS. Interestingly, a 40-kDa form of SYT1 participates in the non-classical export of FGF1, a protein that regulates many cellular processes including angiogenesis, morphogenesis and tumor growth suggesting a potentially key role of SYT1 in cancer [72]. While the precise role of SYT1 in cancer is unclear, the presence of multiple chromosomal rearrangements within this gene suggests that this gene is prone to breakage. Consequently, mechanistic studies are needed to further elucidate the potential role of SYT1 and its chromosomal rearrangements in cancer.
Another amplified gene of interest is DDR2. Mutations and altered expression in DDR2 have been reported in multiple tumor types [41]–[47]. DDR2 is involved in several key roles including proliferation, migration, adhesion and remodeling of the extracellular matrix [48], [49], and is expressed in stromal cells while a relative, DDR1, is expressed in epithelial cells [73]. In vitro studies of hepatic stellate cells and skin fibroblasts have demonstrated a drop in cell proliferation and migration in the absence of DDR2 [74]–[76]. Nude mice that received intrasplenic injections of human melanoma cell line A375 with stably silenced DDR2 had fewer hepatic metastasis than mice receiving mock transfected cells [74]. In contrast DDR2 −/− mice that received intrasplenic injection of murine colon adenocarcinoma cell line MCA38 had more hepatic metastasis than equivalently treated DDR2 +/+ mice [77]. The observed differences in metastatic burden in these mice may be attributable to differing genetic backgrounds, in particular expression of DDR1 that was not reported in either study. DDR1 levels have been demonstrated to be inversely proportional to DDR2 levels, both in vitro and in vivo [47], [78]. While these studies suggest DDR2 may play an important role in liposarcoma, its precise role remains to be elucidated through characterization of additional patients with WDLS and by correlating clinical phenotypes with DDR2 status.
Targeted therapy has enabled tumors with known genetic mutations to be effectively treated by utilizing compounds that exploit a specifically dysregulated molecular pathway. DDR2, which is potentially interesting mechanistically, is also a therapeutic target of interest. Imatinib, nilotinib and dasatinib have been demonstrated to inhibit DDR2 kinase activity [50]. Dasatinib effectively inhibited proliferation in cell lines containing DDR2 mutations [42]. Cells containing PDGFRA amplification or KRAS mutation were less sensitive to dasatinib and imatinib than cells without these genetic aberrations. In addition, a squamous cell lung cancer patient with a DDR2 mutation and no EGFR mutation demonstrated partial response to dasatinib and erolotinib [42] while a second patient with co-occurring CML and squamous cell lung cancer, which possessed a DDR2 mutation, showed a complete metabolic response in the lung tumor after treatment with dasatinib [79]. While this data is preliminary, it does suggest that dasatinib may have been a consideration for this WDLS patient with amplified DDR2, and thus likely amplified DDR2 kinase activity.
A large amplification of MDM2 was identified in this patient and is possibly the result of an unidentified gene fusion or the presence of MDM2 on double minute chromosomes. Interestingly, this patient also had amplification of CPM, which when co-occurring with amplified MDM2 is a unique marker of WDLS [17]. Several MDM2 inhibitors are currently in clinical trials including RO5045337 and RO5503781 (clinicaltrials.gov) of which the first is in a trial targeting liposarcoma. Taken together, the combination of aCGH and WGS allowed the detection of potentially druggable targets in this patient.
While these findings are limited by a sample size of one, this work reveals the value of utilizing multiple technologies to thoroughly interrogate a tumor genome; thus enabling the identification of druggable targets for which therapies are currently available, but are not part of the standard of care for liposarcoma. The cost and time required for next generation sequencing has dropped significantly in recent years along with improvements in variant detection methods, putting work such as this reported here on the brink of clinical application.
