Abstract
Objective
There is much accumulated evidence that EGFR, HER2, and their downstream signaling pathway members such as KRAS, BRAF, and PIK3CA are strongly implicated in cancer development and progression. Recently, mutations in the kinase domains of EGFR and HER2, associated with increased sensitivity to tyrosine kinase inhibitors, have been described.
Methods
To evaluate the mutational status of these genes in intraductal papillary mucinous neoplasm (IPMN)/intraductal papillary mucinous carcinoma (IPMC), EGFR and HER2 were analyzed in 36 IPMN/IPMC, and the results were correlated to the mutational status of the KRAS, BRAF, and PIK3CA genes in the samples.
Results
Together, we identified 1 silent mutation of HER2, 17 (43%) KRAS mutations, 1 (2.7%) BRAF mutation, and 4 (11%) mutations of PIK3CA in the IPMN/IPMC samples.
Conclusions
The EGFR and ERBB2 (HER2) mutations are very infrequent in IPMN/IPMC, suggesting the limited possibility of targeting mutated ERBB2 and EGFR for therapy for these lesions. The KRAS, BRAF, and PIK3CA, however, could represent interesting targets for future therapies in these lesions.
Keywords: Her2, EGFR, PIK3CA, KRAS, BRAF, IPMN
Intraductal papillary mucinous neoplasms (IPMNs) of the pancreas are divided into 3 groups based on increasing nuclear and architectural atypia: intraductal papillary mucinous adenoma, intraductal papillary mucinous borderline, and intraductal papillary mucinous carcinoma (IPMC).1 According to the absence or presence of neoplastic cells invading the pancreatic tissue surrounding the involved ducts, IPMCs are separated into invasive and noninvasive types.2 Most IPMNs are slow growing and less aggressive compared with conventional, pancreatic ductal adenocarcinoma (PDA). The prognosis of patients with noninvasive IPMN consisting of adenoma, adenocarcinoma in situ, or minimally invasive adenocarcinoma is excellent, and the 5-year survival rate was reported to be 77% to 100%.3–6 However, invasive IPMN that macroscopically involves the pancreatic parenchyma comprises 16% to 43% of all IPMN lesions, and the 5-year survival rate for patients with these lesions varied widely from 0% to 64% in several reported series.3–5,7–9 Reported genetic alterations identified in IPMN include mutations in the KRAS,10 TP53,11 STK11/LKB1,12 and PIK3CA13 genes, as well as loss of heterozygosity of several chromosomal loci.12,14 Overexpression of ERBB2 (HER2) has been reported, beginning from the early stage of hyperplasia.15 The ERBB family comprises 4 structurally related receptors: ERBB1 (EGFR), ERBB2 (HER2/neu), ERBB3, and ERBB4. The EGFR kinase domain mutations in lung adenocarcinomas could predict significant clinical responses to orally active epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors.16–18 Furthermore, 1.6% to 9.8% of lung adenocarcinomas also harbored ERBB2 (HER2) kinase domain mutations.19,20 There is much accumulated evidence that EGFR and its family members are strongly implicated in the development and progression of numerous human tumors, including IPMNs, pancreatic intraepithelial neoplasias, and PDA.6,15 Downstream members of the EGFR/ERBB2 signaling pathways, including KRAS, BRAF, and PIK3CA are also frequently mutated in human cancers and act as oncogenic proteins.16,21,22 Previously, we have reported on KRAS, BRAF, and PIK3CA mutations in IPMN/IPMC.13,23 Here, we analyzed mutations in the EGFR and ERBB2 genes in the same cohort and correlated their mutation status to other oncogenic changes.
