Abstract
Using deuterium 2D T1-T2 Inverse Laplace Transform (ILT) NMR we have investigated the distribution, population, and dynamics of waters of hydration in major ampullate N. clavipes and A. aurantia silk as a function of temperature. In both samples studied, correlation times much larger than that of free water are measured and in some cases appear to increase with increasing temperature over the range of 5 to 60 °C(corresponding to reduced tumbling). In addition, the experimental data point to a reduction in the population of water localized in the silk with increasing temperature in the range of 20 to 50°C. Molecular dynamics simulations were performed to probe the thermal characteristics of a variety of repeating motifs found in the two silk samples. The repeating motifs GLGSQ, GAAAAAAG, GPGGY, GPGQQ, GPSG, and GPSGPGS found in N. clavipes, GLGSQ, GYGSG, GPGSG, and GPGSQ found in A. aurantia silk were found to exhibit a thermal property observed in short elastin peptides known as the “inverse temperature transition”. This is a well known characteristic exhibited by short peptides consisting of (VPGXG)n motifs (where X is any amino acid other than proline) found in elastin, a protein responsible for the elasticity of vertebrate tissues. In qualitative agreement with experimental measurements of water in the silks, all the peptides studied in simulation show evidence of an increase in sidechain contacts and peptide hydrogen bonds, concomitant with a decrease in radius of gyration and localized water as the temperature is raised from approximately 5 to 60° C.
I. INTRODUCTION
Silk is a protein fiber spun by spiders composed of complex protein molecules used for various purposes including capturing prey. Spiders have six or seven sets of glands which produce a distinct silk for a specific function.1 Orb weaving spiders such as N. clavipes and A. aurantia are able to synthesize as many as seven different types of silk which include major ampullate, minor ampullate, flagelliform, aggregate, cylindrical, aciniform and piriform silks.2 Major ampullate silk forms the web framework and is more rigid than the flagelliform, which forms the sticky spiral used for capturing prey, and the cylindrical or tubuliform which forms egg cases. The spinning of silk is a complex process which involves linking a mixture of sticky and nonsticky strands and is controlled by the spinnerets at the tip of the abdomen. Dragline spider silk from the N. clavipes and A. aurantia contain two proteins: major ampullate spidroin 1 (MaSp1) and major ampullate spidroin 2 (MaSp2).3–5
The spider silk which has recently received the most attention is that from the major ampullate gland because of its lightweight characteristics and remarkable mechanical properties.6 Each protein has unique motifs that control the tensile strength and elasticity that are believed to be responsible for their extraordinary mechanical properties. For instance MaSp2 has a GPGXX motif, where X can be any one of the small subset amino acids such as serine, tyrosine, leucine, or glutamine. This motif is thought to give the silk elastic characteristics because it is believed to form a β-spiral that is identical to the idealized β-turn of repeating VPGVG motifs present in elastin.7 One major difference between the two major ampullate proteins is the proline content; there is a large quantity of proline in MaSp2 and little or none in MaSp1.5 The percentage of MaSp1 to MaSp2 in N. clavipes dragline silk is approximately 81:19 while in A. aurantia it is approximately 41:59.8 The ratio between MaSp1 and MaSp2 has been shown to vary between different species of spiders8 and these differences have been linked to the spider's diet and habitat.9,10 Table 1 compares the amino acid composition of the major ampullate dragline silk across three different species of spiders and highlights that the glycine and alanine concentrations are relatively abundant while the proline content varies considerably. Gosline and Savage have suggested that the proline concentration may play an important role in silk elasticity11 as well as the formation of network structures.12 Optical birefringence studies were performed to probe these effects from the major ampullate silk of N. clavipes and A. diadematus which have significantly different proline compositions (3.5% for N. clavipes and 16% for A. diadematus). Their data showed that the birefringence reading recorded for both wet and dry silk of N. clavipes is larger than that for the A. diadematus, indicating that the network chains in the dry and wet N. clavipes silk have stiffer fibers than the A. diadematus silk. In N. clavipes, glycine and alanine are the most abundant amino acids found in the major ampullate silk and contribute approximately 47% and 26% of the total composition, respectively. The primary structure of the major ampullate dragline silk of N. clavipes consists of poly-alanine regions which run up to 7 residues in MaSp1 and 10 in MaSp2 and are believed to form β-sheet like structures.3,13 In small peptides consisting of (Ala-Gly-Gly)10 it was determined that Gly-Gly-X region forms a 310 helix14–16, and recent experimental studies of N. clavipes have confirmed that the gly-rich regions form 310 helix type structures, not disordered helices or random coils.17
TABLE I.
Average amino acid composition of major ampullate dragline silk of various spider silks.22
| Amino Acid A. argentata A. aurantia N. clavipes | |||
|---|---|---|---|
| Gly | 44.6 | 46.4 | 47.1 |
| Ala | 19.3 | 17.9 | 26.5 |
| Pro | 10.2 | 9.5 | 1.2 |
| Ser | 6.8 | 5.9 | 3.0 |
| Tyr | 5.4 | 4.8 | 3.6 |
| Leu | 1.0 | 1.5 | 4.2 |
Work and coworkers18,19 have shown that when the major ampullate silk from orb weaver spiders comes in contact with water, the silk absorbs water and shrinks by 55% of its original length and subsequently increases its elastic characteristics. This so called “supercontraction” has been linked to specific repeating motifs20,21 as well as the concentration of proline.22,23 Vollrath and coworkers showed that the Young's modulus of the major ampullate silks from each species used in the study decreases with increasing proline content. During supercontraction, dragline silk reduces in strength and has properties that resemble an elastomer. Thermoelastic measurements performed by Gosline and coworkers on A. diadematus dragline silk immersed in distilled water have shown a reduction in the initial stress of the silk and that its mechanical characteristics are more rubber-like.24 The signatures of supercontraction have also been observed in B. mori silk when the cocoon silk is immersed in a powerful swelling agent20 and in A. diadematus silk when immersed in diferent solvents such as methanol or ethanol.25 One current belief is that this behavior is linked to the observation that residues such as glycine become more mobile when in contact with a solvent, while repeating alanine motifs which are in β-sheet conformations appear unaffected on the NMR timescale.20,21,26
The focus of the present work is to quantify the distribution, population and the dynamical properties of water in N. clavipes and A. aurantia silks. To study the water dynamics in the silk, we have implemented deuterium 2D T1-T2 inverse Laplace transform (ILT) NMR. This experimental method has proven to be powerful for characterizing water in multisite systems and has been recently applied to study the porous properties of water-saturated sedimentary rocks27, potato tissue28, cement pastes29, and to study exchange of water in hydrated elastin.30 Second, we have used molecular dynamics simulations to investigate the temperature dependent characteristics of repeating motifs founds in major ampullate N. clavipes and A. aurantia silks.
II. MATERIALS AND METHODS
A. Sample Preparation
Major ampullate dragline silk was collected from adult female N. clavipes and A. aurantia spiders from central Florida and Arizona, respectively, by winding the silk fiber on a rotating cylinder.31 During the silking process, the spider was first anesthetized to reduce the stress of capture. The spiders were fed a large cricket once a week, and every other day silk was collected from the major ampullate gland located in the abdomen at a rate of 2 cm/s while being watched closely under a microscope to ensure the silk was not contaminated with minor ampullate silk.
B. Deuterium T1-T2 Correlation Experiments
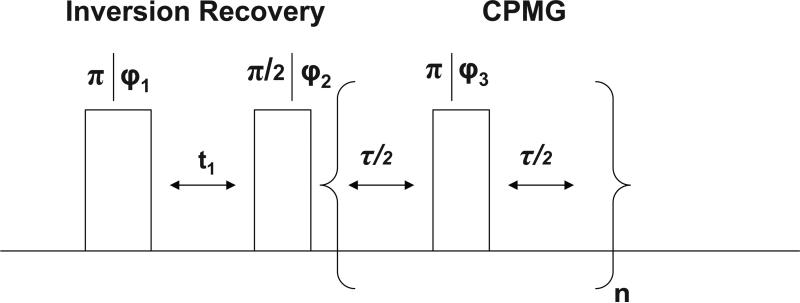
All 2H NMR experiments were performed on a Varian Unity 200 MHz spectrometer with a 5 mm liquid probe; the 90° pulse time was tuned to 26 μs. All the silk samples used for these experiments were soaked in D2O for 24 hours prior to the NMR experiments. During the experiments the samples were suspended in D2O and the temperature was regulated to within 1°C. The NMR pulse sequence for the T1-T2 experiments is illustrated in Figure 1.32 In the experiments the first 180° pulse inverts the magnetization which recovers toward thermal equilibrium with a relaxation time of T1 during delay t1. The second 90° pulse rotates the magnetization into the transverse plane and the NMR signal is detected by a Carr-Purcell-Meiboom-Gill (CPMG) pulse train. We performed measurements by collecting values of t1 ranging logarithmically from 1 ms to 2 s in 68 steps, using a τ spacing of 300 μs and collecting n=6000 points. The two-dimensional T1-T2 NMR relaxation data was processed using an inverse Laplace transform described elsewhere.33,34
FIG. 1.
NMR pulse sequence used in the 2D T1-T2 correlation experiment32. The phase cycling was ϕ1= x, –x, ϕ2= x, x, –x, –x, ϕ3 = y and the receiver phase was ϕReceiver = x, x, –x, –x. In the experiment, the delay t1 is varied from 5 ms to 2 s; the T2 is measured with a CPMG train with n=6000 stroboscopically detected echo peaks79.