In summary, this work is the first to report the entire genome of a WDLS patient utilizing flow cytometry to isolate aneuploid cells prior to aCGH and WGS. We report the identification of a retrotransposon in a hotspot of genomic rearrangement as well as multiple novel structural rearrangements in the genome that likely contribute to the extensive gene amplification observed. In addition, we identified two potential therapeutic targets, MDM2 and DDR2. Further study of these findings in a larger cohort of liposarcoma patients is warranted to estimate the true prevalence of therapeutic targets such as DDR2 and to advance the understanding of the genetic basis of liposarcoma.
Supporting Information
Flow cytometry histogram.
(TIF)
Fusion gene DNA validation primers.
(DOC)
Bacterial Artificial Chromosomes (BACs) utilized in FISH assays.
(DOC)
Summary of identified single nucleotide variants.
(XLS)
Putative fusions identified from whole genome sequencing.
(XLSX)
Putative fusions identified from RNA sequencing fusion analysis.
(XLSX)
Acknowledgments
We would like to thank Dr. Christopher Conley and Leslie Dixon from the Mayo Clinic Biobank for their assistance with sample preparation and pathological evaluation.
Funding Statement
This publication was made possible by CTSA Grant Number UL1 TR000135 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH. Rafael Fonseca is a Clinical Investigator of the Damon Runyon Cancer Research Fund. This work is supported by grants SPORE CA90297052, P01 CA62242, R01 CA83724, ECOG CA 21115T, Predolin Foundation, Mayo Clinic Cancer Center and the Mayo Foundation. This study was supported by NIH Grant Number K12 CA90628. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fletcher CDM, Unni KK, Mertens F (Eds.) (2002) Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon: IARC Press. 415 p.
- 2. Crago AM, Singer S (2011) Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr Opin Oncol 23: 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hameed M (2007) Pathology and genetics of adipocytic tumors. Cytogenet Genome Res 118: 138–147. [DOI] [PubMed] [Google Scholar]
- 4. Conyers R, Young S, Thomas DM (2011) Liposarcoma: molecular genetics and therapeutics. Sarcoma 2011: 483154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boland MV, Quigley HA (2011) Evaluation of a combined index of optic nerve structure and function for glaucoma diagnosis. BMC Ophthalmol 11: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones RL, Fisher C, Al-Muderis O, Judson IR (2005) Differential sensitivity of liposarcoma subtypes to chemotherapy. Eur J Cancer 41: 2853–2860. [DOI] [PubMed] [Google Scholar]
- 7. Laurino L, Furlanetto A, Orvieto E, Dei Tos AP (2001) Well-differentiated liposarcoma (atypical lipomatous tumors). Semin Diagn Pathol 18: 258–262. [PubMed] [Google Scholar]
- 8. Sreekantaiah C, Karakousis CP, Leong SP, Sandberg AA (1992) Cytogenetic findings in liposarcoma correlate with histopathologic subtypes. Cancer 69: 2484–2495. [DOI] [PubMed] [Google Scholar]
- 9. Pedeutour F, Forus A, Coindre JM, Berner JM, Nicolo G, et al. (1999) Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer 24: 30–41. [PubMed] [Google Scholar]
- 10. Dei Tos AP, Doglioni C, Piccinin S, Sciot R, Furlanetto A, et al. (2000) Coordinated expression and amplification of the MDM2, CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol 190: 531–536. [DOI] [PubMed] [Google Scholar]
- 11. Forus A, Weghuis DO, Smeets D, Fodstad O, Myklebost O, et al. (1995) Comparative genomic hybridization analysis of human sarcomas: I. Occurrence of genomic imbalances and identification of a novel major amplicon at 1q21-q22 in soft tissue sarcomas. Genes Chromosomes Cancer 14: 8–14. [DOI] [PubMed] [Google Scholar]
- 12. Mandahl N, Hoglund M, Mertens F, Rydholm A, Willen H, et al. (1994) Cytogenetic aberrations in 188 benign and borderline adipose tissue tumors. Genes Chromosomes Cancer 9: 207–215. [DOI] [PubMed] [Google Scholar]
