Abstract
Purpose
Previously, we reported PIK3CA gene mutations in high-grade intraductal papillary mucinous neoplasms (IPMN). However, the contribution of phosphatidylinositol-3 kinase pathway (PI3K) dysregulation to pancreatic carcinogenesis is not fully understood and its prognostic value unknown. We investigated the dysregulation of the PI3K signaling pathway in IPMN and its clinical implication.
Experimental Design
Thirty-six IPMN specimens were examined by novel mutant-enriched methods for hot-spot mutations in the PIK3CA and AKT1 genes. PIK3CA and AKT1 gene amplifications and loss of heterozygosity (LOH) at the PTEN locus were also evaluated. Additionally, the expression levels of PDPK1/PDK1, PTEN and Ki67 were analyzed by immunohistochemistry.
Results
Three cases carrying the E17K mutation in the AKT1 gene and one case harboring the H1047R mutation in the PIK3CA gene were detected among the 36 cases. PDK1 was significantly overexpressed in the high-grade IPMN vs. low-grade IPMN (p = 0.034) and in pancreatic and intestinal-type of IPMN vs. gastric-type of IPMN (p = 0.020). Loss of PTEN expression was strongly associated with presence of invasive carcinoma and poor survival in these IPMN patients (p = 0.014).
Conclusion
This is the first report of AKT1 mutations in IPMN. Our data indicate that oncogenic activation of the PI3K pathway can contribute to the progression of IPMN, in particular loss of PTEN expression. This finding suggests the potential employment of PI3K pathway-targeted therapies for IPMN patients. The incorporation of PTEN expression status in making surgical decisions may also benefit IPMN patients and should warrant further investigation.
Keywords: PI3K-pathway, AKT1, PTEN, IPMN, mutant-enriched sequencing method
INTRODUCTION
Pancreatic adenocarcinoma is a malignancy of extremely poor prognosis with high mortality and short survival. Methods for its early detection and effective treatments, which will require an understanding of the underlying mechanisms of its development, are urgently needed.
Pancreatic adenocarcinoma is thought to develop from precancerous lesions such as PanIN (pancreatic intraepithelial neoplasia), IPMN, and MCN (mucinous cystic neoplasm) (1). IPMN is the most common cystic precancerous lesion of the pancreas, representing ~20% of surgically resected cystic lesions of the pancreas (2, 3). The prognosis for non-invasive IPMN is excellent and detection and surgical intervention at the preinvasive stage is curative (4, 5). The prognosis is poor for tumors with associated invasive adenocarcinoma, but still more favorable than for conventional pancreatic ductal adenocarcinoma (PDAC) arising from PanIN lesions (6).
By definition, IPMN involve the main pancreatic duct and/or its branches; they are characterized by papillary projections of ductal epithelium and dilatation of the pancreatic duct. Histologically, IPMN are distinguished by the replacement of normal ductal epithelium with a mucinous epithelium showing a broad spectrum of histopathological changes, ranging from IPMN with low grade dysplasia (adenoma), IPMN with moderate dysplasia (borderline tumor) and IPMN with high grade dysplasia/carcinoma in situ (IPMC). IPMN may be associated with invasive adenocarcinoma showing stromal invasion (7). Currently, four histological subtypes of IPMN by immunophenotypic characteristics of the lining epithelium are defined: a gastric foveolar-type (MUC1−, MUC2−, MUC5AC+, MUC6+), an intestinal-type (MUC1−, MUC2+, MUC5AC+, CDX2+), a pancreatobiliary-type (MUC1+, MUC2−, MUC5AC+, CDX2−) and an oncocytic-type (variable expression of MUC1 and MUC2) (8). The intestinal-type usually affects the main pancreatic duct and if an associated invasive carcinoma is present, it is commonly of the mucinous/colloid type with a more favorable prognosis than the conventional PDAC. The pancreatobiliary-type often shows high grade dysplasia with or without associated invasive PDAC, with a tubular morphology showing histologic features of the conventional type of PDAC. The gastric foveolar-type often affects branch ducts and is less commonly associated with an invasive tumor. The oncocytic-type is exceptionally rare. Approximately 20-45% of resected IPMNs have an invasive adenocarcinoma component (9, 10).
The specific mutations leading to the development of various histological grades of IPMN have been partially characterized in previous studies (11-13). Reported genetic alterations identified in IPMN include mutations in KRAS (11, 14, 15), GNAS (16, 17), PIK3CA (12) and BRAF (11). Other changes include the loss of expression of STK11/LKB1 (18-20) and overexpression of TP53 and ERBB2 proteins in the IPMNs (15), and allelic loss of PTEN in the pancreatic cyst fluid DNA of IPMN patients (21). A genetic analysis of microdissected IPMN of different grades within the same tumor has demonstrated early polyclonal epithelia gradually replaced by monoclonal neoplastic cells gaining KRAS mutations as the tumor progress (22). However, the genetic profile of IPMN progression is not yet complete.
The PI3K pathway is genetically deregulated in human cancers at various levels. The first identified genetic mechanism of PI3K pathway activation was the loss of PTEN function by mutation or deletion, leading to the accumulation of the PI3K product phosphatidylinositol 3-phosphate (PIP3). The accumulation of PIP3 activates a signaling cascade starting with the phosphorylation (activation) of the protein serine-threonine kinase AKT by 3-phosphoinositide dependent protein kinase-1 (PDK1/PDPK1). PDK1 is considered a “master kinase” which phosphorylates and is responsible for the activation of all ACG family members, many of them related to cell proliferation, survival or the inhibition of apoptosis, including AKT1, 2 and 3 (23, 24).
Germline mutations in the PTEN gene can cause Cowden syndrome with cancer predisposition, and it is commonly sporadically mutated in prostate cancer, endometrial cancer and glioblastoma among others (25, 26). In mice, PTEN haploinsufficiency has been shown to be sufficient to accelerate the development of metaplasias and PanIN-like lesions in the KrasG12D mutant background. Moreover, in Pdx1-Cre;Ptenlox/lox mice, the metaplastic phenotype shows a more papillary nature than those in Pdx1-Cre;KrasG12D; Ptenlox/lox mice, suggesting that the papillary phenotype is associated with the loss of PTEN (27, 28).
Amplifications of genomic regions containing AKT1 or PIK3CA genes have also been described (29-31). Recent studies have reported high frequencies of somatic mutations in the phosphoinositide-3-kinase catalytic subunit, p110α (PIK3CA) gene, in several cancer types, including colorectal, gastric, thyroid, breast, ovary, certain brain tumors and head and neck squamous cell carcinomas (32-34). In the study by Samuels and colleagues (35), 75% of the mutations found in the PIK3CA gene clustered within the helical (exon 9) and catalytic (exon 20) protein domains and three hot-spot mutations were identified: E542K, E545K (exon 9), and H1047R (exon 20). The three hot-spot mutations have been shown to elevate the PI3K oncogenic activity via Akt signaling pathway, providing transforming properties in vitro and in vivo (36-38). We have previously reported somatic PIK3CA gene mutations in four of 36 (11%) IPMN/IPMCs (12). Consistent to our finding, Lubezky et al. recently described the detection of H1047R hot-spot mutation in two of 27 IPMN cases, one of which was a low-grade IPMN (39).
Rare or absent activating somatic mutations in the AKT1 gene have also been recently described (40-42). The E17K mutation in the pleckstrin homology domain of the AKT1 gene can result in PI3K-independent membrane recruitment of Akt, recapitulating the effects of the AKT8 murine leukemia retrovirus GAG-AKT fusion protein. E17KAKT1 exhibits transforming activity in vitro and in vivo, albeit at lower level than the myristoylated Akt (40, 41). To-date, the mutational status of the AKT1 gene has not been evaluated in IPMN.
The aim of this study was to evaluate the status of the PI3K pathway and its association with clinicopathological variables in 36 IPMN at the molecular level by analyzing the hot-spot mutations in the AKT1 and PIK3CA genes, the LOH status at the PTEN gene locus, and expression levels of PTEN and PDK1. This pathway contains many putative therapeutic targets; knowing the prevalence of alterations in this pathway may be useful to determine its potential diagnostic and therapeutic applications to IPMN patients.