C. Molecular Dynamics Simulations
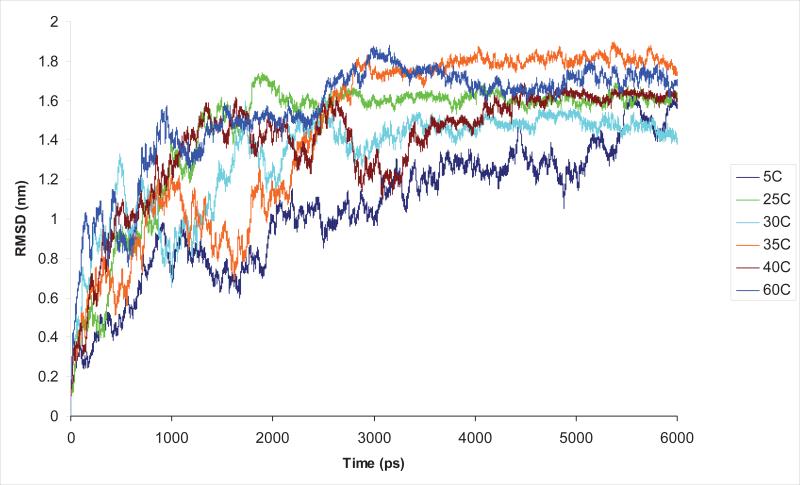
GROMACS35 was used in our molecular dynamics (MD) simulations with a Berendsen thermostat36 and the OPLS-AA/L force field model37 at six temperatures ranging from 5 to 60°C. Each of the simulations began with a single linear-chain of [GLGGQ]5, [GLGSQ]5, [GAAAAAAG]5, [GPGGY]5, [GPGQQ]5, [GPSG]5, [GPSGPGS]5, [GYGSG]5, [GPGSG]5 and [GPGSQ]5. The peptides were placed in a cubic box measuring approximately 128nm3 and solvated with 4048 water molecules using the SPC216 water model38. The N-terminus of the peptides was terminated using the amine (-NH2) group while the C-terminus was terminated by the carboxyl (-COOH) group. An energy minimization was performed on the structures to remove overlapping atoms. The method of steepest descent was used with the maximum step size set to 1 and the force tolerance set to 2000 kJ/mol nm. We performed a short simulation restraining the peptide positions and equilibrating the pressure for 10 ps before executing the full simulations for a duration of 6 ns. Several properties of the peptides were determined by averaging over the last 1 ns of the simulation results (i.e., from 5 to 6 ns). Figure 2 highlights the averaged Cα Root Mean Square Displacement (RMSD) as a function of time at multiple temperatures for [GPGSG]5 and that after approximately 2 ns equilibrium was established. Similar trends were obtained on all peptides studied.
FIG. 2.
Root Mean Square Deviation (RMSD) for (GPGSG)5 averaged over all Cα as a function of time for the temperatures of 5 to 60°C. As discussed in the text, the data shows the peptide equilibrates approximately after 2 ns for all temperatures. Similar results were observed in the other peptides studied, as discussed in the text.
III. RESULTS AND DISCUSSION
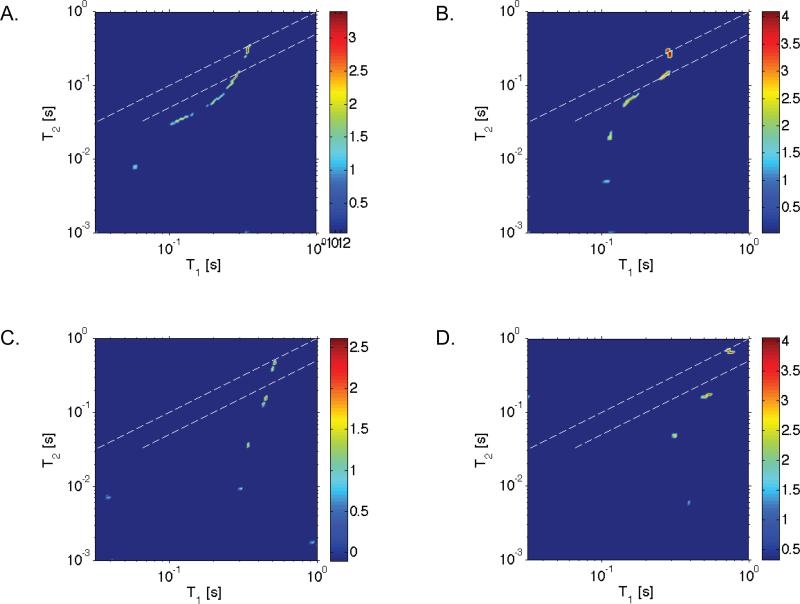
Examples of the acquired T1-T2 2D maps at 15 and 50°C for each silk sample studied are shown in Figure 3. The data, shown on a logarithmic scale in intensity, reveal five peaks corresponding to five reservoirs of water with distinguishable dynamical characteristics at low temperature and four peaks at high temperatures for each of the silk samples. An intriguing observation of the experimental data is that the number of water reservoirs and corresponding relaxation times are remarkably similar in the two silk samples studied.
FIG. 3.
Measured T1-T2 ILT map of water in deuterium hydrated N. clavipes(A,C) and A. aurantia(B,D) silks at 15°C (A,B) and 50°C(C,D). Five peaks are observed corresponding to five reservoirs of water with distinguishable dynamical characteristics at low temperature and four peaks were observed at high temperatures. In the figures, the peaks with the largest T1 and T2 times are bulk water. The dashed lines in the figures represent a range of values where T1 is approximately equal to T2, and the colorbar shown represents the signal intensity on a logarithmic scale.
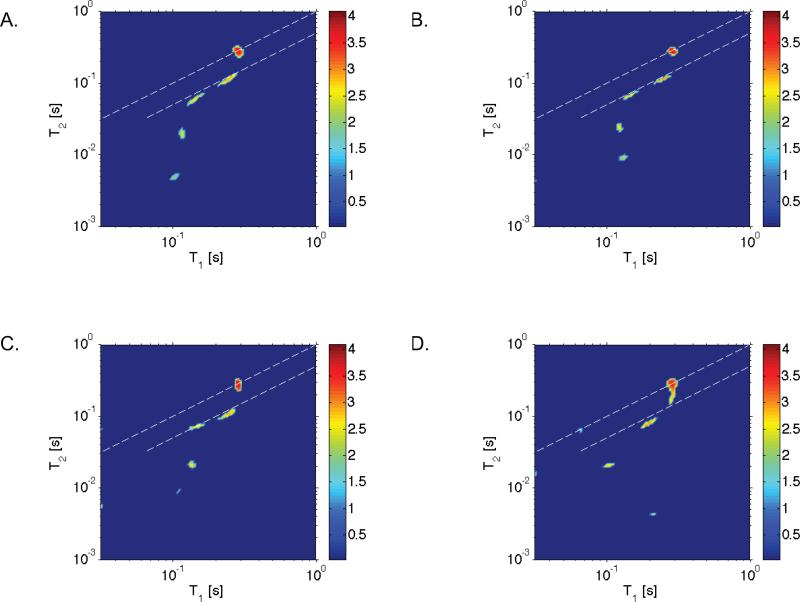
To verify that the ILT algorithm we implemented can faithfully produce both the intensity and the T1 and T2 values that were observed in the experiments, simulations were performed. As an example, taking the A. aurantia silk T1 and T2 values observed at 15°C, we show simulated data in Figure 4A that was generated with Gaussian distributed noise having a standard deviation in intensity equal to that realized in the experiments. Furthermore, in Figure 4B through 4D results of simulations are provided where the standard deviation of the noise intensity was increased to 5, 7 and 10 times that in our experiments. As shown elsewhere32, the ILT routine we implemented is prone to producing artifacts in cases where the signal to noise is low. We find that the algorithm fails when the noise was increased to 10 times that in our experiments; the T1 and T2 values are not correctly measured and the number of reservoirs is also not correct. However, the figure shows that the ILT is capable at resolving the number of reservoirs, T1 and T2 values and relative intensities we observed in the experiments even at 5 times less signal to noise. Similar simulation results were produced for data sets shown in Figure 3, as well as other temperatures that were performed in our study.
FIG. 4.
Simulated ILT maps using the experimental values from A. aurantia silk at 15°C (refer to Figure 3 B) with Gaussian distributed noise where the standard deviation of the noise intensity was A) set equal to that in the experiments or B) 5 times C) 7 times and D) 10 times that in the experiments. As discussed in the text, the simulations reveal that the inverse Laplace algorithm we implemented faithfully reproduces the T1-T2 times as well as the relative signal intensities, even in conditions where the noise is artificially set to 5 times that realized in the experiments. The experimental signal to noise in the these measurements is known to e ect the performance of the inverse Laplace algorithm we implemented, as shown elsewhere32.
Referring again to Figure 3, the two peaks with T1 ≈ T2 in the 2D map have characteristics of isotropic motion in the so-called extreme narrowing limit and denote bulk water and water between the fibers, whereas the peaks closest to the bottom T1 > T2 exhibit much different dynamics. Additional experiments were performed using a T2-T2 correlation experiment39,40 to probe exchange between the sites resolved in the T1-T2 data. Our findings indicated that over time scales from 10μs to 200 ms no exchange was observed. The peaks with T1 > T2 have characteristics reminiscent of the properties of supercooled heavy water41 and heavy water in ice42 where T1 > T2 was observed. Below, we make the assumption that a complex network of hydrogen bonding occurs between water molecules and between water and the protein surface and analyze the relaxation behavior by the same model used in reference41.
For a deuterium nucleus in D2O the deuterium-deuterium dipolar interaction is usually negligible compared with the quadrupolar interaction, and the NMR longitudinal (T1) and transverse (T2) relaxation times may be written as follows:
| (1) |
| (2) |
In the above expressions ω0 is the Larmor frequency of the deuterium nucleus, . The model in the formulation above assumes that the motion of the water molecule may be divided into fast oscillatory motions and slower diffusional processes41. Following Lang et. al., the short time reorientational motions of any given water molecule consists of fast librations about an equilibrium orientation in a random network of surrounding water - in our system the network would also include protein molecules. On a much longer time scale, the orientation of the water molecule will change due to diffusive motion resulting in a rearrangement of the surrounding network of water as well as protein molecules. The librational motion in this model is tacitly assumed to occur at a much shorter time scale than the diffusive motion, and as a consequence the cross-correlation between the two relaxation pathways is assumed to be negligible. The spectral density in these expressions is defined by , with τc denoting the slow diffusion correlation time of the water molecule. As shown in43 the autocorrelation function characterizing the slow and fast motions may be separated, resulting in the expressions for T1 and T2 above. The motional averaging parameter Γ in CQeff is given by
| (3) |
In the above expression the terms D(Ω) denote second rank Wigner rotation matrices, and the subscript IF refers to a transformation from the principle axis (I) of the field gradient tensor to the equilibrium orientation of the oxygen-deuterium bond(F)41. The parameter η = (Vxx – Vyy)/Vzz is the asymmetry parameter of the potential V. For deuterium in bulk water the asymmetry parameter η is approximately zero, however exact knowledge of η is not necessary for determining the correlation time τc and constant CQeff.
