Abstract
In this report, we have designed a simple and efficient green chemistry approach for the synthesis of colloidal silver nanoparticles (b-AgNPs) that is formed by the reduction of silver nitrate (AgNO3) solution using Olax scandens leaf extract. The colloidal b-AgNPs, characterized by various physico-chemical techniques exhibit multifunctional biological activities (4-in-1 system). Firstly, bio-synthesized silver nanoparticles (b-AgNPs) shows enhanced antibacterial activity compared to chemically synthesize silver nanoparticles (c-AgNPs). Secondly, b-AgNPs show anti-cancer activities to different cancer cells (A549: human lung cancer cell lines, B16: mouse melanoma cell line & MCF7: human breast cancer cells) (anti-cancer). Thirdly, these nanoparticles are biocompatible to rat cardiomyoblast normal cell line (H9C2), human umbilical vein endothelial cells (HUVEC) and Chinese hamster ovary cells (CHO) which indicates the future application of b-AgNPs as drug delivery vehicle. Finally, the bio-synthesized AgNPs show bright red fluorescence inside the cells that could be utilized to detect the localization of drug molecules inside the cancer cells (a diagnostic approach). All results together demonstrate the multifunctional biological activities of bio-synthesized AgNPs (4-in-1 system) that could be applied as (i) anti-bacterial & (ii) anti-cancer agent, (iii) drug delivery vehicle, and (iv) imaging facilitator. To the best of our knowledge, there is not a single report of biosynthesized AgNPs that demonstrates the versatile applications (4-in-1 system) towards various biomedical applications. Additionally, a plausible mechanistic approach has been explored for the synthesis of b-AgNPs and its anti-bacterial as well as anti-cancer activity. We strongly believe that bio-synthesized AgNPs will open a new direction towards various biomedical applications in near future.
Keywords: Bio-synthesis, Silver nanoparticle, Green Chemistry, Olax scandens, Multifunctional activities, Antibacterial, anti-cancer.
INTRODUCTION
Nanotechnology dealing with metal nanoparticles has been widely applied in almost every field of science and technology including biomedical sciences due to presence of size and shape dependent unusual physical and chemical properties 1, 2. In the last decade, several groups including ours significantly developed metal nanoparticles based theranostic approaches for the treatment of several diseases 1, 3-11. Among several nanoparticles, recently silver nanoparticles have been extensively used in electronics, engineering, textiles, paints, food industry, cosmetics, bio-sensing, chronic wounds, and even in medicine and biology 10, 12-16. Therefore, design and development of simple, one-step, reliable, low-cost, non-toxic and eco-friendly method for the fabrication of multifunctional silver nanoparticles is of greatest importance to expand their biomedical applications. In this context, synthesis of silver nanoparticles using green chemistry approach 17, 18 has several advantages over conventional chemical methods namely (i) simple, one-step, fast, inexpensive & most reliable method, (ii) environmental friendly due to avoidance of many toxic chemicals, (iii) convenience of bio-resources such as plants, fungi, algae, microorganism that act as reducing as well as stabilizing/capping agent, (iv) universally acceptable solvent like water etc 10, 12, 17, 19-24.
In this present report, we have developed a simple one-step green chemistry approach for the synthesis of colloidal silver nanoparticles using Olax scandens leaf extract due to several reasons such as (i) the plant contains various phytochemicals like, aleanolic acid, oleanolic acid, β-sitosterol, octacosanol, glucosides of β -sitosterol etc, that act as reducing agent as well as stabilizing/capping agentin the synthesis of nanoparticles 25, (ii) Olax scandens is a medicinally important herbal plant useful for the treatment of several diseases such as anti-cancer 26-28, anti-bacterial 29, headache 30, and psoriasis 31, (iii) the phytochemicals like octacosanol, β-sitosterol, and glucosides of β -sitosterol exhibit antiproliferative activity towards cancer cell lines 26-28. Conjugation of these phytochemicals with the as-synthesized b-AgNPs shows enhanced anticancer activity at low concentration compared to free extract (present communication) and (iv) the plant contains fluorescent molecules or proteins that exhibit red fluorescence in the silver nanoconjugates, observed by fluoresence microscope. The fluoresence property helps us to detect the localization of AgNPs inside the cells.