MATERIALS AND METHODS
Patients and Tissue Samples
Surgical paraffin-embedded primary IPMN/IPMC samples of 38 patients were obtained from the archival tissue collection of the Columbia University Medical Center, approved by the institutional review board of the Columbia University Medical Center, and in accordance with Health Insurance Portability and Accountability Act regulations. We analyzed 8 IPMNs, 23 IPMNs with associated IPMCs, 5 IPMCs, and 2 mucinous cystadenomas (see Table 1 for a more detailed register). Invasive carcinoma was associated with IPMN/IPMC in pancreatic resection, but lesion analyzed did not sample invasive carcinoma.
TABLE 1.
Detailed Patient and Sample Data With Observed Mutations of the Lesions Investigated
| Patient No. |
Sex | Age (yrs) |
Lesion Analyzed |
IPMN Nuclear Grade |
Differentiation of Invasive Carcinoma |
Location Within Pancreas |
Maximum Dimension (cm)* |
ERBB2 Mutation |
KRAS Mutation |
BRAF Mutation |
PIK3CA Mutation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 62 | IPMN/border | 2 | N/A | Body | — | A830A | G12D | W551G | |
| 2 | M | 73 | IPMC/inv | 3 | Moderate | Head/body | 4 | G12D | |||
| 3 | M | 67 | IPMC/inv | 3 | Moderate | Head | — | G12V | |||
| 4 | M | 69 | IPMC | 3 | N/A | Head | 3.5 | G12V | S1015F | ||
| 5 | F | 75 | IPMC/inv | 3 | Moderate | Head | 5.5 | T324I | |||
| 6 | F | 68 | IPMC/inv | 3 | Moderate | Head | 5 | G12V | |||
| 7 | M | 65 | IPMN/border | 2 | Poor† | Head | 5 | ||||
| 8 | F | 66 | IPMN/border | 2 | N/A | Unc.proc. | 2 | G12V | |||
| 9 | M | 84 | IPMC/inv. | 3 | Moderate | Head | 5 | ||||
| 10 | M | 53 | IPMN/aden | 1 | N/A | Head | 3.5 | G12V | |||
| 11 | M | 71 | IPMC/inv | 2–3 | N/A | Head | — | ||||
| 12 | M | 81 | IPMC | 3 | N/A | Head | 2.5 | ||||
| 13 | M | 63 | IPMC | 3 | Moderate/poor† | Head | 2.3 | ||||
| 14 | M | 66 | IPMC/inv | 3 | Moderate/poor | Head | 6 | H1047R | |||
| 15 | F | 70 | IPMC/inv | 3 | Moderate/poor | Head/body | 7 | G12R | S615F | ||
| 16 | F | 70 | IPMC/inv | 3 | Moderate | Head | 1.5 | ||||
| 17 | M | 72 | IPMN/border | 2 | N/A | Head | 0.4 | ||||
| 18 | F | 53 | mucin.cystad | 1 | N/A | Head | 3 | ||||
| 19 | M | 79 | IPMC/inv | 3 | Moderate/poor | Head | 6 | G12R | |||
| 20 | M | 63 | IPMC/inv | 3 | Moderate/poor | Head | 3.5 | G12R | |||
| 21 | M | 77 | IPMN/aden | 1 | N/A | Head/body | 2.2 | ||||
| 22 | F | 62 | Mucin.cystad | 1 | N/A | Head | 2 | ||||
| 23 | M | 41 | IPMC/inv | 3 | Moderate/poor | Head | 5 | ||||
| 24 | M | 71 | IPMC/inv | 3 | Moderate/poor | Head | 1.5 | ||||
| 25 | F | 58 | IPMN/aden | 1 | N/A | Head | 1.5 | ||||
| 26 | M | 49 | IPMC/inv | 3 | Moderate | Head | 4.5 | ||||
| 27 | M | 71 | IPMC/inv | 3 | Moderate/poor | Head | 5.5 | G12D | |||
| 28 | M | 74 | IPMC | 3 | Well† | Head/body | — | ||||
| 29 | M | 59 | IPMC | 3 | Poor† | Head | 7 | G12V | |||
| 30 | M | 81 | IPMC/inv | 3 | Moderate/poor | Head | 3 | ||||
| 31 | F | 80 | IPMC/inv | 3 | Moderate/poor | Head | 5 | G12R | |||
| 32 | F | 66 | IPMC/inv | 3 | Poor | Head | 3 | ||||
| 33 | F | 77 | IPMC/inv | 3 | Poor | Head | 3 | ||||
| 34 | M | 73 | IPMC/inv | 3 | Poor | Head | 5.5 | G12D | |||
| 35 | F | 77 | IPMC/inv | 3 | Well | Head | 3.2 | G12D | |||
| 36 | F | 61 | IPMC/inv | 3 | Well | Head | 1 | G12D | |||
| 37 | M | 62 | IPMC/inv | 3 | Moderate | Head | 2.2 | ||||
| 38 | F | 59 | IPMC/inv | 3 | Moderate | Head | 3.4 | G12D |
Maximum tumor size includes both invasive and noninvasive components of tumor.