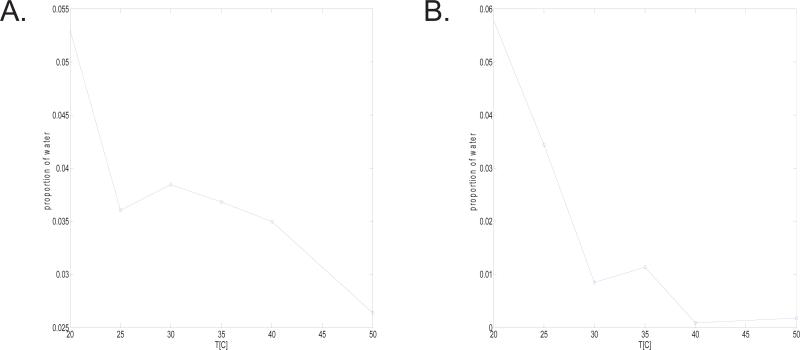
Using Eq. 1 and Eq. 2 we have determined the correlation times and values of CQeff as a function of temperature, and have tabulated our findings in Tables 9-10. The correlation times for the peaks with T1 > T2 are significantly higher than that of free water, which is approximately 5 ps at room temperature. In addition, in both of the silk samples studied (refer to Tables 9-10) two of the three water reservoirs exhibit correlation times that increase with increasing temperature over the range studied. In addition, Figure 4 shows the relative population of these peaks (where T1 > T2) to the total signal intensity observed in the 2D map at every temperature (the population was determined by integrating the signal areas in the 2D maps). The figure indicates that the percentage contribution of the waters of hydration with (T1 > T2) reduce in intensity with increasing temperature.
TABLE IX.
Relaxation times for the components with T1 > T2 in the T1-T2 2D experiment performed on N. clavipes silk.
| Temp [°C] | T1 [s] | T2 [s] | τc × 10−9 [s] | |
|---|---|---|---|---|
| 5 | .04409 ± .0014 | .00392 ± .0003 | 18.4 ± 1.34 | 10.86 ± 1.02 |
| 15 | .05849 ± .0023 | .00785 ± .0007 | 14.2 ± 1.25 | 6.51 ± 0.71 |
| 20 | .04923 ± .0032 | .00784 ± .0006 | 12.7 ± 1.25 | 7.05 ± 0.96 |
| 25 | N/A | N/A | N/A | N/A |
| 30 | N/A | N/A | N/A | N/A |
| 35 | N/A | N/A | N/A | N/A |
| 40 | N/A | N/A | N/A | N/A |
| 50 | N/A | N/A | N/A | N/A |
| Temp [°C] | T1 [s] | T2 [s] | τc × 10−9 [s] | |
|---|---|---|---|---|
| 5 | .05251 ± .0025 | .01645 ± .0004 | 7.45 ± 1.10 | 4.50 ± 0.57 |
| 15 | .12174 ± .0020 | .03575 ± .0065 | 7.89 ± 1.62 | 2.01 ± 0.27 |
| 20 | .10915 ± .0022 | .03073 ± .0036 | 8.21 ± 1.08 | 2.30 ± 0.23 |
| 25 | .11742 ± .0020 | .02663 ± .0030 | 9.82 ± 1.08 | 2.42 ± 0.23 |
| 30 | .08661 ± .0024 | .02077 ± .0018 | 9.38 ± 0.93 | 3.17 ± 0.29 |
| 35 | .07325 ± .0034 | .01462 ± .0009 | 10.8 ± 0.91 | 4.16 ± 0.44 |
| 40 | .07852 ± .0042 | .01223 ± .0007 | 12.9 ± 1.03 | 4.48 ± 0.49 |
| 50 | .30243 ± .0015 | .00933 ± .0005 | 33.1 ± 1.72 | 3.05 ± 0.21 |
| Temp [°C] | T1 [s] | T2 [s] | τc × 10−9 [s] | |
|---|---|---|---|---|
| 5 | .14679 ± .0119 | .04520 ± .0011 | 7.57 ± 1.27 | 1.62 ± 0.27 |
| 15 | .21169 ± .0040 | .06527 ± .0139 | 7.54 ± 1.89 | 1.12 ± 0.17 |
| 20 | .20545 ± .0054 | .05064 ± .0060 | 9.19 ± 1.18 | 1.32 ± 0.15 |
| 25 | .20439 ± .0034 | .04086 ± .0040 | 10.8 ± 0.98 | 1.49 ± 0.13 |
| 30 | .25872 ± .0047 | .04663 ± .0050 | 11.7 ± 1.10 | 1.26 ± 0.11 |
| 35 | .26192 ± .0093 | .04375 ± .0043 | 12.3 ± 1.23 | 1.29 ± 0.14 |
| 40 | .29331 ± .0064 | .04222 ± .0057 | 13.6 ± 1.57 | 1.25 ± 0.14 |
| 50 | .33983 ± .0075 | .03655 ± .0027 | 16.4 ± 1.01 | 1.27 ± 0.10 |
TABLE X.
Relaxation times for the components with T1 > T2 in the T1-T2 2D experiment performed on A. aurantia silk.
| Temp [°C] | T1 [s] | T2 [s] | τc × 10−9 [s] | |
|---|---|---|---|---|
| 5 | .10797 ± .0064 | .00500 ± .0003 | 26.6 ± 1.72 | 6.26 ± 0.71 |
| 15 | .16008 ± .0061 | .00851 ± .0013 | 24.7 ± 2.82 | 3.94 ± 0.55 |
| 20 | .14969 ± .0064 | .00969 ± .0008 | 22.1 ± 1.67 | 3.79 ± 0.41 |
| 25 | .19571 ± .0111 | .00534 ± .0003 | 35.3 ± 2.13 | 4.54 ± 0.50 |
| 30 | N/A | N/A | N/A | N/A |
| 35 | .20546 ± .0084 | .00266 ± .0001 | 52.0 ± 2.49 | 6.32 ± 0.54 |
| 40 | N/A | N/A | N/A | N/A |
| 50 | N/A | N/A | N/A | N/A |
| Temp [°C] | T1 [s] | T2 [s] | τc × 10−9 [s] | |
|---|---|---|---|---|
| 5 | .11415 ± .0044 | .02098 ± .0032 | 11.5 ± 1.72 | 2.81 ± 0.42 |
| 15 | .17859 ± .0073 | .17859 ± .0087 | 10.9 ± 2.75 | 1.72 ± 0.37 |
| 20 | .20411 ± .0083 | .04699 ± .0086 | 9.70 ± 1.94 | 1.38 ± 0.24 |
| 25 | .21991 ± .0098 | .03969 ± .0096 | 11.6 ± 2.82 | 1.47 ± 0.32 |
| 30 | .24590 ± .0106 | .01943 ± .0017 | 19.7 ± 1.59 | 2.07 ± 0.23 |
| 35 | .24562 ± .0083 | .02575 ± .0017 | 16.6 ± 1.20 | 1.78 ± 0.17 |
| 40 | .24855 ± .0055 | .01076 ± .0005 | 27.6 ± 1.10 | 2.82 ± 0.17 |
| 50 | .39090 ± .0095 | .00595 ± .0005 | 47.8 ± 2.18 | 3.06 ± 0.21 |
| Temp [°C] | T1 [s] | T2 [s] | τc × 10−9 [s] | |
|---|---|---|---|---|
| 5 | .16054 ± .0089 | .06553 ± .0143 | 5.66 ± 2.03 | 1.27 ± 0.24 |
| 15 | .20963 ± .0030 | .07735 ± .0190 | 6.32 ± 2.40 | 1.03 ± 0.17 |
| 20 | .29186 ± .0080 | .11745 ± .0263 | 5.76 ± 1.89 | 0.71 ± 0.09 |
| 25 | .27553 ± .0109 | .09885 ± .0243 | 6.52 ± 2.26 | 0.80 ± 0.14 |
| 30 | .26235 ± .0093 | .06864 ± .0097 | 8.72 ± 1.45 | 1.00 ± 0.14 |
| 35 | .29548 ± .0156 | .07884 ± .0107 | 8.60 ± 1.45 | 0.88 ± 0.13 |
| 40 | .29080 ± .0110 | .06237 ± .0050 | 10.2 ± 0.98 | 1.00 ± 0.11 |
| 50 | .31266 ± .0111 | .04821 ± .0035 | 13.0 ± 1.01 | 1.13 ± 0.11 |
The tables also highlight the correlation time τc and constant CQeff as a function of temperature.77
Dielectric relaxation methods have been recently used to investigate water in restricted environments, such as micelles and microemulsions, and have shown that bulk water experiences fast solvation dynamics at picosecond timescales while confined water may experience slow nanosecond solvation dynamics.44–46 The origin of the slow dynamics in this, and in many other systems such as lipids47 and DNA molecules48, is currently under debate in the community. Nevertheless, molecular dynamics simulations of water in simplified geometries, for example, have revealed a slowing down of solvation dynamics upon confinement.49,50 These results are reminiscent of what we have experimentally observed in the hydrated silk of N. clavipes and A. aurantia where the tumbling correlation times of water with T1>T2 range from 5 to 35 ns. Interestingly, similar values for the correlation times have been observed in experimental studies of water in elastin.51
The static quadrupolar coupling constant for deuterium in heavy water at room temperature is approximately 214 kHz41. In the N. clavipes and A. aurantia silk samples we found that ranged from approximately 1 × 1010[s−2] to 10 × 1010[s−2], thus CQeff ranges from approximately 10 to 50 kHz. This effective quadrupolar coupling constant is different than the static or instantaneous value and includes librational motion parametrized by Γ defined by equation 3. The constant CQeff appears to vary for some of the components as a function of temperature, which may be a result of the thermally induced changes in the density of water within each reservoir, or the extent of hydrogen bonding between water molecules and water molecules and silk. Simulation studies have shown that the quadrupolar coupling constant reduces by approximately 30% from its static value and the asymmetry parameter η by increases %20 when the O-D bond length in heavy water changes by only %642. However, the differences observed in the static quadrupolar coupling constant and that measured in the water in silk would suggest a larger difference, which may include variations in the O-D bond length as well as anisotropic motion that was not included in the formulation of the model for equations 1 and 2. The observation of a reduced quadrupolar coupling constant on the order of 120 kHz in D20 has been reported in recent work of structural water in helical peptides52.