- 13. Italiano A, Bianchini L, Keslair F, Bonnafous S, Cardot-Leccia N, et al. (2008) HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int J Cancer 122: 2233–2241. [DOI] [PubMed] [Google Scholar]
- 14. Pilotti S, Della Torre G, Lavarino C, Sozzi G, Minoletti F, et al. (1998) Molecular abnormalities in liposarcoma: role of MDM2 and CDK4-containing amplicons at 12q13-22. J Pathol 185: 188–190. [DOI] [PubMed] [Google Scholar]
- 15. Tap WD, Eilber FC, Ginther C, Dry SM, Reese N, et al. (2011) Evaluation of well-differentiated/de-differentiated liposarcomas by high-resolution oligonucleotide array-based comparative genomic hybridization. Genes Chromosomes Cancer 50: 95–112. [DOI] [PubMed] [Google Scholar]
- 16. Italiano A, Bianchini L, Gjernes E, Keslair F, Ranchere-Vince D, et al. (2009) Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clin Cancer Res 15: 5696–5703. [DOI] [PubMed] [Google Scholar]
- 17. Erickson-Johnson MR, Seys AR, Roth CW, King AA, Hulshizer RL, et al. (2009) Carboxypeptidase M: a biomarker for the discrimination of well-differentiated liposarcoma from lipoma. Mod Pathol 22: 1541–1547. [DOI] [PubMed] [Google Scholar]
- 18. Rubin BP, Dal Cin P (2001) The genetics of lipomatous tumors. Semin Diagn Pathol 18: 286–293. [PubMed] [Google Scholar]
- 19. Ruiz C, Lenkiewicz E, Evers L, Holley T, Robeson A, et al. (2011) Advancing a clinically relevant perspective of the clonal nature of cancer. Proc Natl Acad Sci U S A 108: 12054–12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Asmann YW, Middha S, Hossain A, Baheti S, Li Y, et al. (2011) TREAT: a bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics 28: 277–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goya R, Sun MG, Morin RD, Leung G, Ha G, et al. (2010) SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics 26: 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 24. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chin SF, Daigo Y, Huang HE, Iyer NG, Callagy G, et al. (2003) A simple and reliable pretreatment protocol facilitates fluorescent in situ hybridisation on tissue microarrays of paraffin wax embedded tumour samples. Mol Pathol 56: 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. (2002) The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 15: 403–410. [DOI] [PubMed] [Google Scholar]
- 29. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 30. Jurka J, Klonowski P, Dagman V, Pelton P (1996) CENSOR – a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem 20: 119–121. [DOI] [PubMed] [Google Scholar]
- 31. Wheeler TJ, Clements J, Eddy SR, Hubley R, Jones TA, et al. (2013) Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res 41: D70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pickeral OK, Makałowski W, Boguski MS, Boeke JD (2000) Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res 10: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goodier JL, Ostertag EM, Kazazian HHJ (2000) Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9: 653–657. [DOI] [PubMed] [Google Scholar]
- 34. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) ClustalW and ClustalX version 2. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 35. Wang L, Wang S, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28: 2184–2185. [DOI] [PubMed] [Google Scholar]
- 36. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim D, Salzberg SL (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, et al. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39: D945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haines DS (1997) The mdm2 proto-oncogene. Leuk Lymphoma 26: 227–238. [DOI] [PubMed] [Google Scholar]
- 41. Willenbrock K, Kuppers R, Renne C, Brune V, Eckerle S, et al. (2006) Common features and differences in the transcriptome of large cell anaplastic lymphoma and classical Hodgkin's lymphoma. Haematologica 91: 596–604. [PubMed] [Google Scholar]