Invasive carcinoma was associated with IPMN/IPMC in pancreatic resection, but lesion analyzed did not sample invasive carcinoma.
F indicates female; IPMN/aden, IPMN adenoma; IPMN/border, IPMN borderline; IPMC/inv, IPMC with invasion; M, male; mucin.cystad, mucinous cystadenomas; N/A, not applicable; —, not available; Unc.proc, uncinate process.
DNA Samples and Mutation Analysis
Paraffin-embedded tumor samples were reviewed by the same pathologists (H.E.R. and N. T. C.) and microdissected by hand. Genomic DNA was extracted using QIAmp DNA Mini Kit (Qiagen, Valencia, Calif). The ERBB2 (HER2) (exon 20), EGFR (exons 19 and 21), KRAS (exon 1), BRAF (exons 11 and 15), and PIK3CA (exons 4, 9, and 20) were analyzed by polymerase chain reaction (PCR) amplification of genomic DNA and direct sequencing of the PCR products. Genomic DNA (40 ng per sample) was amplified with primers covering the coding region and the exon/intron boundaries of each exon analyzed. Before sequencing, all PCR products were purified (QIAquick PCR Purification Kit; Qiagen). Sequencing was performed with ABI’s 3100 capillary automated sequencers at the DNA core facility of Columbia University Medical Center. All samples found to have a genetic alteration in the target gene were subsequently sequenced in the reverse direction to confirm the mutation. The mutation was further verified by sequencing of a second PCR product derived independently from the original template. Corresponding normal tissues derived from surrounding nontumorous tissue or from a tumor-free block served as the normal control for each patient.
RESULTS
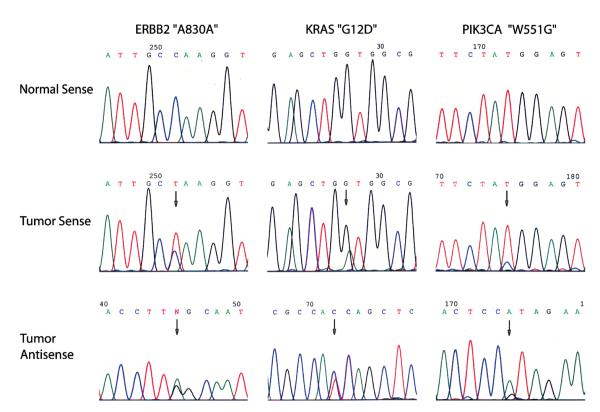
One sample (2.8%), a borderline IPMN, nuclear grade 2, harbored alterations in 3 different genes: a silent ERBB2 (HER2) mutation (A830A), a G12D mutation of KRAS, and an exon 9 mutation of PIK3CA (W551G) (Fig. 1). Two samples (5.6%) harbored mutations in 2 different genes. In an IPMC without invasion sample (nuclear grade 3), we found a KRAS G12V mutation and a nonhotspot mutation in the exon 20 of PIK3CA (S1015F). In an IPMC with invasion sample, (nuclear grade 3, differentiation moderate to poor), a KRAS G12R and an exon 15 mutation of BRAF (S615F) were found coin-hibited. In 16 (44%) of 36 samples, we identified single mutations in only 1 gene: 14 (38.8%) of 36 cases harbored a KRAS mutation in exon 1 (4 [33.3%]/12 IPMNs without associated invasive carcinoma [1 nuclear grade 1, 1 nuclear grade 2, and 2 nuclear grade 3] and 10 [41.7%]/24 IPMCs with associated invasive carcinoma [all nuclear grade 3]); 2 (5.5%) of 36 cases showed a single mutation of PIK3CA (1 exon 4 mutation in an IPMC with invasion [nuclear grade 3, differentiation moderate] and 1 exon 20 hotspot mutation [H1047R] in an IPMC with invasion [nuclear grade 3, differentiation moderate to poor]) (Table 1). None of the mutations was detected in the matching normal tissues.