MATERIALS AND METHODS
Patients and tissue samples
Thirty-six formalin fixed paraffin embedded IPMN were obtained from the archival tissue collection at the CUMC. The 36 cases were chosen based on their histological typing for a balanced presentation of IPMN-1-3 for a tissue microarray. We chose to study this set of patients for the same reason, and also the availabilities of the tissue microarray and clinicopathological findings. The acquisition of the tissue specimens was approved by the Institutional Review Board and performed in accordance with Health Insurance Portability and Accountability Act (HIPAA) regulations. All samples were selected from pancreatic resections performed at Columbia University Presbyterian Hospital between January 2007 and June 2008. By definition, all the IPMN included in the study involved the main pancreatic duct and/or branches. The patients consisted of 16 males and 20 females with ages ranging from 44 to 85 years (median age: 70.0). Histological typing of the tumors was performed according to the recommendations in the WHO classification (43). The IPMN in this study were all positive for MUC5A immunolabeling (data not shown) and included gastric, intestinal and pancreatobiliary histological subtypes. Detailed clinicopathological findings of the 36 IPMN cases are presented in Table 1. Patients with pancreatic neoplasms were recruited to our institutional prospective longitudinal outcomes study. Patients being followed for premalignant cystic neoplasms were evaluated with cross sectional imaging and / or endoscopic ultrasound every 6 to 12 months depending on patient specific demographics. Patients who have undergone pancreatic resection are evaluated and managed by our multidisciplinary group at regular intervals for the remainder of their lives. Patients who underwent resection of a benign IPMN lesion were evaluated yearly with magnetic resonance imaging/magnetic resonance cholangiopancreatography (MRI/MRCP) to screen the remnant pancreas for new or recurrent disease. For patients with benign IPMN, no evidence of disease (NED) was defined as the absence of cystic changes within the remnant pancreas on follow-up MRI/MRCP imaging. Patients who underwent resection of a malignant IPMN were offered chemotherapy to reduce the risk of recurrence. Once this was completed, patients were seen every three months with repeat surveillance imaging at six-month intervals. For patients with malignancies, recurrences were detected by imaging studies and serum tumor markers, and then confirmed by tissue biopsy. Patients were considered as NED (no evidence of disease) when there was no elevation in serum tumor markers and no new findings on surveillance imaging. Length of follow-up in this study was calculated from the day of surgery to the last clinic appointment on record at our center.
Table 1.
Summarized report of the 36 patient samples and genetic alterations identified in the PI3K/AKT/PTEN pathway.
| Patient No. |
Gender (Age) | Cyst size (cm) |
Location Within Pancreas |
Main duct or Mixed (MD) versus Branch duct (BD) |
IPMN Lesion Analyzed Histologic Subtype Gastric = G Intestinal = I Pancreatobiliary = P and Nuclear Grade (1-3) |
Resection Specimen (Highest Grade of IPMN) |
Resection Specimen (Stage of Invasive Cancer) |
Outcome | PIK3CA mutant |
AKT1 mutant |
PIK3CA amp |
AKT amp |
PTEN LOH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F (70) | 3.5 | Head | Main | G (1) | IPMN-2 | - | NED | No | No | No | ||
| 2 | F (79) | 2.2 | Body | Main | P (3) | IPMN-3 | pT1N0 | DOD | No | No | Yes | ||
| 3 | M (64) | 2.0 | Head | Main | I (2) | IPMN-3 | - | Recurrence | No | No | No | ||
| 4 | M (64) | 1.2 | Head | Main | G (1) | IPMN-3 | - | Recurrence | No | No | Yes | ||
| 5 | F (66) | 2.1 | Head | Main | G (1) | IPMN-1 | - | NED | No | No | NA | ||
| 6 | M (78) | 4.5 | Head | Branch | I (2) | IPMN-3 | - | NED | Yes | No | No | ||
| 7 | M (85) | 5.5 | Head | N/A | I (2) | IPMN-3 | pT3N1 | DOD | E17K | No | No | Yes | |
| 8 | F (44) | 1.0 | Head | Main | I (3) | IPMN-3 | - | NED | No | No | Yes | ||
| 9 | F (70) | 3.0 | Head | Main | P (3) | IPMN-3 | - | NED | No | No | No | ||
| 10 | M (75) | 3.0 | Head | Branch | P (3) | IPMN-3 | - | Recur/ DOD | No | No | No | ||
| 11 | F (79) | 2.0 | Head | Main | G (3) | IPMN-3 | - | NED | No | No | No | ||
| 12 | F (75) | 2.0 | Tail | Branch | G (2) | IPMN-2 | - | DOD | No | No | No | ||
| 13 | F (85) | 4.3 | Body | Main | I (3) | IPMN-3 | - | NED | No | No | No | ||
| 14 | F (36) | 4.8 | Head | Main | G (2) | IPMN-2 | - | NED | Yes | No | No | ||
| 15 | F (66) | 2.1 | Head | Main | I (3) | IPMN-3 | pT3N1 | Recur/ DOD | Yes | No | No | ||
| 16 | F (68) | 1.7 | Tail | Main | G (2) | IPMN-2 | - | NED | H1047R | No | No | No | |
| 17 | F (59) | 5.0 | Head | Branch | I (2) | IPMN-3 | NED | No | No | Yes | |||
| 18 | M (70) | 5.2 | Head | Branch | P (3) | IPMN-3 | NED | No | No | No | |||
| 19 | M (73) | 5.5 | Body | Main | G (2) | IPMN-2 | NED | No | No | No | |||
| 20 | M (78) | 1.6 | Head | Main | G (1) | IPMN-3 | pT1N0 | NED | E17K | No | No | No | |
| 21 | F (58) | 1.0 | Body | Main | G (1) | IPMN-1 | - | NED | Yes | No | No | ||
| 22 | M (79) | 5.1 | Tail | Main | P (3) | IPMN-3 | pT3N1 | Recur/DOD | No | Yes | Yes | ||
| 23 | F (58) | 4.0 | Body | Main | I (3) | IPMN-3 | NED | No | No | NA | |||
| 24 | M (71) | 5.0 | Head | Main | I (3) | IPMN-3 | NED | No | No | Yes | |||
| 25 | F (55) | 1.4 | Head/neck | Branch | G (3) | IPMN-3 | NED | No | No | NA | |||
| 26 | F (58) | 1.2 | Body | Branch | G (2) | IPMN-2 | NED | Yes | No | Yes | |||
| 27 | M (53) | 3.6 | Tail | Main | P (3) | IPMN-3 | NED | No | No | No | |||
| 28 | M (61) | 1.5 | Head | Main | I (2) | IPMN-3 | pT1N0 | NED | No | No | No | ||
| 29 | M (74) | 0.5 | Body/tail | Main | P (3) | IPMN-3 | - | Recurrence | No | No | Yes | ||
| 30 | F (78) | 4.5 | Distal | Main | G (2) | IPMN-2 | NED | No | No | NA | |||
| 31 | F (67) | 2.6 | Body | Main | G (2) | IPMN-2 | NED | No | No | NA | |||
| 32 | M (66) | 1.3 | Distal | Branch | I (3) | IPMN-3 | NED | Yes | No | No | |||
| 33 | F (82) | 2.0 | Head | Main | P (3) | IPMN-3 | pT3N1 | DOD | No | No | NA | ||
| 34 | M (68) | 2.0 | Distal | Main | I (3) | IPMN-3 | - | NED | No | No | NA | ||
| 35 | M (72) | 3.0 | Distal | N/A | I (3) | IPMN-3 | NED | E17K | Yes | No | NA | ||
| 36 | F (78) | 1.1 | Head/body | Branch | I (2) | IPMN-2 | NED | No | No | Yes |
IPMN-1 (low grade dysplasia) = IPMN, adenoma; IPMN-2 (moderate dysplasia) = IPMN, borderline tumor; IPMN-3 (high grade dysplasia) = IPMC, carcinoma in situ; NED: No evidence of diseases; DOD: Death of disease; NA: not informative.
Preparation of DNA extracts
To enrich the number of neoplastic cells procured from each sample, Laser Capture Microdissection (LCM) was performed on the IPMN. The regions containing the IPMN neoplastic cell populations were microscopically defined by the pathologists on our team (A.T.T. and H.E.R). Five to ten 5-μm serial sections were microdissected for each case. Paraffin-embedded tumor samples were deparaffinized by incubating the slides in xylene for two minutes and rehydrated in 99.9% ethanol for 2 × 10 minutes, in 96% ethanol for 2 × 10 minutes, and in 70% ethanol for 2 × 10 minutes. Slides were stained with hematoxylin and eosin (H&E). Microdissection was carried out using a laser microdissection microscope (P.A.L.M., Bernried, Germany). Approximately between 10,000 and 14,000 cells were collected into 50 μl of ATL buffer, (Lysis buffer from QIAamp DNA Mini Kit; QIAGEN, Valencia, CA). Surrounding non-neoplastic tissues were treated, microscopically defined, and dissected the same as the tumors, and served as the corresponding normal control for each sample. DNA extraction was performed according to manufacturer’s instructions.