- 42. Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, et al. (2011) Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov 1: 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chua HH, Yeh TH, Wang YP, Huang YT, Sheen TS, et al. (2008) Upregulation of discoidin domain receptor 2 in nasopharyngeal carcinoma. Head Neck 30: 427–436. [DOI] [PubMed] [Google Scholar]
- 44. Bai Y, Li J, Fang B, Edwards A, Zhang G, et al. (2012) Phosphoproteomics identifies driver tyrosine kinases in sarcoma cell lines and tumors. Cancer Res 72: 2501–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee NO, Park JW, Lee JA, Shim JH, Kong SY, et al. (2012) Dual action of a selective cyclooxygenase-2 inhibitor on vascular endothelial growth factor expression in human hepatocellular carcinoma cells: novel involvement of discoidin domain receptor 2. J Cancer Res Clin Oncol 138: 73–84. [DOI] [PubMed] [Google Scholar]
- 46. Rodrigues R, Roque L, Espadinha C, Pinto A, Domingues R, et al. (2007) Comparative genomic hybridization, BRAF, RAS, RET, and oligo-array analysis in aneuploid papillary thyroid carcinomas. Oncol Rep 18: 917–926. [PubMed] [Google Scholar]
- 47. Ford CE, Lau SK, Zhu CQ, Andersson T, Tsao MS, et al. (2007) Expression and mutation analysis of the discoidin domain receptors 1 and 2 in non-small cell lung carcinoma. Br J Cancer 96: 808–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruiz PA, Jarai G (2012) Discoidin domain receptors regulate the migration of primary human lung fibroblasts through collagen matrices. Fibrogenesis Tissue Repair 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vogel WF, Abdulhussein R, Ford CE (2006) Sensing extracellular matrix: an update on discoidin domain receptor function. Cell Signal 18: 1108–1116. [DOI] [PubMed] [Google Scholar]
- 50. Day E, Waters B, Spiegel K, Alnadaf T, Manley PW, et al. (2008) Inhibition of collagen-induced discoidin domain receptor 1 and 2 activation by imatinib, nilotinib and dasatinib. Eur J Pharmacol 599: 44–53. [DOI] [PubMed] [Google Scholar]
- 51. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kanehisa MaG (2000) S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, et al. (1999) UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science 284: 1372–1376. [DOI] [PubMed] [Google Scholar]
- 54. Szak ST, Pickeral OK, Landsman D, Boeke JD (2003) Identifying related L1 retrotransposons by analyzing 3′ transduced sequences. Genome Biol 4: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pfeiffer MJ, Smit FP, Sedelaar JP, Schalken JA (2011) Steroidogenic enzymes and stem cell markers are upregulated during androgen deprivation in prostate cancer. Mol Med 17: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, et al. (2012) Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res 22: 2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, et al. (2001) Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 21: 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aksoy O, Chicas A, Zeng T, Zhao Z, McCurrach M, et al. (2012) The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence. Genes Dev 26: 1546–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carvajal LA, Hamard PJ, Tonnessen C, Manfredi JJ (2012) E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev 26: 1533–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jung H, Kim WK, Kim do H, Cho YS, Kim SJ, et al. (2009) Involvement of PTP-RQ in differentiation during adipogenesis of human mesenchymal stem cells. Biochem Biophys Res Commun 383: 252–257. [DOI] [PubMed] [Google Scholar]
- 61. Koso H, Takeda H, Yew CC, Ward JM, Nariai N, et al. (2012) Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells. Proc Natl Acad Sci USA 109: E2998–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kool J, Berns A (2009) High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer 9: 389–399. [DOI] [PubMed] [Google Scholar]