Tables 2 and 3 highlight the known primary structures for N. clavipes and A. aurantia silks. Glycine and alanine make up the majority of the composition any one of the samples, and one point to take note of is the similarity in the repeating motifs found in each silk. N. clavipes and A. aurantia major ampullate silks are made up of similar repetitions such as GLGSQ, GYGGQ, GPSG and GPGQQ. Additionally, there are blocks of alanine regions in the spider silks which form β-sheets which would be expected to not allow for the penetration of water.53 Results from the molecular dynamics simulations of the peptides studied in this work are tabulated in Table 4 through Table 8. The general trends in all of the simulations we performed point to an increase in sidechain contacts and peptide hydrogen bonds, concomitant with a decrease in the number of peptide-water hydrogen bonds, radii of gyration, solvent accessible surface areas (SASA)s, non-polar SASAs and number of proximal water molecules as the temperature is raised from 5 to 60°C. This finding is reminiscent of an observation made in simulation based studies of short elastin mimetic peptides VPGVG and LGGVG.54 One peculiarity in our experimental data, shown in Figure 4, is that the population of water decreases from 20 to 25 °C, but then increases slightly when the temperature is raised to 30 °C and additional heating of the samples results in further reduction of the population of water with T1 > T2. Some of the peptides we simulated also exhibit this characteristic near 35 °C; for example the repeating motifs (GYGSG)5,(GLGSQ)5, (GLGGQ)5, and (GPGGY)5 show a clear decrease in the number of proximal water molecules as the temperature is raised from 5 to 25 °C, however at 30 °C the number of water molecules within 0.6nm of the peptide atoms increases, and additional heating of the peptide to 60 °C results in further expulsion of water.
TABLE II.
Repeating motifs (shown in color) found in MaSp1 and MaSp2 silks from N. clavipes studied in the MD simulations of this work.75
| N.clavipes |
|---|
| MaSp1 |
| QGAGAAAAAAGGAGQGGYGGLGGQGAGQGGYGGLGGQGAGQGAGAAAAAAAGGAGQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQGGYGGLGSQG |
| AGRGGLGGQGAGAAAAAAAGGAGQGGYGGLGNQGAGRGGQGAAAAAAGGAGQGGYGGLGSQGARGGLGGQGAGAAAAAAGGAGQGGYGGLGGQ |
| GAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAGGAGQGGLGGQGAGQGAGASAAAAGGAGQGGYGGLGSQGAGRGGEGAGAAAAAAGGAGQGGY |
| GGLGGQGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAGGAGQGGLGGQGAGQGAGAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAVAA |
| AAAGGAGQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQRGYGGLGNQGAGRGGLGGQGAGAAAAAAAGGAGQGGYGGLGNQGAGRGGQG |
| AAAAAGGAGQGGYGGLGSQGAGRGGQGAGAAAAAAVGAGQEGIRGQGAGQGGYGGLGSQGSGRGGLGGQGAGAAAAAAGGAGQGGLGGQGAGQGA |
| GAAAAAAGGVRQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQGGYGGLGGQGVGRGGLGGQGAGAAAAGGAGQGGYGGVGSGASAASAAASRLSSPQA |
| SSRLSSAVSNLVATGPTNSAALSSTISNVVSQIGASILVFLDVMSSFKLFSRLFLLLSRS |
| MaSp2 |
| PGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAAAGPGGYGPGQQGPGGYGPGQQGPGRYGPGQQGPSGPGSAAAAAAGSGQQGPGGYGPRQQ |
| GPGGYGQGQQGPSGPGSAAAASAAASAESGQQGPGGYGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAASGPGQQGPGGYGPGQQ |
| GPGGYGPGQQGPSGPGSAAAAAAAASGPGQQGPGGYGPGQQGPGGYGPGQQGLSGPGSAAAAAAAGPGQQGPGGYGPGQQGPSGPGS |
| AAAAAAAAAGPGGYGPGQQGPGGYGPGQQGPSGAGSAAAAAAAGPGQQGLGGYGPGQQGPGGYGPGQQGPGGYGPGSASAAAAAAGPGQQ |
| GPGGYGPGQQGPSGPGSASAAAAAAAAGPGGYGPGQQGPGGYAPGQQGPSGPGSASAAAAAAAAGPGGYGPGQQGPGGYAPGQQGPSG |
| PGSAAAAAAAAAGPGGYGPAQQGPSGPGIAASAASAGPGGYGPAQQGPAGYGPGSAVAASAGAGSAGYGPGSQASAAASRLASPDS |
| GARVASAVSNLVSSGPTSSAALSSVISNAVSQIGASNPGLSGCDVLIQALLEIVSACVTILSSSSIGQVNYGAASQFAQVVGQSVLSAF |
TABLE III.
Repeating motifs (shown in color) found in MaSp1 and MaSp2 silks from A. aurantia76 studied in the MD simulations of this work.
| A.aurantia |
|---|
| MaSp1 |
| AGQGGAGAAAAAAAAGGAGGAGRGGLGAGGAGQGYGSGLGGQGGAGGGAAAAAAAAAGGQGGQGGYGGLGSQGAGQGGYGAGQ |
| GGAGAAAAAAAAGGAGGAGRGGLGAGGAGQGYGSGLGGQGGAGQGGAAAAAAAAGGQGGQGGYGGLGSQGAGQGGAGRGAAAA |
| AAAAGGQGGRGGYGGLGSQGAGQGGYGAGQGGAGAAAAAAAAGGAGEGGLGAGGAGQGYGSGLGGQGGAGQGG |
| AAAAAAAAGGQGGHGGYGGLGSQGAGQGGAGRGAAAAAAAAGGQGGQGGYGGLGSQGAGQGGYGAGQGGAAAAA |
| AAAAAGGAGGAGRGELGAGGAGQGYGXGLGGQGGAGQRGAASVAALAGGQGGQGGFGGFSSQGAGQGAYGGGAYSGQGAAASVSAASAAASRLS |
| SPGAASRVSSAVTSLVSSGGPTNPAALSNTISXVVSQISE |
| MaSp2 |
| PGGAGQQGPGGQGPYGPGAAAAAAAAGGYGPGAGQQGPXGAGQQGPGSQGPGGAGQQGPGGQGPYGPGAAAAAAAVGGYGPGAGQQ |
| GPGSQGPGSGGQQGPGGQGPYGPSAAAAAAAAGGYGPGAGQQGPGSQGPGSGGQQGPGGLGPYGPSAAAAAAAAGGYGPGAGQQGPGSQGPGSG |
| GQQRPGGLGPYGPSAAAAAAAAGGYGPGAGQQGPGSQGPGSGGQQRPGGLGPYGPSAAAAAAAAGGYGPGAGQQGPGSQAPVASAAASRLSSPQA |
| SSRVSSAVSTLVSSGPTNPAALSNAISSVVSQVSASNPGLSGCDVLVQALLELVSALVHILGSSSIGQINYAAS |
TABLE IV.
Molecular dynamics based findings for [GPGSG]5 (top) and [GPGSQ]5 (bottom) from 5°C to 60°C in A. aurantia (MaSp2) silk
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2 )B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 0.88 ± 0.09 | 19.0 ± 0.97 | 10.1 ± 0.63 | 271.7 ± 21.3 | 12257.3 ± 566.2 | 4.50 ± 1.21 | 52.0 ± 3.60 |
| 25 | 0.76 ± 0.05 | 18.4 ± 0.71 | 9.90 ± 0.43 | 274.8 ± 15.4 | 11611.3 ± 503.6 | 4.24 ± 1.38 | 49.6 ± 3.90 |
| 30 | 0.70 ± 0.03 | 16.5 ± 0.74 | 9.25 ± 0.36 | 332.5 ± 13.6 | 10237.0 ± 434.2 | 8.99 ± 2.03 | 40.7 ± 4.08 |
| 35 | 0.79 ± 0.02 | 17.7 ± 0.56 | 9.85 ± 0.34 | 292.1 ± 9.15 | 11037.0 ± 366.1 | 7.67 ± 1.87 | 43.3 ± 4.03 |
| 40 | 0.75 ± 0.03 | 16.7 ± 0.47 | 9.22 ± 0.33 | 326.7 ± 11.6 | 10428.5 ± 278.4 | 7.18 ± 1.43 | 43.2 ± 3.66 |
| 60 | 0.73 ± 0.03 | 17.0 ± 0.55 | 9.51 ± 0.36 | 306.7 ± 10.2 | 10311.8 ± 374.0 | 7.48 ± 1.32 | 42.5 ± 3.72 |
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 1.64 ± 0.09 | 24.4 ± 0.66 | 11.4 ± 0.31 | 260.4 ± 16.4 | 15652.9 ± 438.3 | 2.17 ± 1.45 | 71.8 ± 4.55 |
| 25 | 1.67 ± 0.09 | 23.0 ± 0.63 | 11.2 ± 0.30 | 272.8 ± 11.9 | 14696.4 ± 423.4 | 4.71 ± 1.30 | 65.0 ± 4.42 |
| 30 | 0.85 ± 0.07 | 21.6 ± 0.85 | 9.85 ± 0.54 | 322.1 ± 19.8 | 13664.9 ± 609.5 | 4.32 ± 1.36 | 64.7 ± 4.32 |
| 35 | 0.90 ± 0.02 | 19.7 ± 0.49 | 9.55 ± 0.35 | 368.5 ± 10.5 | 12304.9 ± 330.3 | 9.27 ± 1.72 | 53.6 ± 4.21 |
| 40 | 0.82 ± 0.03 | 20.3 ± 0.81 | 10.0 ± 0.45 | 333.4 ± 16.7 | 12775.3 ± 496.2 | 6.58 ± 1.79 | 58.3 ± 4.64 |
| 60 | 0.85 ± 0.06 | 20.5 ± 0.85 | 9.68 ± 0.46 | 335.2 ± 23.2 | 12291.5 ± 609.7 | 6.37 ± 1.78 | 56.6 ± 5.04 |
radius of gyration averaged over all Cα
solvent accessible surface area (SASA) was computed with the minimum distance set to 0.14nm
side-chain contacts were counted when two neighboring aliphatic carbon atoms from neighboring residues were within 0.65nm of each other
number of water molecules were counted when a water molecule is within 0.6nm of any protein atoms, and (E) hydrogen bonds are defined as when the hydrogen and the acceptor atoms are within 0.35nm.
hydrogen bonds are defined as when the hydrogen and the acceptor atoms are within 0.35nm.