Mutational Analysis of the PIK3CA gene
Mutations in exons 9 and 20 of the PIK3CA gene were analyzed by direct genomic sequencing methods and confirmed by our previously described mutant-enriched sequencing method (44). Polymerase chain reaction (PCR) amplification of genomic DNA (40 ng each) and direct sequencing of the PCR products were performed using the same primers and conditions as previously described (44). All PCR fragments were purified using ExoSAP-IT kit (Affymetrix, Santa Clara, CA) and sequencing was performed with ABI Prism 3730xl DNA analyzers by Genewiz, Inc (South Plainfield, NJ) using the PCR primers (44). Any alteration detected was further verified by sequencing of a second PCR product derived independently from the original DNA template.
Conventional direct genomic sequencing of the AKT1 gene
The point mutation G > A at nucleotide 49 of the AKT1 gene (E17K) was first examined by direct genomic sequencing. Genomic DNA was amplified with primers designed to amplify exclusively the hot-spot (AKT1-F 5′ - ACATCTGTCCTGGCACAC – 3′: AKT1-R 5′ -GCCAGTGCTTGTTGCTTG - 3′) (40). All PCR fragments were purified using Invitrogen PureLinkTM PCR purification kit (Life Technologies, Carlsbad, CA) and sequencing was carried out with ABI 3730x/DNA analyzers by Genewiz, Inc.
Mutant-enriched sequencing for detecting AKT1 mutation E17K
Based on the same principles that we have employed to design the mutant-enriched sequencing method for PIK3CA (44), here we developed a sensitive mutant-enriched sequencing method specific for the E17K hot-spot mutation of the AKT1 gene (Fig. 1A). A mismatched primer was designed to create a unique restriction enzyme site for EcoRI in the AKT1 exon 2 region in the first round of PCR. The mismatched primer AKT1ME-R (5′ - GGCCGCCAGGTCTTGATGAATT - 3′) was used as the reverse primer for both rounds of PCR. The forward primers for the first and second PCR were respectively AKT1ME-F1 (5′ - GGCTGTGCAGACTGGCCCAG - 3′) and AKT1ME-F2 (5′ - ACACAGCTCGGGGTGGCTCT - 3′). EcoRI digestion was performed at 37°C overnight. The forward PCR primer AKT1ME-F2 was also used as the DNA sequencing primer (Fig. 1A). Any alteration detected was further verified at least twice by repeating the process starting with a second PCR product derived independently from the original DNA template.
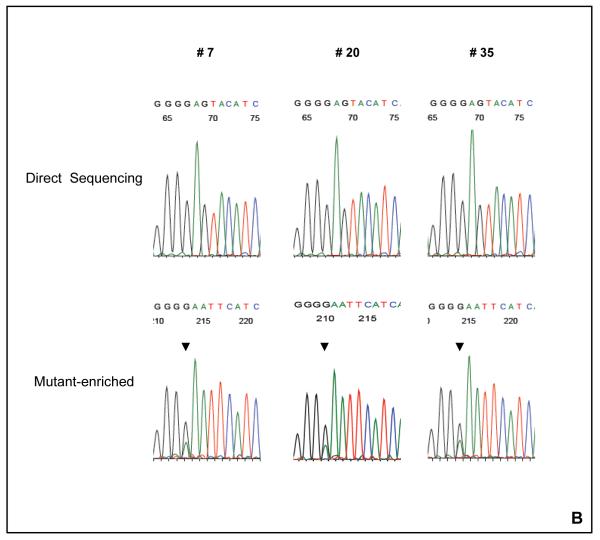
Figure 1. The detection of AKT1 E17K mutation with the mutant-enriched sequencing method.
(A) The schematic for the mutant-enriched sequencing method for AKT1 E17K. Briefly, a unique restriction enzyme site EcoRI was introduced by mismatch PCR to the wild-type DNA strand but not the mutant sequence in the first round of PCR. The mismatch primer (AKT1ME-R) has two nucleotide substitutions (G → A and A→ T). Subsequent EcoRI digestion of the wild-type DNA strands would allow preferential amplification of the mutant templates in the second round of PCR. (B) Detection of AKT1 E17K mutation by mutant-enriched method. AKT1 E17K mutation was not detectable by the conventional direct sequencing method in cases # 7, # 20 and # 35, but was enriched and visible with the mutant-enriched sequencing method.
The PCR condition for all the PCR reactions is 95°C, 5 minutes; (95°C, 30 seconds; 60°C, 30 seconds; 72°C, 30 seconds) × 25 and 40 cycles for first and second PCR respectively; 72°C, 7 minutes.
To develop the method, a 583bp product containing the E17K hot-spot mutation was amplified using a known mutant DNA sample as a template, using primers AKT1G49AFwd 5′- ACATCTGTCCTGGCACAC -3′ and AKT1G49ARvs 5′-GCCAGTGCTTGTTGCTTG – 3′. The PCR product was subcloned into a pcDNA™3.3-TOPO expression vector using the pcDNA™3.3-TOPO cloning kit (Life Technologies). Mutant and wild-type colonies were sequenced and selected. To determine the approximate sensitivity of our assay, we made serial dilutions of the mutant plasmid with the wild-type plasmid. When the ratio of the mutant to wild-type DNA copies reached 1:100, the mutant-enriched sequencing result still contained a recognizable mutant peak. However, conventional direct genomic sequencing, the mutant peak disappeared when the ratio of mutant and wild-type DNA was lower than 1:10. This indicated that the mutant-enriched sequencing method was at least 10 times more sensitive than the conventional sequencing method.
AKT1 E17K Taqman® Mutation Detection Assay
AKT1 E17K mutation was further confirmed using a Competitive Allele-Specific Taqman® PCR (castPCR™) (Life Technologies). Fresh DNA from each of the potential positive cases was extracted as follows: Five to ten 10-μm serial sections were microdissected for each case using a clean sterile scalpel for each of the three samples. After paraffin containing tissue was scrapped, DNA was extracted using QIAamp DNA FFPE Mini Kit (QIAGEN, Valencia, CA) following manufacturer’s instructions.
Each specific AKT1 E17K mutant allele assay (Assay ID: Hs0000986_mu) contains: an allele-specific primer that detects the mutant allele, a MGB oligonucleotide blocker that suppresses the wild-type allele, and a locus-specific Taqman® FAM™ dye-labeled MGB probe. A gene reference assay (Assay ID: Hs0001010_rf) designed to amplify a mutation-free and polymorphism-free region of the target gene was used in parallel. Each assay contains: a locus-specific pair of forward and reverse primers and a locus-specific Taqman® FAM™ dye-labeled MGB probe.
AKT1 E17K mutation detection experiments were performed in ABI 7500 Sequence Detector (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. In brief, after amplification, the Ct values were determined by the Applied Biosystems real-time PCR instrument software. Using four different normal (wild-type) gDNA samples in triplicates, a mutation detection ΔCt cutoff value was determined [(Detection ΔCt cutoff = Average ΔCt – (3 × the standard deviation)]. Data files containing the samples Ct values were imported into Life Technologies Mutation Detector Software. In the analysis calculations, the difference between the Ct value of the mutant allele assay and the Ct value of the gene reference assay was calculated; this ΔCt value represents the quantity of the specific mutation allele detected within each sample. Values less than the Detection ΔCt cutoff value (ΔCt = 7.17) were considered as positive for the mutation.
PIK3CA and AKT1 gene amplification
Gene amplification was evaluated by quantitative real-time PCR (Q-PCR), performed in ABI Prism 7500 Sequence Detector (Applied Biosystems, Foster City, CA, USA) using Power SyBr Green PCR Master Mix and the following oligonucleotides: for the PIK3CA gene (Chr. 3q.26.3), Fwd (5′ - ATCTTTTCTCAATGATGCTTGGCT - 3′), Rvs (5′ - CTAGGGTGTTTCGAATGTATG - 3′); COL7A1 (collagen, type VII, alpha 1; Chr. 3p21.1) as the reference gene, Fwd (5′-ACCCAGTACCGCATCATTGTG -3′), Rvs (5′- TCAGGCTGGAACTTCAGTGTG - 3′). For the AKT1 gene (Chr. 14q32.3), Fwd (5′- ACGGGCACATTAAGATCACA - 3′), Rvs (5′- TGCCGCAAAAGGTCTTCATG -3′) and DHSR4 (dehydrogenase/reductase (SDR family) member 4; Chr.14q11.2) as the reference gene, Fwd (5′- GGTAGTCTAGGGCAGGTCCA - 3′), Rvs (5′-ATGATTTGGGCCAGAAGGGG - 3′). Optimal primer concentrations were determined using optimization protocols from Applied Biosystems SYBR Green PCR master mix manual.