FIGURE 1.
The oncogenic mutation spectrum of patient no. 1. Our data showed that KRAS, BRAF, and PIK3CA mutations can often coexist in the same patient, whereas mutations of ERBB2 and EGFR are rare in IPMN/IPMC.
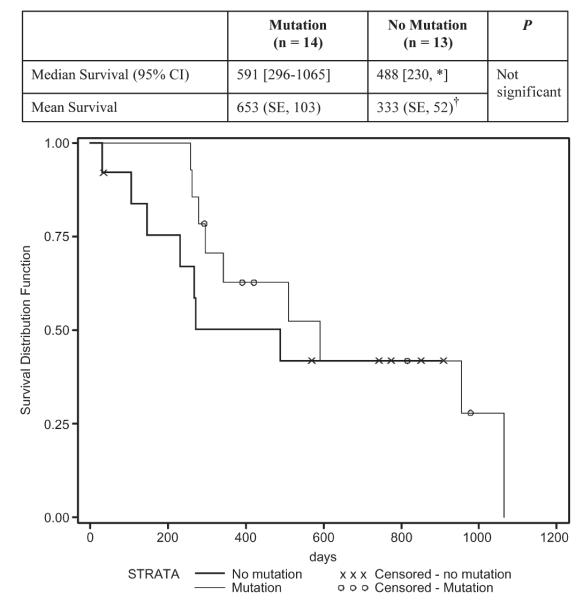
To investigate whether oncogenic activation impacts patient survival, we compared IPMC/invasive cancer patients with any of the 3 oncogenic mutations to those without any mutation. The median survival of patients with mutation(s) is 591 days and of those without any mutation is 488 days in our small collection (Fig. 2). Although a larger study is necessary in the future to obtain a significant P value, our results suggested that patients with any oncogenic mutation may have a better median survival than those without.
FIGURE 2.
Survival curves of IPMC/invasive carcinoma patients with or without any of the oncogenic mutations. The survival was calculated by the Kaplan-Meier method, and differences in survival between the mutation groups were tested with the log-rank test. Our data suggested that patients with any oncogenic mutation may have a better survival median, although a larger study is needed in the future to obtain a significant P value. *The upper limit was a censored observation (i.e., the patient was still alive so that survival time was calculated to the date of last follow-up). †The mean survival time and its SE were underestimated because the largest observation was censored and the estimation was restricted to the largest event time. CI indicates confidence interval.