- 63. Peng PD, Cohen CJ, Yang S, al e (2009) Efficient nonviral Sleeping Beauty transposon based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther 16: 1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jin Z, Maiti S, Huls H, al e (2011) Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective Lentiviral vectors in primary human cells. Mol Ther 19: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jones RB, Garrison KE, Wong JC, Duan EH, Nixon DF, et al. (2008) Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PloS one 3: e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Carlini F, Ridolfi B, Molinari A, Parisi C, Bozzuto G, et al. (2010) The reverse transcription inhibitor abacavir shows anticancer activity in prostate cancer cell lines. PloS one 5: e14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Panagopoulos I, Mandahl N, Mitelman F, Aman P (1995) Two distinct FUS breakpoint clusters in myxoid liposarcoma and acute myeloid leukemia with the translocations t(12;16) and t(16;21). Oncogene 11: 1133–1137. [PubMed] [Google Scholar]
- 68. Crozat A, Aman P, Mandahl N, Ron D (1993) Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363: 640–644. [DOI] [PubMed] [Google Scholar]
- 69. Ichikawa H, Shimizu K, Hayashi Y, Ohki M (1994) An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res 54: 2865–2868. [PubMed] [Google Scholar]
- 70. Rabbitts TH, Forster A, Larson R, Nathan P (1994) Chromosomal translocations in human cancer. Nature 372: 143–149. [DOI] [PubMed] [Google Scholar]
- 71. Shimizu K, Ichikawa H, Tojo A, Kaneko Yea (1993) An ets-related gene, ERG, is rearranged in human myeloid leukemia with t(16;21) chromosomal translocation. Proc Natl Acad Sci USA 90: 10280–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kathir KM, Gao L, Rajalingam D, Daily AE, Brixey S, et al. (2010) NMR characterization of copper and lipid interactions of the C2B domain of synaptotagmin I-relevance to the non-classical secretion of the human acidic fibroblast growth factor (hFGF-1). Biochim Biophys Acta 1798: 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Alves F, Vogel W, Mossie K, Millauer B, Hofler H, et al. (1995) Distinct structural characteristics of discoidin I subfamily receptor tyrosine kinases and complementary expression in human cancer. Oncogene 10: 609–618. [PubMed] [Google Scholar]
- 74. Badiola I, Villace P, Basaldua I, Olaso E (2011) Downregulation of discoidin domain receptor 2 in A375 human melanoma cells reduces its experimental liver metastasis ability. Oncol Rep 26: 971–978. [DOI] [PubMed] [Google Scholar]
- 75. Olaso E, Ikeda K, Eng FJ, Xu L, Wang LH, et al. (2001) DDR2 receptor promotes MMP-2-mediated proliferation and invasion by hepatic stellate cells. J Clin Invest 108: 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Olaso E, Labrador JP, Wang L, Ikeda K, Eng FJ, et al. (2002) Discoidin domain receptor 2 regulates fibroblast proliferation and migration through the extracellular matrix in association with transcriptional activation of matrix metalloproteinase-2. J Biol Chem 277: 3606–3613. [DOI] [PubMed] [Google Scholar]
- 77. Badiola I, Olaso E, Crende O, Friedman SL, Vidal-Vanaclocha F (2011) Discoidin domain receptor 2 deficiency predisposes hepatic tissue to colon carcinoma metastasis. Gut 61: 1465–1472. [DOI] [PubMed] [Google Scholar]
- 78. Maeyama M, Koga H, Selvendiran K, Yanagimoto C, Hanada S, et al. (2008) Switching in discoid domain receptor expressions in SLUG-induced epithelial-mesenchymal transition. Cancer 113: 2823–2831. [DOI] [PubMed] [Google Scholar]
- 79. Pitini V, Arrigo C, Di Mirto C, Mondello P, Altavilla G (2013) Response to dasatinib in a patient with SQCC of the lung harboring a discoid-receptor-2 and synchronous chronic myelogenous leukemia. Lung Cancer 82: 171–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry histogram.
(TIF)
Fusion gene DNA validation primers.
(DOC)
Bacterial Artificial Chromosomes (BACs) utilized in FISH assays.
(DOC)
Summary of identified single nucleotide variants.
(XLS)
Putative fusions identified from whole genome sequencing.
(XLSX)
Putative fusions identified from RNA sequencing fusion analysis.
(XLSX)