The errors shown in the table were determined by the standard deviation of the fluctuations observed over the 2 ns of data used in the analysis, as discussed in the text.
TABLE VIII.
Molecular dynamics based findings for [GPSG]5 (top) and [GPSGPGS]5 (bottom) from 5°C to 60°C in N. clavipes (MaSp2) silk
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 0.78 ± 0.04 | 16.4 ± 0.50 | 8.81 ± 0.24 | 210.1 ± 8.59 | 11012.4 ± 322.2 | 1.61 ± 1.01 | 46.2 ± 3.67 |
| 25 | 0.90 ± 0.03 | 15.1 ± 0.41 | 8.44 ± 0.31 | 241.2 ± 7.90 | 10196.1 ± 307.7 | 4.40 ± 1.16 | 38.8 ± 3.50 |
| 30 | 0.66 ± 0.02 | 13.5 ± 0.49 | 7.24 ± 0.34 | 264.3 ± 11.5 | 9137.8 ± 351.9 | 3.70 ± 1.41 | 38.5 ± 3.59 |
| 35 | 1.00 ± 0.06 | 16.6 ± 0.69 | 9.18 ± 0.45 | 208.9 ± 17.5 | 10654.7 ± 438.4 | 3.36 ± 1.39 | 41.1 ± 3.59 |
| 40 | 0.66 ± 0.04 | 14.1 ± 0.57 | 7.88 ± 0.30 | 264.4 ± 11.5 | 9200.0 ± 381.2 | 5.65 ± 1.65 | 35.6 ± 3.60 |
| 60 | 0.69 ± 0.02 | 14.6 ± 0.45 | 7.92 ± 0.28 | 251.4 ± 10.2 | 9275.9 ± 345.9 | 2.69 ± 1.50 | 38.2 ± 3.82 |
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 1.07 ± 0.03 | 27.9 ± 0.53 | 15.3 ± 0.39 | 414.6 ± 10.4 | 19284.7 ± 408.1 | 2.99 ± 0.73 | 80.9 ± 4.39 |
| 25 | 0.98 ± 0.03 | 26.1 ± 1.12 | 14.8 ± 0.70 | 465.1 ± 27.2 | 17800.0 ± 662.8 | 5.37 ± 1.77 | 72.5 ± 5.06 |
| 30 | 0.95 ± 0.03 | 24.0 ± 0.71 | 13.6 ± 0.45 | 493.3 ± 13.5 | 16643.4 ± 487.9 | 5.71 ± 1.53 | 68.1 ± 4.91 |
| 35 | 0.84 ± 0.02 | 22.2 ± 0.44 | 13.4 ± 0.33 | 524.2 ± 11.1 | 15355.4 ± 373.2 | 11.1 ± 1.57 | 60.2 ± 3.84 |
| 40 | 0.86 ± 0.01 | 22.6 ± 0.58 | 12.8 ± 0.35 | 558.8 ± 12.8 | 15678.0 ± 410.2 | 5.93 ± 1.36 | 67.5 ± 4.22 |
| 60 | 0.99 ± 0.04 | 24.4 ± 1.53 | 14.0 ± 0.77 | 475.7 ± 30.6 | 16068.1 ± 894.0 | 7.48 ± 2.09 | 62.3 ± 5.72 |
radius of gyration averaged over all Cα
solvent accessible surface area (SASA) was computed with the minimum distance set to 0.14nm
side-chain contacts were counted when two neighboring aliphatic carbon atoms from neighboring residues were within 0.65nm of each other
number of water molecules were counted when a water molecule is within 0.6nm of any protein atoms
hydrogen bonds are defined as when the hydrogen and the acceptor atoms are within 0.35nm.
The errors shown in the table were determined by the standard deviation of the fluctuations observed over the 2 ns of data used in the analysis, as discussed in the text.
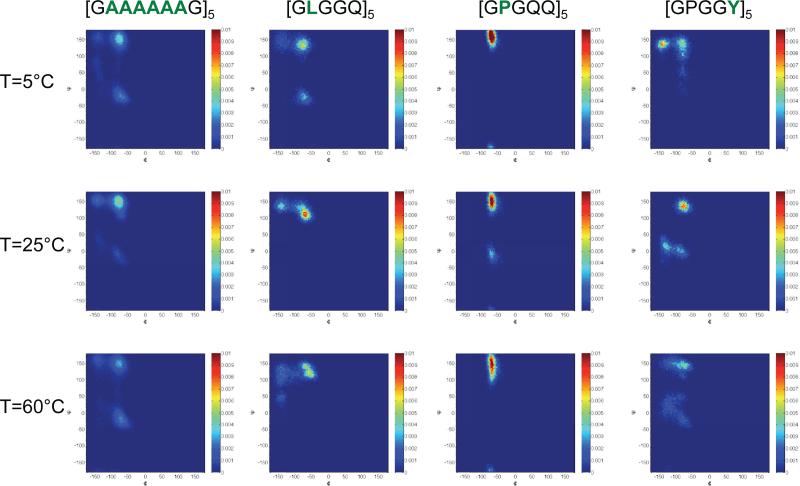
Examples of the Ramachandran maps from our simulations for the various motifs we investigated are shown in Figure 6. The figures were produced by accounting for the dihedral angles in the last 1 ns of any given simulation. In each figure we have normalized the integrated Ramachandran space to unity so that the results across temperatures may be compared. Overall, we find that the general features in any of the Ramachandran maps are that the motifs occupy a larger region of the dihedral space at 60°C compared with 5°C. This general feature appears to be reminiscent of the simulation findings of the elastin mimetic peptide (VPGVG)1855. To our knowledge NMR experiments on hydrated N. clavipes or A. aurantia have been performed only at or near 25°C, however there is good agreement between these experimental results and our short time simulations which we discuss below.
FIG. 6.
Ramachandran maps resulting from simulations of the various short peptide motifs at 5°C, 25°C and 60°C. The figure highlights the backbone conformations for alanine ([GAAAAAAG]5, leucine( [GLGGQ)5], proline([GPGQQ]5) and tyrosine([GPGGY5]). As discussed in the text, changes in temperature are observed pointing to a broadening of the dihedral angle range that is taken up by the motifs between the low and high temperatures. In the figure, the color scale denotes the number of occurrences a particular dihedral angle was measured over 1 ns of simulation time; the integrated area of the Ramachandran space was normalized to unity for ease of comparison between the temperatures.
Referring to Figure 6, at 25°C the dihedral angles (Φ, Ψ)=(150,−80) and (−150,150) in the alanine motifs in (GAAAAAAG)5 which appears in N. clavipes reveals the strong presence of β-sheet motifs. The observation of β sheet structural features in the repeating alanine segments of this silk is well known in both hydrated3 and dry silk samples56. Our simulations also appear to indicate a very small population of structures that occupied the region of space near (Φ, Ψ)=(−80,−20) which would appear to be α-helical. Experiments on a short MaSp1 N. clavipes peptide which include the motif -GAAAAAAG- have shown that the alanine in close proximity to glycine adopted an α-helical structure when exposed to various solvents57. Additionally, experiments performed on 13C labeled silk from N. edulis revealed that approximately seventy-three percent of all the torsion angles for alanine motifs occupied the β-sheet region, with the remainder being α-helical58. Referring again to Figure 6, our simulations indicate that leucine in the repeating motif (GLGGQ)5 shows a large population in the dihedral range near (Φ, Ψ)=(−90,120) which we interpret to be a distorted 31 helix at 25°C; this finding is in agreement with recently reported experimental work performed by J. Ashida and coworkers on short model peptides consisting of AGG and LGG motifs59. In recent simulation studies of MaSp1 and MaSp2 proteins from the N. clavipes spider dragline silk the proline motifs were observed to take up type I and type II β-turn conformations with a small α-helical component60. Our results presented in Figure 6 for the proline residues in (GPGQQ)5 agree well with these findings, and experiments on 13C/15N-proline labeled A. aurantia dragline silk which provide evidence for an elastin-like β-turn structure for the repetitive Gly-Pro-Gly-X-X motif prevalent in major ampullate spidroin 2 (MaSp2)61. Lastly, we show examples of the Ramachandran maps for the tyrosine residues in (GPGGY)5 at 5°C, 25°C and 60°C. The simulations at 25°C indicate what may be interpreted as either a 31-helix or type II β-turn in addition to an almost equal population of an α-helical motif. In experiments on uniformly 13C labeled hydrated N. clavipes, tyrosine exhibited random coil characteristics on the NMR time scale3. The distribution of dihedral angles taken up by tyrosine in (GPGGY)5 in our short time simulations may account for this observation. To experimentally probe the structural changes with temperature is beyond the scope of the present work, however, we believe that studies on short peptides using well known two dimensional NMR methodology, such as a spin diffusion experiment, should be feasible (see reference57 for example).