DISCUSSION
The discovery of EGFR and ERBB2 (HER2) kinase domain mutations predominantly in lung adenocarcinomas, but also in gastric, ovarian, and brain tumors16,19,20,24 led us to analyze their genetic status in IPMN/IPMC. Overexpression of ERBB2 (HER2) in IPMNs has been reported to be 55% in the early stage of hyperplasia to up to 80% in the stage of carcinoma in situ.6,15 We identified 1 missense mutation in exon 20 of the ERBB2 (HER2) kinase domain at nucleotide 2640 (A830A) and none for EGFR, indicating that mutations of these genes are very infrequent in IPMNs/IPMCs. Downstream members of the EGFR/ERBB2 signaling pathways, including, KRAS, BRAF, and PIK3CA are frequently mutated in human cancers and act as oncogenic proteins.16,21,22 Previous studies have found KRAS mutations in 31% to 86% of IPMNs (47% in the present study, without any correlation to tumor size, stage, or differentiation).10,23,25 This is unlike PDA where KRAS is mutated at a frequency close to 100%.26 This suggests that in a large percentage of IPMNs/IPMCs, the Ras–Raf–message encrypton key–extracellular signal regulated kinase–mitogen-activated protein kinase pathway might be activated differently, other than by KRAS mutation. We identified 1 somatic BRAF mutation out of 36 cases of IPMN/IPMC examined (2.7%) in an IPMC with invasive carcinoma. Although located at exon 15, the S615F mutation is not the previously described hot-spot mutation at exon 15 (V600E) of the BRAF gene21,27 and was found to coexist with a G12R mutation of KRAS in the same sample. BRAF mutations, other than BRAF V600E, have been previously reported to coexist with RAS mutations.21,28 PIK3CA gene mutations were identified in several human tumors.22,29 They predominantly occur within exons 9 and 20, affecting the functionally important helical and kinase domains of the protein,22 and they are oncogenic as shown in the functional studies.22,30,31 Although 2 of the 3 mutations in exons 9 and 20 are not hot-spot mutations, they are likely to affect the kinase activity of PIK3CA.
Our data show that EGFR and ERBB2 (HER2) kinase domain mutations are very infrequent in IPMN/IPMC, suggesting the limited possibility of targeting mutated ERBB2 and EGFR for therapy for these lesions. PIK3CA is the first gene reported to be mutated in IPMN/IPMC, but not in PDA.13,32 According to our data, PIK3CA and BRAF gene mutations play an important role in the tumorigenesis of IPMN/IPMC, and in comparison to KRAS gene mutations, occur later during their transition to malignancy. They could represent interesting targets for future targeted therapies in IPMC. There is limited treatment option available for IPMN/IPMC patients currently, other than surgery. Our data suggest KRAS, BRAF, and PIK3CA as potential targets for future therapeutic development.
Acknowledgments
This work was supported by the National Cancer Institute Temin Award CA95434 and the National Cancer Institute R01 CA109525.
REFERENCES
- 1.Hruban RH, Takaori K, Klimstra DS, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–987. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 2.Kloppel G, Solcia E, Longnecker DS. Histological Typing of Tumours of the Exocrine Pancreas. Springer; New York: 1996. [Google Scholar]
- 3.Raimondo M, Tachibana I, Urrutia R, et al. Invasive cancer and survival of intraductal papillary mucinous tumors of the pancreas. Am J Gastroenterol. 2002;97:2553–2558. doi: 10.1111/j.1572-0241.2002.06022.x. [DOI] [PubMed] [Google Scholar]
- 4.Maire F, Hammel P, Terris B, et al. Prognosis of malignant intraductal papillary mucinous tumours of the pancreas after surgical resection. Comparison with pancreatic ductal adenocarcinoma. Gut. 2002;51:717–722. doi: 10.1136/gut.51.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohn TA, Yeo CJ, Cameron JL, et al. Intraductal papillary mucinous neoplasms of the pancreas: an updated experience. Ann Surg. 2004;239:788–797. doi: 10.1097/01.sla.0000128306.90650.aa. [discussion 797–799] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermanova M, Lukas Z, Nenutil R, et al. Amplification and over expression of HER-2/neu in invasive ductal carcinomas of the pancreas and pancreatic intraepithelial neoplasms and the relationship to the expression of p21(WAF1/CIP1) Neoplasma. 2004;51:77–83. [PubMed] [Google Scholar]