Dissociation curve analysis of all the PCR products showed a simple peak and the correct size of each product was confirmed by agarose gel electrophoresis. The relative copy number for PIK3CA and AKT1 was calculated using the 2−ΔΔCt method. ΔΔCt represents the difference between the paired tissue samples (ΔCt Tumor - ΔCT of matched normal), with ΔCt being the average Ct for the target gene (PIK3CA and AKT1) minus the average Ct for the reference gene (COL7A1 or DHSR4 respectively). Values greater than 2.0 were considered positive for gene amplification.
SNP-PCR-RFLP PTEN Loss of Heterozygosity (LOH) Analysis
LOH analyses of the PTEN locus were assessed by Single Nucleotide Polymorphism-PCR-Restriction Fragment Length Polymorphism (SNP-PCR-RFLP) analysis using seven SNP markers on the tumor and matched non-neoplastic DNA from 36 IPMN samples. All SNPs were intragenic to the PTEN gene. To select SNP markers, genotypic and allelic frequency of 680 SNPs present in the PTEN gene was obtained from the Ensembl database. SNPs reported to have high heterozygosity indices were chosen. Twenty-seven SNPs with high heterozygosity indices were checked for the presence of restriction endonuclease site using the SNP cutter program (available at http://bioinfo.bsd.uchicago.edu/SNP_cutter.htm.) as described previously (45). We identified eight SNPs; each harbors a unique restriction site. Primers were designed for each SNP to generate a PCR product of 200–350 bp. Of the eight SNPs, seven yielded bands of distinguishable unequal sizes upon restriction digestion that were detectable on an agarose gel. The selected seven SNPs were then employed for the LOH analyses. Digestion of the PCR products was carried out with appropriate restriction enzymes (New England Biolabs Inc. Ipswich, MA) and resolved on 1% agarose gel. One SNP (rs34421660) was described as a deletion/insertion with a 32bp difference between both alleles, allowing the analysis without restriction enzyme digestion. All the primers were ordered from Sigma-Aldrich (St. Louis, MO).
For LOH analyses, only those sample pairs in which non-neoplastic DNA showed a heterozygous pattern were considered informative. Non-neoplastic DNA with a homozygous pattern was considered not informative for that SNP marker and excluded from the study. Allelic loss was recorded if the intensity of the signal from one allele was reduced at least 50% in the tumor DNA when compared to that in the non-neoplastic DNA for a given SNP marker on visual inspection by 2 independent observers (46) (A.T.T. and D.G.C.). PTEN oligonucleotides, locus and restriction enzymes are listed in Table 2.
Table 2.
The list of primers and restriction enzymes for the SNP-PCR-RFLP analyses at the PTEN locus. Note that SNP rs34421660 is an insertion/deletion of 32-bp and it doesn’t require restriction enzyme digestion to differentiate the two alleles.
| Ref SNP# | Restriction Enzyme |
Fwd Primer (5′-3′) | Rvs Primer (5′-3′) |
|---|---|---|---|
| rs1903858 | HindIII | TTTCTGCAGGAAATCCCATAGC | TAGCCAGCTCTTAAATCTGACTTCC |
| rs1234225 | PvuII | AACAAATGTTGAAAGGTGCTCAAA | TGATGGTGTTCCACAGGTGTCT |
| rs10490920 | NcoI | TCAAGAAGTCCAAGAGCATT | AGACAAGACAAGCCACCTAA |
| rs2735343 | HhaI | AGTGGAGACAGACTGACCTG | CTGTAGACATCAATGCTTGG |
| rs701848 | HaeIII | TCCTACATGTGCTTTATTGATTTGC | TTTGAAGACACCAAATTTCTGGA |
| rs1234224 | AsiI;SsiI | GCAAGTGTCGGGAAGTGTAACC | CCACTGTGCTCTCTATCCCACC |
| rs34421660 | No enzyme | AGAAAGTGACTCTGATTTACCTAAT | ATTGCTCCTGTTGAAACCT |
Immunohistochemistry
Tissue microarrays (TMAs) containing the 36 IPMN cases (three 1.5mm cores/case) were constructed. The sections were deparaffinized with standard xylene and hydrated through graded alcohols into water. Antigen retrieval was performed using Envision Flex Target Retrieval solution, high pH (Dako, Carpinteria, CA). Antibody incubation was done at room temperature on an automatic immunodetection workstation (Dako Autostainer Plus) using the Dako EnVision Flex + Visualization System (Dako Autostainer) with the following antibodies: PTEN (138G6; 1:50 dilution; Cell Signaling Technology, Danvers, MA), PKB Kinase (E-3) (1:2000; dilution; Santa Cruz Biotechnology, Dallas, TX) and monoclonal Ki67 (1:100 dilution, Dako). Counterstaining with hematoxylin for 1 minute was the final step. Level of protein expression refers to the intensity of mucinous epithelium only, and was analyzed as follows: PTEN IHC was scored as normal (2), decreased (1) or negative (0). Benign pancreatic tissues sampled in the TMA served as positive and negative control. Pancreatic ducts and centroacinar cells display normal PTEN expression (2) and served as positive internal controls; while acinar cells do not express PTEN and served as negative control (0). PKB kinase IHC was scored as weak (1) or strong (2) based on the cytoplasmic intensity of the neoplastic epithelium. Non-neoplastic pancreatic ducts served as positive control for weak expression (1), pancreatic acinar cells served as positive control for strong expression (2). Human normal alveolar lung tissue served as negative control. Ki67 IHC was scored as percentage of neoplastic epithelial cells showing nuclear expression. All the TMAs included a negative control. The pathologists who performed the scoring (H.E.R. and A.T.T.) were blinded with regard to clinical data and genetic data until the completion of our study.
Statistical analysis
For statistical purposes, clinicopathological features were dichotomized as: IPMN lesion: IPMN (IPMN-1/2); IPMC (IPMN-3). Nuclear Grade: Low (1-2); High (3). PTEN expression level was dichotomized as Low/absent (≤1) or Normal (>1). The expression of PDK1 was adopted by the median as Normal (<2) or High (≥2). Age and cyst size were dichotomized by the median value (age = 70 years; size = 2.1 cm). The χ2 test (Fisher’s exact test for small cell counts) was used for comparison between categorical variables and Student’s t-test for parametric continuous variables. Multivariable logistic regression models were conducted to calculate the odds ratio and 95% confidence interval for the association between PTEN expression and the presence of invasive carcinoma; the model adjusted for PDK1 expression. Survival curves were calculated using Kaplan-Meier product-limit estimate. Differences between survival times were analyzed by the log-rank method. Multivariable Cox proportional hazards models were used to calculate hazard ratios and 95% confidence interval of the association between risk factors and survival. We included variables in our multivariable model that were statistically significant in the univariate analysis; these variables include: PTEN expression, presence of invasive carcinoma and age). All tests were two sided. P values ≤ 0.05 were considered statistically significant. All statistical analyses were performed using SPSS statistical software version 17.0 (SPSS Inc. Chicago, IL, USA).
RESULTS
Mutations in PIK3CA and AKT1
IPMN were examined for the presence of the hot-spot mutations in the AKT1 and PIK3CA genes. We used both direct genomic sequencing and mutant-enriched sequencing methods on the DNA samples extracted from 36 microdissected IPMN specimens. One patient with borderline IPMN (moderate dysplasia) harbored the H1047R (A3140G) hot-spot mutation of the PIK3CA gene in the IPMN (data not shown). The E17K (A49G) AKT1 gene mutation was detected in three IPMN; two of the three were intestinal-type and one was gastric-type IPMN. While the PIK3CA mutation was detected by direct genomic sequencing, the three AKT1 mutations were identified only when the newly developed mutant-enriched sequencing method was employed (Fig.1). Using Taqman® Mutation Detection Assay for AKT1 E17K, we confirmed the presence of the AKT1 E17K mutation in two of the three positive cases. The mutations were somatic because they were not observed in the corresponding normal tissues of the same patients.
Analysis of AKT1 and PIK3CA gene amplification in IPMN
The AKT1 and PIK3CA gene loci were investigated for potential amplification in the 36 IPMN specimens by Q-PCR. Gene amplification of the AKT1 locus was detected in one IPMC specimen (2.8%). PIK3CA gene amplification was detected in seven (19.4%) out of the 36 IPMN. The case with the AKT1 amplification did not show co-amplification with PIK3CA. The amplification of either of these two genes did not correlate with any of the clinicopathological parameters.