Gosline and coworkers reported on an experimental measurement of the relative length versus temperature on major ampullate silk of A. diadematus between 10 and ~30°C suspended in distilled water24,62. Their results showed a decrease in the initial length of the silk with increasing temperature from 10 to 30°C. Over this temperature range, the linear coefficient of expansion was found to be −1.8×10−4°C−1 and above 30°C the linear coefficient was observed to markedly change to −3.9×10−4°C−1. The observed negative thermal expansion coefficient correlates well with what we observe in our simulations of short silk motifs and in our experimental studies of water in the silk samples. During supercontraction, the expulsion of water and reduction in size should be interpreted as entropic in origin; increase in temperature results in an further increase in configurational entropy. The change in configurational entropy in hydrated silks allows for rubber-like characteristics-hydrated silk elasticity is driven by an entropic force (FEnt=-T∂S/∂L) familiar in the classical treatment of rubber elasticity.
D.W. Urry and coworkers have experimentally demonstrated the so-called “inverse temperature transition” in an elastin polypentapeptide [VPGVG]n.63 In one study, by measuring the carbonyl 13C NMR relaxation times as a function of temperature, a transition in the mobility of the backbone was observed; the experimental data showed an increase in mobility of the protein upon raising the temperature between 10 to 25°C, then a reduction in mobility was observed from 25 to 40°C. The simulation studies of V. Daggett and coworkers on the elastin mimetic peptide55 [VPGVG]18 have given further insights into the inverse temperature transition of this peptide. Similar to the findings reported in this work, their simulations showed an increase in protein hydrogen bonds and side-chain contacts, as well as a decrease in SASA, radius of gyration, and number of localized water molecules as the temperature is raised from 5 to 60°C. The unique thermal behavior of short elastin peptides, usually having the repeat [VPGXG]n (where X is any amino acid except proline), has recently gained interest for a wide range of applications. In drug delivery, for example, elastin based peptides have been used to target a tumor site due to the fact that these peptides undergo an inverse temperature transition which is sensitive to pH and/or temperature64. Chilkoti and others65–70 have used modified elastin like peptides as drug carriers to the sites where tumors are located; in one study involving a mouse model71, it was found that this method of drug delivery slows down the progression of the tumor significantly. Additionally, elastin like peptides that exhibit an inverse temperature transition are currently at the heart of many research endeavors relating to tissue engineering72 for suture and cartilage repairs, and in other biotechnology applications73,74. Our simulation findings indicate that many motifs found in silks exhibit thermal characteristics similar to the well studied VPGVG motif of elastin and may be of use in these and other applications.
IV. CONCLUSION
In this work we have reported on measurements of the relaxation times of water hydrated N. clavipes and A. aurantia silks as a function of temperature by deuterium 2D T1-T2 ILT NMR. This experimental approach was used to enumerate the number of water reservoirs observable on the NMR time scale and to characterize their relative populations, and dynamics. In all of the silk samples studied we observed the most restricted components of water (where T1 > T2) reducing in population with increasing temperature between 20°C and 50°C. These components of water had correlation times much larger than that of free water, suggesting restricted and/or anisotropic dynamics. While the experimental method has allowed for distinguishing the number of water reservoirs based on their dynamical characteristics, this approach does not allow one to establish their location in proximity to any structural motifs of the silk. Molecular dynamics simulations were performed to probe the thermal characteristics of a variety of repeating motifs found in N. clavipes and A. aurantia silks. In simulations, we find that the peptides exhibit characteristics reminiscent of the so-called inverse temperature transition found in short elastin like motifs as the temperature is raised from 5°C to 60°C. In [GLGGQ]5, [GLGSQ]5, [GAAAAAAG]5, [GPGGY]5, [GPGQQ]5, [GPSG]5, [GPSGPGS]5, [GYGSG]5, [GPGSG]5 and [GPGSQ]5 peptides, we observed a decrease in the radius of gyration, SASA, non-polar SASA as well as an increase in the number of side-chain contacts and the number of peptide-peptide hydrogen bonds as the temperature is raised from 5 to 60°C. In addition, the simulations also reveal a decrease in number of water molecules similar to what was observed in the experimental measurements using 2D T1-T2 ILT NMR methodology.
FIG. 5.
Relative population of the waters of hydration exhibiting T1 > T2 as a function of temperature in A) N. clavipes and B) A. aurantia silks. As discussed in the text, the graphs show that these components reduce in signal intensity as the temperature is increased from 20 to 50°C. A similar trend as a function of temperature was observed in the MD based studies of several short repeating motifs found in each silk sample.
TABLE V.
Molecular dynamics based findings for [GYGSG]5 (top) and [GLGSQ]5 (bottom) from 5°C to 60°C in A. aurantia (MaSp1) silk (the repeating motif [GLGSQ]5 also appears in N. Clavipes MaSp1)
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 1.27 ± 0.14 | 25.7 ± 0.90 | 13.2 ± 0.60 | 228.8 ± 10.9 | 16039.4 ± 467.6 | 0.43 ± 0.60 | 75.8 ± 4.48 |
| 25 | 0.87 ± 0.04 | 18.3 ± 0.64 | 9.75 ± 0.35 | 354.9 ± 13.8 | 11854.2 ± 380.1 | 8.08 ± 1.39 | 56.6 ± 4.21 |
| 30 | 0.76 ± 0.02 | 19.2 ± 0.61 | 10.2 ± 0.36 | 346.4 ± 11.7 | 12461.3 ± 398.6 | 4.53 ± 1.14 | 61.8 ± 4.07 |
| 35 | 0.77 ± 0.05 | 18.9 ± 0.52 | 10.3 ± 0.37 | 340.1 ± 10.8 | 11914.2 ± 331.9 | 8.33 ± 1.89 | 55.0 ± 4.29 |
| 40 | 0.90 ± 0.06 | 21.3 ± 0.74 | 10.9 ± 0.57 | 313.0 ± 14.4 | 13236.9 ± 431.2 | 3.44 ± 1.51 | 64.5 ± 4.51 |
| 60 | 0.72 ± 0.02 | 17.4 ± 0.58 | 9.69 ± 0.44 | 377.3 ± 11.7 | 10812.1 ± 378.2 | 9.96 ± 1.96 | 49.4 ± 4.04 |
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 1.21 ± 0.05 | 25.9 ± 0.60 | 13.4 ± 0.35 | 257.6 ± 12.1 | 17174.6 ± 354.9 | 4.28 ± 1.24 | 74.5 ± 4.20 |
| 25 | 0.81 ± 0.05 | 21.0 ± 0.84 | 10.9 ± 0.48 | 331.1 ± 12.1 | 14177.2 ± 494.9 | 7.65 ± 1.75 | 62.3 ± 4.46 |
| 30 | 0.96 ± 0.04 | 21.3 ± 0.57 | 11.4 ± 0.46 | 316.8 ± 13.1 | 14274.2 ± 401.2 | 7.45 ± 2.20 | 61.9 ± 5.06 |
| 35 | 1.15 ± 0.11 | 23.2 ± 1.08 | 12.3 ± 0.59 | 259.8 ± 11.4 | 15311.1 ± 589.2 | 5.92 ± 2.36 | 65.1 ± 5.04 |
| 40 | 0.79 ± 0.05 | 20.3 ± 1.07 | 10.3 ± 0.56 | 306.4 ± 19.9 | 13473.2 ± 670.2 | 5.57 ± 1.53 | 60.3 ± 5.09 |
| 60 | 0.74 ± 0.01 | 20.2 ± 0.72 | 10.9 ± 0.34 | 330.0 ± 11.0 | 12991.5 ± 456.8 | 8.63 ± 2.08 | 54.0 ± 4.39 |
radius of gyration averaged over all Cα
solvent accessible surface area (SASA) was computed with the minimum distance set to 0.14nm
side-chain contacts were counted when two neighboring aliphatic carbon atoms from neighboring residues were within 0.65nm of each other
number of water molecules were counted when a water molecule is within 0.6nm of any protein atoms
hydrogen bonds are defined as when the hydrogen and the acceptor atoms are within 0.35nm.
The errors shown in the table were determined by the standard deviation of the fluctuations observed over the 2 ns of data used in the analysis, as discussed in the text.
TABLE VI.
Molecular dynamics based findings for [GLGGQ]5 (top) and [GAAAAAAG]5 (bottom) from 5°C to 60°C in N. clavipes (MaSp1) silk (the motif [GLGGQ]5 also appears in the MaSp1 sequence of A. aurantia)
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 1.24 ± 0.12 | 24.0 ± 0.64 | 11.6 ± 0.41 | 248.5 ± 12.8 | 15653.7 ± 393.0 | 1.93 ± 0.86 | 69.2 ± 4.19 |
| 25 | 0.69 ± 0.02 | 17.3 ± 0.74 | 9.03 ± 0.39 | 372.9 ± 14.3 | 11573.5 ± 521.6 | 9.12 ± 1.84 | 48.7 ± 4.84 |
| 30 | 0.72 ± 0.03 | 18.3 ± 0.78 | 9.20 ± 0.46 | 381.2 ± 14.5 | 12036.9 ± 504.2 | 8.31 ± 1.69 | 51.1 ± 4.21 |
| 35 | 0.94 ± 0.08 | 21.3 ± 0.89 | 11.1 ± 0.53 | 291.6 ± 22.8 | 13750.3 ± 523.6 | 6.58 ± 1.88 | 56.3 ± 5.08 |
| 40 | 0.80 ± 0.03 | 18.5 ± 0.78 | 9.23 ± 0.48 | 370.2 ± 18.7 | 11958.9 ± 460.0 | 6.99 ± 1.56 | 51.6 ± 4.42 |
| 60 | 0.68 ± 0.02 | 17.8 ± 0.51 | 9.41 ± 0.35 | 376.9 ± 12.7 | 11306.5 ± 388.0 | 9.41 ± 1.86 | 46.1 ± 4.21 |
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 2.44 ± 0.10 | 32.6 ± 0.56 | 19.7 ± 0.44 | 387.2 ± 10.9 | 21032.4 ± 423.6 | 3.87 ± 1.31 | 82.1 ± 4.62 |
| 25 | 0.88 ± 0.03 | 25.4 ± 0.80 | 15.7 ± 0.57 | 509.2 ± 15.4 | 17061.2 ± 511.2 | 9.04 ± 1.72 | 68.6 ± 4.49 |
| 30 | 0.87 ± 0.02 | 24.5 ± 0.69 | 15.0 ± 0.57 | 540.6 ± 14.4 | 16100.4 ± 499.0 | 8.20 ± 1.84 | 63.2 ± 4.52 |
| 35 | 2.76 ± 0.10 | 29.8 ± 1.84 | 18.3 ± 0.10 | 428.8 ± 32.5 | 19146.5 ± 975.9 | 5.87 ± 1.75 | 73.9 ± 5.61 |
| 40 | 1.22 ± 0.07 | 28.0 ± 0.79 | 17.0 ± 0.48 | 464.4 ± 15.7 | 17825.1 ± 498.1 | 5.61 ± 1.57 | 69.9 ± 4.67 |
| 60 | 1.21 ± 0.09 | 27.8 ± 0.92 | 17.1 ± 0.54 | 466.7 ± 16.4 | 17209.0 ± 569.5 | 6.65 ± 1.76 | 65.3 ± 5.10 |
radius of gyration averaged over all Cα
solvent accessible surface area (SASA) was computed with the minimum distance set to 0.14nm
side-chain contacts were counted when two neighboring aliphatic carbon atoms from neighboring residues were within 0.65nm of each other
number of water molecules were counted when a water molecule is within 0.6nm of any protein atoms
hydrogen bonds are defined as when the hydrogen and the acceptor atoms are within 0.35nm.