- 7.Yamao K, Ohashi K, Nakamura T, et al. The prognosis of intraductal papillary mucinous tumors of the pancreas. Hepatogastroenterology. 2000;47:1129–1134. [PubMed] [Google Scholar]
- 8.Chari ST, Yadav D, Smyrk TC, et al. Study of recurrence after surgical resection of intraductal papillary mucinous neoplasm of the pancreas. Gastroenterology. 2002;123:1500–1507. doi: 10.1053/gast.2002.36552. [DOI] [PubMed] [Google Scholar]
- 9.Nakagohri T, Konishi M, Inoue K, et al. Invasive carcinoma derived from intraductal papillary mucinous carcinoma of the pancreas. Hepatogastroenterology. 2004;51:1480–1483. [PubMed] [Google Scholar]
- 10.Z’Graggen K, Rivera JA, Compton CC, et al. Prevalence of activating K-ras mutations in the evolutionary stages of neoplasia in intraductal papillary mucinous tumors of the pancreas. Ann Surg. 1997;226:491–498. doi: 10.1097/00000658-199710000-00010. [discussion 498–500] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sessa F, Solcia E, Capella C, et al. Intraductal papillary-mucinous tumours represent a distinct group of pancreatic neoplasms: an investigation of tumour cell differentiation and K-ras, p53 and c-erbB-2 abnormalities in 26 patients. Virchows Arch. 1994;425:357–367. doi: 10.1007/BF00189573. [DOI] [PubMed] [Google Scholar]
- 12.Sato N, Rosty C, Jansen M, et al. STK11/LKB1 Peutz-Jeghers gene inactivation in intraductal papillary-mucinous neoplasms of the pancreas. Am J Pathol. 2001;159:2017–2022. doi: 10.1016/S0002-9440(10)63053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schonleben F, Qiu W, Ciau NT, et al. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12:3851–3855. doi: 10.1158/1078-0432.CCR-06-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii H, Inagaki M, Kasai S, et al. Genetic progression and heterogeneity in intraductal papillary-mucinous neoplasms of the pancreas. Am J Pathol. 1997;151:1447–1454. [PMC free article] [PubMed] [Google Scholar]
- 15.Moriya T, Kimura W, Semba S, et al. Biological similarities and differences between pancreatic intraepithelial neoplasias and intraductal papillary mucinous neoplasms. Int J Gastrointest Cancer. 2005;35:111–119. doi: 10.1385/IJGC:35:2:111. [DOI] [PubMed] [Google Scholar]
- 16.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 17.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 20.Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 21.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 22.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 23.Schonleben F, Qiu W, Bruckman KC, et al. BRAF and KRAS gene mutations in intraductal papillary mucinous neoplasm/carcinoma (IPMN/IPMC) of the pancreas. Cancer Lett. 2006;8:8. doi: 10.1016/j.canlet.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JW, Soung YH, Kim SY, et al. ERBB2 kinase domain mutation in a gastric cancer metastasis. Apmis. 2005;113:683–687. doi: 10.1111/j.1600-0463.2005.apm_284.x. [DOI] [PubMed] [Google Scholar]
- 25.Satoh K, Shimosegawa T, Moriizumi S, et al. K-ras mutation and p53 protein accumulation in intraductal mucin-hypersecreting neoplasms of the pancreas. Pancreas. 1996;12:362–368. doi: 10.1097/00006676-199605000-00007. [DOI] [PubMed] [Google Scholar]
- 26.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- 27.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 28.Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62:6451–6455. [PubMed] [Google Scholar]
- 29.Qiu W, Schonleben F, Li X, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:1441–1446. doi: 10.1158/1078-0432.CCR-05-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikenoue T, Kanai F, Hikiba Y, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 31.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallmeier E, Calhoun ES, Kern SE. No mutations in PIK3CA identified in pancreatic carcinoma. NOGO. 2004;8:2. [Google Scholar]