Analysis of PTEN LOH
Of the 36 cases, 28 (77.8%) showed heterozygosity at the PTEN locus by SNPPCR-RFLP, and eight were not informative (as defined in the Material and Methods) for all the seven SNPs analyzed. Of the 28 heterozygous cases, 10 displayed LOH (10/28, 35.7%) (Table 1).
PTEN, PDK1 and Ki67 protein expression in IPMN
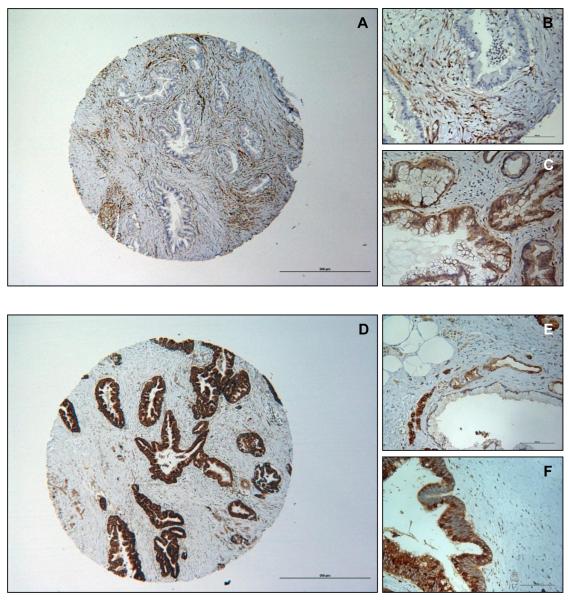
The protein expression levels of PTEN, PDK1 and Ki67 were studied by immunohistochemistry. PTEN expression was low or absent in 11 (30.6%) of the 36 cases. PDK1 was overexpressed in 20 (55.6%) of the 36 cases. Ki67 score was high in 18 (50%) of the 36 cases; 16 out of the 18 (88.8%) were IPMC and two were IPMN without carcinoma. In this set of IPMN samples, strong immunolabeling of PDK1 was associated with higher Ki67 labeling index (Fisher’s exact, p = 0.018). Representative staining of the IPMN samples expressing different levels of PTEN and PDK1 are shown in Figure 2.
Figure 2. Loss of PTEN expression and overexpression of PDK1 in IPMN.
(A) Representative absent immunopositivity of PTEN (40X); (B) Example of absent PTEN expression in IPMN with strong immunolabeling in stroma (200X). (C) Example of normal/strong cytoplasmic expression of PTEN in IPMN (200X). (D) Representative strong immunopositivity of PDK1 (40X). (E) Detail of weak cytoplasmic PDK1 immunolabeling in IPMN (200X). (F) Example of strong cytoplasmic PDK1 immunopositivity in IPMN (200X). Scale bars are 500um and 100um.
Associations of PTEN and PDK1 expression with clinicopathological features and patient outcome
Downregulation of PTEN at protein level was associated with the three following clinicopathological parameters: nuclear grade, presence of invasive carcinoma, and disease outcome. PDK1 overexpression, on the other hand, showed association with IPMN subtypes and progression (Table 3). PTEN protein expression was more frequently downregulated in the IPMN with nuclear grade 3 vs. IPMN with nuclear grade 1 and/or 2 (Fisher’s Exact, p = 0.027).
Table 3.
Associations of reduced PTEN protein expression and PDK1 overexpression with clinicopathological features, type, lesion, nuclear grade, and patient outcome.
| Characteristics | No. | Weak/Absent PTEN expression (%) |
p* | Strong PDK1 expression (%) |
p* |
|---|---|---|---|---|---|
| Mean Age, Years | 68.9 | 46 | 0.863† | 89 | 0.667† |
| Mean Cyst Size, cm | 2.8 | 2.8 | 0.979† | 3.3 | 0.044† |
| IPMN Type | |||||
| Gastric | 14 | 2 (14) | 0.060 | 4 (29) | 0.020** |
| Pancreatic | 8 | 5 (62) | 7 (87) | ||
| Intestinal | 14 | 4 (29) | 9 (64) | ||
| IPMN Lesion | |||||
| IPMN | 11 | 2 (18) | 0.439 | 3 (27) | 0.034** |
| IPMC | 25 | 9 (36) | 17 (68) | ||
| Nuclear Grade | |||||
| Low (1-2) | 18 | 2 (11) | 0.027** | 9 (50) | 0.738 |
| High (3) | 18 | 9 (50) | 11 (61) | ||
| Disease Status | |||||
| Alive | 29 | 6 (21) | 0.018** | 15 (52) | 0.426 |
| Deceased | 7 | 5 (71) | 5 (71) | ||
| Recurrence | |||||
| Yes | 6 | 2(33) | 0.871 | 4 (67) | 1.000 |
| No | 30 | 9(30) | 16 (53) |
The following variables were dichotomized for statistical analysis: nuclear grade was dichotomized as low (nuclear grades 1-2) and high (nuclear grade 3); IPMN lesion was grouped as IPMN (IPMN-1 or IPMN, adenoma and IPMN-2 or IPMN, borderline tumor) and IPMC (IPMN-3 or IPMC, carcinoma in situ); disease status was grouped as alive (no evidence of disease or recurrence) and deceased (death of disease).
χ2 test/Fisher’s exact test.
Student’s t-test.
Statistically significant.
PDK1 protein expression was detected in each IPMN subtype but was more frequently overexpressed in intestinal and pancreatic-type IPMN vs. gastric-type IPMN (Chi-Square, p = 0.020; Table 3). It was also observed that PDK1 was more commonly overexpressed in IPMC than IPMN adenoma and borderline tumor (Fisher’s exact, p = 0.034), showing an increasing trend with IPMN progression: Adenoma (0/2; 0%); Borderline (3/9; 33.3%); In situ (17/25; 68%). All the cases with PDAC associated, (7/7; 100%) showed strong PDK1 expression. Moreover, PDK1 overexpression showed a linear correlation with the maximum cyst diameter detected (Chi-square, p = 0.044).
A striking difference in survival was observed between patients with normal expression of PTEN and those with reduced or absent PTEN expression; the latter group experienced shortened disease-specific survival (Median survival time: PTEN normal= 60.6 months, PTEN low = 48.9 months; log-rank test, p = 0.014, Fig. 3). As expected IPMN patients with an invasive carcinoma had poorer survival than those without invasive carcinoma; a 12 fold increase risk of mortality was observed in multivariable models adjusting for PTEN expression and presence of invasive carcinoma (hazard ratio = 11.9, 95% CI = 1.7-83.5, p = 0.012); addition of age to the model, did not affect the result. This result may indicate that the association of PTEN expression with patient survival was only due to the presence of invasive carcinoma (Fisher’s exact, p = 0.018). When we examined the factors associated with the presence of invasive carcinoma, absent or weak PTEN expression was associated with a 8-fold increase in the odds controlling for PDK1 expression (odd ratio = 8.3, 95% CI = 1.034-67.142, p = 0.046). However, these results are based on a small sample size.
Figure 3. PTEN down-regulation is associated with poor prognosis in patients with IPMN.
The survival was calculated by the Kaplan–Meier method comparing patients with normal vs. low PTEN expression. The data show that low PTEN expression is associated with poor survival (Median survival time: PTEN normal= 60.6 months, PTEN low = 48.9 months; p = 0.014, hazard ratio = 6.1). There was no stratification.
DISCUSSION
In this study, we conducted a comprehensive evaluation of the PI3K signaling pathway in 36 well-characterized IPMN samples to understand the extent of its dysregulation in IPMN and the potential impact on clinical outcome.
We identified the H1047R mutation of PIK3CA in one (2.7%) of the 36 IPMN. We also detected the E17K AKT1 gene mutation in three (8.3%) of the 36 IPMN cases. The H1047R mutation of PIK3CA is known to affect the functionally important kinase domain of the protein, conferring an increased lipid kinase activity, which can lead to the activation of PI3K signaling pathway in the absence of growth factors. The mutation has been shown to be sufficient to induce oncogenic cell transformation of chicken embryo fibroblasts (CEFs) and NIH3T3 cells (36, 37, 47). It has also been described that p110α mutants induce in vivo angiogenesis and malignant cell growth in chorioallantoic membrane of chick embryo and cause hemangiosarcomas in young chickens (36).