The errors shown in the table were determined by the standard deviation of the fluctuations observed over the 2 ns of data used in the analysis, as discussed in the text.
TABLE VII.
Molecular dynamics based findings for [GPGQQ]5 (top) and [GPGGY]5 (bottom) from 5°C to 60°C in N. clavipes (MaSp2) silk
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 1.55 ± 0.10 | 25.7 ± 0.66 | 10.5 ± 0.41 | 259.7 ± 13.9 | 16850.3 ± 450.3 | 1.60 ± 1.13 | 81.5 ± 4.84 |
| 25 | 0.88 ± 0.08 | 20.3 ± 0.64 | 8.63 ± 0.39 | 367.8 ± 18.3 | 13742.6 ± 570.8 | 5.66 ± 2.10 | 66.7 ± 5.56 |
| 30 | 0.79 ± 0.01 | 19.1 ± 0.46 | 8.33 ± 0.35 | 364.7 ± 10.7 | 12965.5 ± 361.8 | 8.49 ± 1.72 | 61.3 ± 4.49 |
| 35 | 0.91 ± 0.02 | 20.6 ± 0.57 | 8.68 ± 0.28 | 333.5 ± 15.2 | 13736.8 ± 407.3 | 7.13 ± 1.71 | 64.2 ± 4.58 |
| 40 | 1.00 ± 0.03 | 21.4 ± 0.57 | 8.75 ± 0.36 | 315.5 ± 11.6 | 14093.3 ± 398.5 | 5.09 ± 1.29 | 67.5 ± 4.69 |
| 60 | 0.95 ± 0.10 | 21.4 ± 0.81 | 9.07 ± 0.36 | 327.5 ± 13.3 | 13685.1 ± 484.5 | 5.75 ± 2.08 | 64.2 ± 5.41 |
| Temp (°C) | Rg (nm)A | SASA (nm2)B | Non Polar SASA (nm2)B | Side-Chain ContactsC | Number of Water MoleculesD | Peptide - Peptide H BondsE | Peptide - Water H BondsE |
|---|---|---|---|---|---|---|---|
| 5 | 1.16 ± 0.05 | 21.2 ± 0.43 | 13.2 ± 0.36 | 377.5 ± 10.4 | 14073.8 ± 331.3 | 3.85 ± 1.13 | 52.1 ± 3.42 |
| 25 | 0.68 ± 0.01 | 17.3 ± 0.49 | 10.6 ± 0.35 | 462.7 ± 18.1 | 11570.6 ± 343.3 | 6.91 ± 1.20 | 44.3 ± 3.18 |
| 30 | 1.97 ± 0.04 | 23.1 ± 0.60 | 13.9 ± 0.50 | 364.3 ± 15.4 | 14778.2 ± 381.5 | 2.32 ± 0.78 | 54.4 ± 3.89 |
| 35 | 0.77 ± 0.02 | 19.5 ± 0.53 | 12.1 ± 0.41 | 457.1 ± 25.7 | 12649.5 ± 383.1 | 5.21 ± 1.25 | 47.4 ± 3.90 |
| 40 | 0.79 ± 0.03 | 18.3 ± 0.54 | 11.3 ± 0.42 | 485.8 ± 31.4 | 11852.5 ± 412.5 | 5.91 ± 1.57 | 43.2 ± 3.95 |
| 60 | 0.68 ± 0.02 | 18.1 ± 0.53 | 11.1 ± 0.39 | 512.5 ± 19.9 | 11202.7 ± 402.6 | 6.40 ± 1.44 | 40.9 ± 3.85 |
radius of gyration averaged over all Cα
solvent accessible surface area (SASA) was computed with the minimum distance set to 0.14nm
side-chain contacts were counted when two neighboring aliphatic carbon atoms from neighboring residues were within 0.65nm of each other
number of water molecules were counted when a water molecule is within 0.6nm of any protein atoms
hydrogen bonds are defined as when the hydrogen and the acceptor atoms are within 0.35nm.
The errors shown in the table were determined by the standard deviation of the fluctuations observed over the 2 ns of data used in the analysis, as discussed in the text.
ACKNOWLEDGEMENTS
This research was supported, in part, by a grant of computer time from the City University of New York High Performance Computing Center under NSF Grants CNS-0855217, CNS-0958379 and ACI-1126113. G.S. Boutis acknowledges support from award No. SC1GM086268-06 from the National Institute of General Medical Sciences and funding from the PSC-CUNY. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health (NIH). O. T. Ukpebor acknowledges supported from the RISE program at Hunter College under Grant GM060665. The authors thank Gregory P. Holland for providing the N. clavipes and A. aurantia dragline silk samples and Tetsuo Asakura for useful discussions. In addition we thank Moshe C. Silverstein, Steven Morgan and Anthony Papaioannou for useful discussions and help with the simulations and Yi-Qiao Song for the use of the ILT code. Lastly we thank Noel Goddard for useful comments relating to this work.
REFERENCES
- 1.Murad AM, Rech EL. Journal of Molecular Modeling. 2010;17:1183–1189. doi: 10.1007/s00894-010-0823-4. [DOI] [PubMed] [Google Scholar]
- 2.Hinman MB, Jones JA, Lewis RV. Trends in Biotechnology. 2000;18:374–379. doi: 10.1016/s0167-7799(00)01481-5. [DOI] [PubMed] [Google Scholar]
- 3.Holland GP, Jenkins JE, Creager MS, Lewis RV, Yarger JL. Biomacromolecules. 2008;9:651–657. doi: 10.1021/bm700950u. [DOI] [PubMed] [Google Scholar]
- 4.Xu M, Lewis RV. Proceedings of the National Academy of Sciences. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinman MB, Lewis RV. Journal of Biological Chemistry. 1992;267:19320–19324. [PubMed] [Google Scholar]
- 6.Gosline J, Guerette P, Ortlepp C, Savage K. Journal of Experimental Biology. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 7.Urry DW, Ohnishi T, Long MM, Mitchell LW. International Journal of Peptide and Protein Research. 1975;7:367–378. doi: 10.1111/j.1399-3011.1975.tb02455.x. [DOI] [PubMed] [Google Scholar]
- 8.Brooks AE, Steinkraus HB, Nelson SR, Lewis RV. Biomacromolecules. 2005;6:3095–3099. doi: 10.1021/bm050421e. [DOI] [PubMed] [Google Scholar]
- 9.Craig CL, Riekel C, Herberstein ME, Weber RS, Kaplan D, Pierce NE. Molecular Biology and Evolution. 2000;17:1904–1913. doi: 10.1093/oxfordjournals.molbev.a026292. [DOI] [PubMed] [Google Scholar]
- 10.Tso I-M, Wu H-C, Hwang I-R. Journal of Experimental Biology. 2005;208:1053–1061. doi: 10.1242/jeb.01437. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi CY, Shipley NH, Lewis RV. International Journal of Biological Macromolecules. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 12.Savage KN, Gosline JM. Journal of Experimental Biology. 2008;211:1937–1947. doi: 10.1242/jeb.014217. [DOI] [PubMed] [Google Scholar]
- 13.Simmons A, Ray E, Jelinski LW. Macromolecules. 1994;27:5235–5237. [Google Scholar]
- 14.Ashida J, Ohgo K, Komatsu K, Kubota A, Asakura T. Journal of Biomolecular NMR. 2003;25:91–103. doi: 10.1023/a:1022220428948. [DOI] [PubMed] [Google Scholar]
- 15.Holland GP, Creager MS, Jenkins JE, Lewis RV, Yarger JL. Journal of the American Chemical Society. 2008;130:9871–9877. doi: 10.1021/ja8021208. [DOI] [PubMed] [Google Scholar]
- 16.van Beek JD, Hess S, Vollrath F, Meier BH. Proceedings of the National Academy of Sciences. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bratzel G, Buehler MJ. Journal of the Mechanical Behavior of Biomedical Materials. 2012;7:30–40. doi: 10.1016/j.jmbbm.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Work RW. Journal of Experimental Biology. 1985;118:379–404. [Google Scholar]
- 19.Work RW. Journal of Arachnology. 1981;9:299–308. [Google Scholar]
- 20.Yang Z, Liivak O, Seidel A, LaVerde G, Zax DB, Jelinski LW. Journal of the American Chemical Society. 2000;122:9019–9025. [Google Scholar]
- 21.Jelinski LW, Blye A, Liivak O, Michal C, LaVerde G, Seidel A, Shah N, Yang Z. International Journal of Biological Macromolecules. 1999;24:197–201. doi: 10.1016/s0141-8130(98)00085-3. [DOI] [PubMed] [Google Scholar]
- 22.Creager MS, Jenkins JE, Thagard-Yeaman LA, Brooks AE, Jones JA, Lewis RV, Holland GP, Yarger JL. Biomacromolecules. 2010;11:2039–2043. doi: 10.1021/bm100399x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Sponner A, Porter D, Vollrath F. Biomacromolecules. 2008;9:116–121. doi: 10.1021/bm700877g. [DOI] [PubMed] [Google Scholar]
- 24.Gosline JM, Denny MW, DeMont EM. Nature. 1984;309:551–552. [Google Scholar]
- 25.Shao Z, Young RJ, Vollrath F. International Journal of Biological Macromolecules. 1999;24:295–300. doi: 10.1016/s0141-8130(98)00093-2. [DOI] [PubMed] [Google Scholar]
- 26.Holland GP, Lewis RV, Yarger JL. Journal of the American Chemical Society. 2004;126:5867–5872. doi: 10.1021/ja031930w. [DOI] [PubMed] [Google Scholar]
- 27.Kleinberg R, Farooqui S, Horsfield M. Journal of Colloid and Interface Science. 1993;158:195–198. [Google Scholar]