We have previously reported PIK3CA gene mutations in 11% of IPMN, suggesting that the PIK3CA gene and its pathway may have a role in IPMN but not PanIN/PDA tumorigenesis (12). In our previous study, PIK3CA gene mutations were detected in three high-grade IPMN, and one IPMN, borderline, with the caveat that the majority of the cases examined had high-grade dysplasia. One of the four mutations detected in the previous study was H1047R in a IPMC with invasion, the other three were not hot-spot mutations (12). In the current study more IPMN with low-grade and borderline dysplasia were included and the hot-spot mutation H1047R was detected in an IPMN, borderline. The mutation frequency is slightly lower in the current study probably due to the facts that more low-grade cases were included and only hot-spot mutation sites were examined. Nevertheless, a number of recent studies encompassing different histological grades of IPMN are in agreement with our findings, supporting an important role for PIK3CA in IPMN (12, 39, 48). There is one exception in which PIK3CA mutations were not detected in IPMN but were identified in intraductal tubulopapillary neoplasms of the pancreas (ITNP), a rare variant of intraductal neoplasm of the pancreas recently recognized as a new class of pancreatic tumor (13). Consistent expression of MUC5A among our samples definitively confirmed them as IPMN and ruled out ITNP (data not shown) (9).
The mutational status of AKT1 gene has not been examined before in IPMN. Here we show that by using a novel mutant-enriched method, we were able to detect the E17K mutation in three (8.3%) of the 36 IPMN cases; two of those were IPMC associated with invasive components. The detection of the AKT1 E17K mutation in IPMN but not PDA further signified the unique importance of the PI3K signaling pathway in IPMN progression. While presence of false positive mutations could be possible due to a low-quantity DNA extracted from the tumors and/or a potential non-template extension of theAKT1ME-R primer used in the mutant-enriched assay, the absence of a mutant peak on any of the paired normal samples examined suggests that this a rather infrequent event. Furthermore, each positive sample was verified from an independent PCR of the original genomic DNA at least two more times. To further rule out false positivity, new DNA from the same potential positive cases was extracted, the presence of AKT1 E17K mutations was then analyzed using the AKT1 E17K Taqman® Mutation Detection Assay. By using this method, we were able to confirm two of the three potential positive cases. The apparent discrepancy could be due to a number of reasons: (1) different mutant to wild-type ratio between the different DNA samples from the same patients; (2) different sensitivity of the two methods applied; or (3) lack of calibration ΔCt value for the AKT1_33765_mu assay, could lead to a smaller detection ΔCt cutoff calculated with our samples, which could limit the detection of mutant allele.
Intriguingly, PIK3CA and AKT1 genes mutations were not detected in PanIN/PDA (35) (49) (41, 50). This suggests that, although IPMN and PanIN share overlapped molecular alterations, such as KRAS, p16, p53, and SMAD4 (9), pancreatic carcinoma associated with IPMN may arise through a different molecular pathway from PanIN/PDA. While STK11/LKB1 gene is inactivated more frequently in IPMN than in PanIN/PDA (18-20), PIK3CA and AKT1 are the only genes with mutations unique to IPMN and not detectable in PanIN/PDA thus far (12, 35, 41, 48-50). Direct comparisons of IPMN and PanIN progression might be instructive in characterizing the divergent molecular and histological pathways of pancreatic cancer evolution between the two and reveal additional unique alterations in IPMN.
To further examine the status of the PI3K pathway at the genomic level, we investigated the LOH at the PTEN locus and it was found in 10 of the 28 (35.7%) IPMN analyzed. Albeit the small sample number, LOH frequency at PTEN steadily increased as IPMN progressed (0/1 or 0% IPMN, adenoma; 2/7 or 28.6% IPMN, borderline; 8/20 or 40% IPMC, in situ). The same increasing frequency could be observed with the nuclear grade (1/4 or 25% nuclear grade 1; 4/12 or 33.3% nuclear grade 2; 5/12 or 41.7% nuclear grade 3). There was no tumor size, gender or age bias associated with the LOH status at PTEN. PIK3CA was amplified in 19.4% of the IPMN analyzed, while AKT1 was amplified in only one IPMN associated with invasive carcinoma. These events did not correlate with the clinical variables.
Although we found PDK1 to be frequently overexpressed in our IPMN samples (20/36 or 55.6%), this overexpression was not associated with patient survival. However, PDK1 expression was significantly higher in intestinal and pancreatic-type IPMN vs. gastric-type IPMN (p = 0.020). The three different IPMN subtypes included in this study possess distinct histopathologic features. Similarly to previous reports, in our series, gastric-type IPMN was the least associated with invasive tumor (4/14 or 28.6%) than both pancreatic (8/8 or 100%) and intestinal (13/14 or 92.8%) (51). PDK1 immunolabeling was more frequently upregulated in IPMC vs. IPMN (p = 0.034). Both LOH at PTEN and PDK1 overexpression patterns further depict an increasing aberrant PI3K signaling accompanying IPMN progression. Future studies are needed to define the mechanisms leading to PDK1 overexpression on IPMN samples and whether or not this correlates with PDK1 activation; this could help understand the contribution of PDK1 to PI3K signaling in these patients.
Weak or absent PTEN expression was observed in 30.6% of the IPMN analyzed by IHC. This reduced expression was significantly associated with higher nuclear grade. Furthermore, we found that weak or loss of PTEN expression was associated with reduced survival rate among our IPMN patients (p = 0.014). However, multivariate Cox analysis showed that the presence of invasive carcinoma was the only independent factor of poor prognosis. The low number of IPMN samples with associated invasive carcinoma included in the study (n = 6) and the strong association between the presence of invasive carcinoma and low/absent PTEN expression in this set of IPMN render it difficult to separate the effect of these two variables on survival. Keeping this limitation in mind, our findings suggest that PTEN expression level may have potential diagnostic and prognostic values, and may need to be taken into consideration when determining treatment options for IPMN patients. Future larger studies are needed to further evaluate the clinical values of PTEN as an independent prognostic marker distinct from invasive cancer. In addition, determining if expression levels of PTEN in cells isolated from cyst fluid or biopsy and within IPMN are correlated with the presence of invasive cancer and patient prognosis will be key to preoperative risk assessment and stratification. These studies will be crucial next-steps in determining the utility of PTEN expression in making surgical decisions.
Within this manuscript, we examined a large number of associations, and we observed a limited number of significant associations. Thus, due to the issues of multiple comparisons, we are cautious in our interpretations of the results; some of the statistically significant associations we observed may be explained by chance. Using a conservative approach of Bonferroni correction, we would require a p-value of <0.003 to achieve statistical significance. Based on this p-value, none of our results would be statistically significant, and may all be a result of chance. However, these statistically significant findings are supported by well-defined biologic rationale (e.g. PDK1 overexpression and PTEN loss in IPMN progression are logical) (26, 31) . Thus, these results require replication in larger studies.
Taken together, these data indicate that aberrations of the PI3K pathway are common in IPMN and suggest an important role for this signaling pathway in cystic tumors of the pancreas. This is the first report of AKT1 mutations in IPMN tumors. Although a larger scale study designed to determine if PTEN is an independent variable from invasive carcinoma is needed, nevertheless, these results indicate that dysregulated PI3K signaling pathway is involved in the malignant progression of IPMN. If inhibition of the PI3K signaling pathway can slow progression of IPMN and development of invasive cancer, patients may benefit from novel therapeutics specifically targeting PDK1, mTOR, and PIK3CA as neoadjuvant or adjuvant therapies.
STATEMENT OF RELEVANCE.
IPMN is a common subtype of precancerous lesions of the pancreas that can advance to invasive carcinoma if left untreated. Although patients with IPMN associated with invasive cancer face better prognosis than those with conventional pancreatic ductal adenocarcinoma, the 5-year survival is poor and surgical resection remains the most effective treatment option for the patients. Currently there is no effective way of monitoring the malignant transformation of IPMN to invasive cancer and there is no established consensus in making the surgical decision. Considering the high-risk natures of malignant transformation and surgical resections, it is therefore crucial to establish a protocol balancing the risks for both. Here we report the potential uses of PTEN expression as a prognostic/diagnostic marker for IPMN patients. In addition, the extensive dysregulation of the PI3K signaling pathway in IPMN suggest that targeted therapies to this pathway may benefit patients with IPMN.
ACKNOWLEDGMENTS
We thank Dr. Maria Victoria Gonzalez Mean for excellent technical and statistical assistance and helpful discussions; Dr. Juana Maria Garcia-Pedrero for providing technical advice and reagents/antibodies; we thank Dr. Roger Vaughan for helpful discussion and support with statistical analysis.
Funding sources: This work was supported by the Stewart Trust Awards for Pilot Projects in Cancer Research, NCI R56CA109525, and NCI R01CA109525
REFERENCES
- 1.Hruban RH, Takaori K, Canto M, Fishman EK, Campbell K, Brune K, et al. Clinical importance of precursor lesions in the pancreas. Journal of hepato-biliary-pancreatic surgery. 2007;14(3):255–63. doi: 10.1007/s00534-006-1170-9. doi: DOI 10.1007/s00534-006-1170-9. PubMed PMID: ISI:000246734400006. [DOI] [PubMed] [Google Scholar]
- 2.Kloppel G, Kosmahl M. Cystic lesions and neoplasms of the pancreas. The features are becoming clearer. Pancreatology. 2001;1(6):648–55. doi: 10.1159/000055876. doi: 10.1159/000055876. PubMed PMID: 12120249. [DOI] [PubMed] [Google Scholar]
- 3.Belyaev O, Seelig MH, Muller CA, Tannapfel A, Schmidt WE, Uhl W. Intraductal papillary mucinous neoplasms of the pancreas. Journal of clinical gastroenterology. 2008;42(3):284–94. doi: 10.1097/MCG.0b013e3180500761. doi: 10.1097/MCG.0b013e3180500761. PubMed PMID: 18223495. [DOI] [PubMed] [Google Scholar]
- 4.Andrejevic-Blant S, Kosmahl M, Sipos B, Kloppel G. Pancreatic intraductal papillary-mucinous neoplasms: a new and evolving entity. Virchows Arch. 2007;451(5):863–9. doi: 10.1007/s00428-007-0512-6. Epub 2007/09/28. doi: 10.1007/s00428-007-0512-6. PubMed PMID: 17899180; PubMed Central PMCID: PMC2063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crippa S, Salvia R, Warshaw AL, Dominguez I, Bassi C, Falconi M, et al. Mucinous cystic neoplasm of the pancreas is not an aggressive entity: lessons from 163 resected patients. Annals of surgery. 2008;247(4):571–9. doi: 10.1097/SLA.0b013e31811f4449. Epub 2008/03/26. doi: 10.1097/SLA.0b013e31811f4449. PubMed PMID: 18362619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohn TA, Yeo CJ, Cameron JL, Hruban RH, Fukushima N, Campbell KA, et al. Intraductal papillary mucinous neoplasms of the pancreas: an updated experience. Ann Surg. 2004;239(6):788–97. doi: 10.1097/01.sla.0000128306.90650.aa. discussion 97-9. PubMed PMID: 15166958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haugk B. Pancreatic intraepithelial neoplasia-can we detect early pancreatic cancer? Histopathology. 2010;57(4):503–14. doi: 10.1111/j.1365-2559.2010.03610.x. Epub 2010/09/30. doi: 10.1111/j.1365-2559.2010.03610.x. PubMed PMID: 20875068. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa T, Kloppel G, Volkan Adsay N, Albores-Saavedra J, Fukushima N, Horii A, et al. Classification of types of intraductal papillary-mucinous neoplasm of the pancreas: a consensus study. Virchows Arch. 2005;447(5):794–9. doi: 10.1007/s00428-005-0039-7. Epub 2005/08/10. doi: 10.1007/s00428-005-0039-7. PubMed PMID: 16088402. [DOI] [PubMed] [Google Scholar]
- 9.Shi C, Hruban RH. Intraductal papillary mucinous neoplasm. Hum Pathol. 2012;43(1):1–16. doi: 10.1016/j.humpath.2011.04.003. Epub 2011/07/23. doi: S0046-8177(11)00157-2 [pii] 10.1016/j.humpath.2011.04.003. PubMed PMID: 21777948. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M, Chari S, Adsay V, Fernandez-del Castillo C, Falconi M, Shimizu M, et al. International consensus guidelines for management of intraductal papillary mucinous neoplasms and mucinous cystic neoplasms of the pancreas. Pancreatology. 2006;6(1-2):17–32. doi: 10.1159/000090023. doi: 10.1159/000090023. PubMed PMID: 16327281. [DOI] [PubMed] [Google Scholar]
- 11.Schonleben F, Qiu W, Bruckman KC, Ciau NT, Li X, Lauerman MH, et al. BRAF and KRAS gene mutations in intraductal papillary mucinous neoplasm/carcinoma (IPMN/IPMC) of the pancreas. Cancer letters. 2007;249(2):242–8. doi: 10.1016/j.canlet.2006.09.007. PubMed PMID: 17097223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schonleben F, Qiu W, Ciau NT, Ho DJ, Li X, Allendorf JD, et al. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12(12):3851–5. doi: 10.1158/1078-0432.CCR-06-0292. PubMed PMID: 16778113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi H, Kuboki Y, Hatori T, Yamamoto M, Shiratori K, Kawamura S, et al. Somatic mutations in PIK3CA and activation of AKT in intraductal tubulopapillary neoplasms of the pancreas. The American journal of surgical pathology. 2011;35(12):1812–7. doi: 10.1097/PAS.0b013e31822769a0. Epub 2011/09/29. doi: 10.1097/PAS.0b013e31822769a0. PubMed PMID: 21945955. [DOI] [PubMed] [Google Scholar]
- 14.Z’Graggen K, Rivera JA, Compton CC, Pins M, Werner J, Fernandez-del Castillo C, et al. Prevalence of activating K-ras mutations in the evolutionary stages of neoplasia in intraductal papillary mucinous tumors of the pancreas. Ann Surg. 1997;226(4):491–8. doi: 10.1097/00000658-199710000-00010. discussion 8-500. PubMed PMID: 9351717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sessa F, Solcia E, Capella C, Bonato M, Scarpa A, Zamboni G, et al. Intraductal papillary-mucinous tumours represent a distinct group of pancreatic neoplasms: an investigation of tumour cell differentiation and K-ras, p53 and c-erbB-2 abnormalities in 26 patients. Virchows Arch. 1994;425(4):357–67. doi: 10.1007/BF00189573. PubMed PMID: 7820300. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Matthaei H, Maitra A, Dal Molin M, Wood LD, Eshleman JR, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3(92):92ra66. doi: 10.1126/scitranslmed.3002543. Epub 2011/07/22. doi: 3/92/92ra66 [pii] 101126/scitranslmed.3002543. PubMed PMID: 21775669; PubMed Central PMCID: PMC3160649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(52):21188–93. doi: 10.1073/pnas.1118046108. Epub 2011/12/14. doi: 1118046108 [pii] 10.1073/pnas.1118046108. PubMed PMID: 22158988; PubMed Central PMCID: PMC3248495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato N, Rosty C, Jansen M, Fukushima N, Ueki T, Yeo CJ, et al. STK11/LKB1 Peutz-Jeghers gene inactivation in intraductal papillary-mucinous neoplasms of the pancreas. The American journal of pathology. 2001;159(6):2017–22. doi: 10.1016/S0002-9440(10)63053-2. Epub 2001/12/06. doi: S0002-9440(10)63053-2 [pii] 10.1016/S0002-9440(10)63053-2. PubMed PMID: 11733352; PubMed Central PMCID: PMC1850608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su GH, Hruban RH, Bansal RK, Bova GS, Tang DJ, Shekher MC, et al. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. The American journal of pathology. 1999;154(6):1835–40. doi: 10.1016/S0002-9440(10)65440-5. Epub 1999/06/11. doi: S0002-9440(10)65440-5 [pii] 10.1016/S0002-9440(10)65440-5. PubMed PMID: 10362809; PubMed Central PMCID: PMC1866632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsubayashi H, Sato N, Fukushima N, Yeo CJ, Walter KM, Brune K, et al. Methylation of cyclin D2 is observed frequently in pancreatic cancer but is also an age-related phenomenon in gastrointestinal tissues. Clin Cancer Res. 2003;9(4):1446–52. Epub 2003/04/10. PubMed PMID: 12684418. [PubMed] [Google Scholar]
- 21.Schoedel KE, Finkelstein SD, Ohori NP. K-Ras and microsatellite marker analysis of fine-needle aspirates from intraductal papillary mucinous neoplasms of the pancreas. Diagn Cytopathol. 2006;34(9):605–8. doi: 10.1002/dc.20511. Epub 2006/08/11. doi: 10.1002/dc.20511. PubMed PMID: 16900481. [DOI] [PubMed] [Google Scholar]