- 28.Hills B, Costa A, Marigheto N, Wright K. Applied Magnetic Resonance. 2005;28:13–27. [Google Scholar]
- 29.McDonald PJ, Korb J-P, Mitchell J, Monteilhet L. Physical Review E. 2005;72:011409. doi: 10.1103/PhysRevE.72.011409. [DOI] [PubMed] [Google Scholar]
- 30.Sun C, Boutis GS. New Journal of Physics. 2011;13:025026. doi: 10.1088/1367-2630/13/2/025026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Work RW, Emerson PD. Journal of Arachnology. 1982;10:1–10. [Google Scholar]
- 32.Song Y-Q, Venkataramanan L, Hurlimann M, Flaum M, Frulla P, Straley C. Journal of Magnetic Resonance. 2002;154:261–268. doi: 10.1006/jmre.2001.2474. [DOI] [PubMed] [Google Scholar]
- 33.Venkataramanan L, Song Y-Q, Hurlimann MD. IEEE Transactions on Signal Processing. 2002;50:1017–1026. [Google Scholar]
- 34.Neudert O, Stapf S, Mattea C. Journal of Magnetic Resonance. 2011;208:256–261. doi: 10.1016/j.jmr.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 35.Hess B, Kutzner C, van der Spoel D, Lindahl E. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 36.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. The Journal of Chemical Physics. 1984;81:3684–3690. [Google Scholar]
- 37.Jorgensen WL, Tirado-Rives J. Journal of the American Chemical Society. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 38.Teleman O, Jönsson B, Engström S. Molecular Physics: An International Journal at the Interface Between Chemistry and Physics. 1987;60:193–203. [Google Scholar]
- 39.Lee JH, Labadie C, Springer CS, Jr, Harbison GS. Journal of the American Chemical Society. 1993;115:7761–7764. [Google Scholar]
- 40.Washburn K, Callaghan P. Physical review letters. 2006;97:175502. doi: 10.1103/PhysRevLett.97.175502. [DOI] [PubMed] [Google Scholar]
- 41.Lang E, Lüdemann H-D. Berichte der Bunsengesellschaft für physikalische Chemie. 1980;84:462–470. [Google Scholar]
- 42.Cummins PL, Bacskay GB, Hush NS, Halle B, Engström S. The Journal of chemical physics. 1985;82:2002. [Google Scholar]
- 43.Lang E, Piculell L, Lüdemann H-D. The Journal of Chemical Physics: J. Chem. Phys. 1984;81:3820–3827. [Google Scholar]
- 44.Bhattacharyya K, Bagchi B. The Journal of Physical Chemistry A. 2000;104:10603–10613. [Google Scholar]
- 45.Nandi N, Bhattacharyya K, Bagchi B. Chemical Reviews. 2000;100:2013–2046. doi: 10.1021/cr980127v. [DOI] [PubMed] [Google Scholar]
- 46.D'Angelo M, Fioretto D, Onori G, Palmieri L, Santucci A. Phys. Rev. E. 1996;54:993–996. doi: 10.1103/physreve.54.993. [DOI] [PubMed] [Google Scholar]
- 47.Datta A, Pal SK, Mandal D, Bhattacharyya K. The Journal of Physical Chemistry B. 1998;102:6114–6117. [Google Scholar]
- 48.Brauns EB, Madaras ML, Coleman RS, Murphy CJ, Berg MA. Journal of the American Chemical Society. 1999;121:11644–11649. [Google Scholar]
- 49.Senapati S, Chandra A. The Journal of chemical physics. 1999;111:1223. [Google Scholar]
- 50.Faeder J, Ladanyi B. The Journal of Physical Chemistry B. 2000;104:1033–1046. [Google Scholar]
- 51.Sun C, Mitchell O, Huang J, Boutis GS. The Journal of Physical Chemistry B. 2011;115:13935–13942. doi: 10.1021/jp207607r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pometun MS, Gundusharma UM, Richardson JF, Wittebort RJ. Journal of the American Chemical Society. 2002;124:2345–2351. doi: 10.1021/ja017364r. [DOI] [PubMed] [Google Scholar]
- 53.Izdebski T, Akhenblit P, Jenkins JE, Yarger JL, Holland GP. Biomacromolecules. 2010;11:168–74. doi: 10.1021/bm901039e. [DOI] [PubMed] [Google Scholar]
- 54.Huang J, Sun C, Mitchell O, Ng N, Wang ZN, Boutis GS. The Journal of Chemical Physics. 2012;136:085101. doi: 10.1063/1.3685454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li B, Alonso DO, Daggett V. Journal of Molecular Biology. 2001;305:581–592. doi: 10.1006/jmbi.2000.4306. [DOI] [PubMed] [Google Scholar]
- 56.Simmons A, Ray E, Jelinski LW. Macromolecules. 1994;27:5235–5237. [Google Scholar]
- 57.Asakura T, Yang M, Kawase T, Nakazawa Y. Macromolecules. 2005;38:3356–3363. [Google Scholar]
- 58.Van Beek J, Hess S, Vollrath F, Meier B. Proceedings of the National Academy of Sciences. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashida J, Ohgo K, Komatsu K, Kubota A, Asakura T. Journal of biomolecular NMR. 2003;25:91–103. doi: 10.1023/a:1022220428948. [DOI] [PubMed] [Google Scholar]
- 60.Keten S, Buehler MJ. Journal of the Royal Society Interface. 2010;7:1709–1721. doi: 10.1098/rsif.2010.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jenkins JE, Creager MS, Butler EB, Lewis RV, Yarger JL, Holland GP. Chemical Communications. 2010;46:6714–6716. doi: 10.1039/c0cc00829j. [DOI] [PubMed] [Google Scholar]
- 62.Savage KN, Gosline JM. Journal of Experimental Biology. 2008;211:1948–1957. doi: 10.1242/jeb.014225. [DOI] [PubMed] [Google Scholar]
- 63.Urry DW, Trapane TL, Iqbal M, Venkatachalam CM, Prasad KU. Biochemistry. 1985;24:5182–5189. doi: 10.1021/bi00340a034. [DOI] [PubMed] [Google Scholar]
- 64.Meyer DE, Chilkoti A. Biomacromolecules. 2004;5:846–51. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 65.Chilkoti A, Dreher MR, Meyer DE. Advanced Drug Delivery Reviews. 2002;54:1093–1111. doi: 10.1016/s0169-409x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 66.Massodi I, Thomas E, Raucher D. Molecules. 2009;14:1999–2015. doi: 10.3390/molecules14061999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacKay AJ, Chen M, McDaniel JR, Liu W, Simnick AJ, Chilkoti A. Nature Materials. 2009;8:993–999. doi: 10.1038/nmat2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herrero-Vanrell R, Rincón a. C., Alonso M, Reboto V, Molina-Martinez IT, Rodríguez-Cabello JC. Journal of Controlled Release. 2005;102:113–22. doi: 10.1016/j.jconrel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Bidwell GL, Davis AN, Fokt I, Priebe W, Raucher D. Investigational New Drugs. 2007;25:313–26. doi: 10.1007/s10637-007-9053-8. [DOI] [PubMed] [Google Scholar]
- 70.McDaniel JR, Callahan DJ, Chilkoti A. Advanced Drug Delivery Reviews. 2010;62:1456–67. doi: 10.1016/j.addr.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu W, MacKay JA, Dreher MR, Chen M, McDaniel JR, Simnick AJ, Callahan DJ, Zalutsky MR, Chilkoti A. Journal of Controlled Release. 2010;144:2–9. doi: 10.1016/j.jconrel.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nettles DL, Chilkoti A, Setton LA. Advanced Drug Delivery Reviews. 2010;62:1479–1485. doi: 10.1016/j.addr.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banki MR, Feng L, Wood DW. Nature Methods. 2005;2:659–661. doi: 10.1038/nmeth787. [DOI] [PubMed] [Google Scholar]
- 74.Megeed Z, Winters RM, Yarmush ML. Biomacromolecules. 2006;7:999–1004. doi: 10.1021/bm0507002. [DOI] [PubMed] [Google Scholar]
- 75.Jenkins JE, Creager MS, Lewis RV, Holland GP, Yarger JL. Biomacromolecules. 2010;11:192–200. doi: 10.1021/bm9010672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jenkins JE, Creager MS, Butler EB, Lewis RV, Yarger JL, Holland GP. Chemical Communications. 2010;46:6714–6716. doi: 10.1039/c0cc00829j. [DOI] [PubMed] [Google Scholar]
- 77.Abragam A. The Principles of Nuclear Magnetism. Clarendon Press; 1961. [Google Scholar]
- 78.Bloembergen N, Purcell EM, Pound RV. Physical Review. 1948;73:679–712. [Google Scholar]
- 79.Duer M. Introduction to solid-state NMR spectroscopy. Blackwell; 2004. [Google Scholar]