- 22.Wada K, Takada T, Yasuda H, Amano H, Yoshida M, Sugimoto M, et al. Does “clonal progression” relate to the development of intraductal papillary mucinous tumors of the pancreas? J Gastrointest Surg. 2004;8(3):289–96. doi: 10.1016/j.gassur.2003.09.027. PubMed PMID: 15019925. [DOI] [PubMed] [Google Scholar]
- 23.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15(2):161–70. doi: 10.1016/j.semcdb.2003.12.022. Epub 2004/06/24. PubMed PMID: 15209375. [DOI] [PubMed] [Google Scholar]
- 24.Bayascas JR. Dissecting the role of the 3-phosphoinositide-dependent protein kinase-1 (PDK1) signalling pathways. Cell cycle (Georgetown, Tex. 2008;7(19):2978–82. doi: 10.4161/cc.7.19.6810. Epub 2008/09/20. doi: 6810 [pii]. PubMed PMID: 18802401. [DOI] [PubMed] [Google Scholar]
- 25.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nature reviews. 2002;2(7):489–501. doi: 10.1038/nrc839. PubMed PMID: 12094235. [DOI] [PubMed] [Google Scholar]
- 26.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22(14):2954–63. doi: 10.1200/JCO.2004.02.141. PubMed PMID: 15254063. [DOI] [PubMed] [Google Scholar]
- 27.Hill R, Calvopina JH, Kim C, Wang Y, Dawson DW, Donahue TR, et al. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer research. 2010;70(18):7114–24. doi: 10.1158/0008-5472.CAN-10-1649. Epub 2010/09/03. doi: 0008-5472.CAN-10-1649 [pii] 10.1158/0008-5472.CAN-10-1649. PubMed PMID: 20807812; PubMed Central PMCID: PMC2940963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, Wang Y, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer cell. 2005;8(3):185–95. doi: 10.1016/j.ccr.2005.07.015. PubMed PMID: 16169464. [DOI] [PubMed] [Google Scholar]
- 29.Bellacosa A, de Feo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. International journal of cancer. 1995;64(4):280–5. doi: 10.1002/ijc.2910640412. PubMed PMID: 7657393. [DOI] [PubMed] [Google Scholar]
- 30.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nature genetics. 1999;21(1):99–102. doi: 10.1038/5042. PubMed PMID: 9916799. [DOI] [PubMed] [Google Scholar]
- 31.Pedrero JM, Carracedo DG, Pinto CM, Zapatero AH, Rodrigo JP, Nieto CS, et al. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer. 2005;114(2):242–8. doi: 10.1002/ijc.20711. PubMed PMID: 15543611. [DOI] [PubMed] [Google Scholar]
- 32.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell cycle (Georgetown, Tex. 2004;3(10):1221–4. doi: 10.4161/cc.3.10.1164. PubMed PMID: 15467468. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Rostan G, Costa AM, Pereira-Castro I, Salvatore G, Hernandez R, Hermsem MJ, et al. Mutation of the PIK3CA gene in anaplastic thyroid cancer. Cancer research. 2005;65(22):10199–207. doi: 10.1158/0008-5472.CAN-04-4259. PubMed PMID: 16288007. [DOI] [PubMed] [Google Scholar]
- 34.Qiu W, Schonleben F, Li X, Ho DJ, Close LG, Manolidis S, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(5):1441–6. doi: 10.1158/1078-0432.CCR-05-2173. PubMed PMID: 16533766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science (New York, NY. 2004;304(5670):554. doi: 10.1126/science.1096502. PubMed PMID: 15016963. [DOI] [PubMed] [Google Scholar]
- 36.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(5):1475–9. doi: 10.1073/pnas.0510857103. PubMed PMID: 16432179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(3):802–7. doi: 10.1073/pnas.0408864102. PubMed PMID: 15647370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuels Y, Diaz LA, Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer cell. 2005;7(6):561–73. doi: 10.1016/j.ccr.2005.05.014. PubMed PMID: 15950905. [DOI] [PubMed] [Google Scholar]
- 39.Lubezky N, Ben-Haim M, Marmor S, Brazowsky E, Rechavi G, Klausner JM, et al. High-throughput mutation profiling in intraductal papillary mucinous neoplasm (IPMN) J Gastrointest Surg. 2011;15(3):503–11. doi: 10.1007/s11605-010-1411-8. Epub 2011/01/13. doi: 10.1007/s11605-010-1411-8. PubMed PMID: 21225475. [DOI] [PubMed] [Google Scholar]
- 40.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448(7152):439–44. doi: 10.1038/nature05933. PubMed PMID: 17611497. [DOI] [PubMed] [Google Scholar]
- 41.Bleeker FE, Felicioni L, Buttitta F, Lamba S, Cardone L, Rodolfo M, et al. AKT1(E17K) in human solid tumours. Oncogene. 2008 doi: 10.1038/onc.2008.170. PubMed PMID: 18504432. [DOI] [PubMed] [Google Scholar]
- 42.Brugge J, Hung MC, Mills GB. A new mutational AKTivation in the PI3K pathway. Cancer cell. 2007;12(2):104–7. doi: 10.1016/j.ccr.2007.07.014. PubMed PMID: 17692802. [DOI] [PubMed] [Google Scholar]
- 43.Hruban RH, Takaori K, Klimstra DS, Adsay NV, Albores-Saavedra J, Biankin AV, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. The American journal of surgical pathology. 2004;28(8):977–87. doi: 10.1097/01.pas.0000126675.59108.80. Epub 2004/07/15. doi: 00000478-200408000-00001 [pii]. PubMed PMID: 15252303. [DOI] [PubMed] [Google Scholar]
- 44.Qiu W, Tong GX, Manolidis S, Close LG, Assaad AM, Su GH. Novel mutant-enriched sequencing identified high frequency of PIK3CA mutations in pharyngeal cancer. International journal of cancer. 2008;122(5):1189–94. doi: 10.1002/ijc.23217. PubMed PMID: 17990317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang R, Zhu Z, Zhu H, Nguyen T, Yao F, Xia K, et al. SNP Cutter: a comprehensive tool for SNP PCR-RFLP assay design. Nucleic Acids Res. 2005;33(Web Server issue):W489–92. doi: 10.1093/nar/gki358. Epub 2005/06/28. doi: 33/suppl_2/W489 [pii] 10.1093/nar/gki358. PubMed PMID: 15980518; PubMed Central PMCID: PMC1160119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan E, Li CM, Yamashiro DJ, Kandel J, Thaker H, Murty VV, et al. Genomic profiling maps loss of heterozygosity and defines the timing and stage dependence of epigenetic and genetic events in Wilms’ tumors. Molecular Cancer Research: MCR. 2005;3(9):493–502. doi: 10.1158/1541-7786.MCR-05-0082. [DOI] [PubMed] [Google Scholar]
- 47.Ikenoue T, Kanai F, Hikiba Y, Tanaka Y, Imamura J, Ohta M, et al. Functional consequences of mutations in a putative Akt phosphorylation motif of B-raf in human cancers. Molecular carcinogenesis. 2005;43(1):59–63. doi: 10.1002/mc.20102. PubMed PMID: 15791648. [DOI] [PubMed] [Google Scholar]
- 48.Mohri D, Asaoka Y, Ijichi H, Miyabayashi K, Kudo Y, Seto M, et al. Different subtypes of intraductal papillary mucinous neoplasm in the pancreas have distinct pathways to pancreatic cancer progression. J Gastroenterol. 2012;47(2):203–13. doi: 10.1007/s00535-011-0482-y. Epub 2011/11/02. doi: 10.1007/s00535-011-0482-y. PubMed PMID: 22041919. [DOI] [PubMed] [Google Scholar]
- 49.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, NY. 2008;321(5897):1801–6. doi: 10.1126/science.1164368. PubMed PMID: 18772397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohamedali A, Lea NC, Feakins RM, Raj K, Mufti GJ, Kocher HM. AKT1 (E17K) mutation in pancreatic cancer. Technology in cancer research & treatment. 2008;7(5):407–8. doi: 10.1177/153303460800700509. PubMed PMID: 18783292. [DOI] [PubMed] [Google Scholar]
- 51.Ban S, Naitoh Y, Mino-Kenudson M, Sakurai T, Kuroda M, Koyama I, et al. Intraductal papillary mucinous neoplasm (IPMN) of the pancreas: its histopathologic difference between 2 major types. The American journal of surgical pathology. 2006;30(12):1561–9. doi: 10.1097/01.pas.0000213305.98187.d4. Epub 2006/11/24. doi: 10.1097/01.pas.0000213305.98187.d4 00000478-200612000-00009 [pii]. PubMed PMID: 17122512. [DOI] [PubMed] [Google Scholar]