The biosynthesized silver nanoparticles (b-AgNPs) exhibit multifunctional activities (4-in-1 system) such as biocompatibility, anti-cancer & anti-bacterial activity and fluorescence imaging. To the best of our knowledge, there is not a single report of bio-synthesized silver nanoparticles (b-AgNPs) that demonstrates the versatile applications (4-in-1 system) towards various biomedical applications. Additionally, a plausible mechanistic approach has been investigated for anti-bacterial & anti-cancer activity of b-AgNPs. The results together will provide the basic foundation for the development of versatile applications of biosynthesized AgNPs towards biomedical technology in near future.
2. EXPERIMENTAL PROCEDURES
2.1. Materials
Silver nitrate (AgNO3), osmium tetroxide (OsO4), glutardehyde, dihydroethidium (DHE), 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate (DCFDA), Glutathione (GSH), Hydrogen peroxide (H2O2), Potassium permanganate (KMnO4) were purchased from Sigma Aldrich Chemicals, St. Louis, MO, USA and used without further purification. Initially, 10-2 (M) of AgNO3 stock solution was prepared in sterile Millipore water and used for the biosynthesis of silver nanoparticles (b-AgNPs) and further cell culture experiments. Lung cancer cells A549), mouse melanoma cell line (B16), human breast cancer cell lines (MCF7), rat cardiomyoblast cell line (H9C2), and human umbilical vein endothelial cells (HUVEC) were purchased from ATCC, USA.
2.2. Preparation of Olax scandens leaf extract in methanol (MeOH)
300 grams of fresh Olax scandens leaves were thoroughly washed with distilled water (3 times) followed by sterile Millipore water in a 500 mL beaker. The cleaned leaves were allowed to dry for 2 weeks and then 50 g of dried leaf powder were placed in Soxhlet apparatus for MeOH extraction. The dark green crude extract was dried by hot water bath and finally 1 g of Olax scandens leaf powder was obtained. 10 mg of this crude powder was dissolved in 1 mL of DMSO: H2O (20: 80; v/v) mixture in order to make a 10 mg/ mL-1 stock solution (denoted as 'OX'). This 'OX' has been used for the synthesis & characterizations of AgNPs and other experiments (cell cultures, anti-bacterial, imaging etc.).
2.3. Biosynthesis of silver nanoparticles (b-AgNPs)
In this present article, we have used Olax scandens leaf extract as reducing as well as stabilizing/capping agent for the formation of silver nanoparticles. The color, shape, size and stability of silver nanoparticles depend on the concentration/volume of the reducing agent (here Olax scandens extract). Therefore, in order to get optimize AgNPs, we have carried out a series of reactions using the various concentrations of Olax leaf extract (150-750 µL) (Table 1) keeping constant concentration of AgNO3 (150 µL of 10-2 M) in total 5 mL solution. This green chemistry approach for the synthesis of AgNPs is simple, efficient, economically cheap and environmentally friendly method that does not need any special conditions such as sophisticated instrument, vacuum condition, catalyst, template etc. Finally, the reaction was carried out under ambient conditions in water (universally accepted solvent).
Table 1.
Reaction conditions of biosynthesis of silver nanoparticles (b-AgNPs) using Olax Scandens leaf extract.
| Exp. No | OX-Extract (μL) | AgNO3 (μL) | Water (mL) | *Time (hour) | Size from DLS (nm) | Size from TEM (nm) | Zeta potential (charge mV) |
|---|---|---|---|---|---|---|---|
| 1. | 150 | 150 | 4.70 | ~ 7 | ~ 85-130 | 20-110 | - 11.8±0.2 |
| 2. | 400 | 150 | 4.45 | ~4.5 | ~ 75-140 | 10-85 | - 10.1±0.7 |
| 3. | 500 | 150 | 4.35 | ~ 2 | ~ 55-100 | 10-55 | - 15.2±0.5 |
| 4. | 750 | 150 | 4.10 | ~1.5 | ~ 60-140 | 10-90 | - 13.2±0.4 |
[a] *Time was required to appear the yellow coloration. Total volume of reaction mixture was maintained at 5 mL (formation of b-AgNPs).
2.4. Chemical synthesis silver nanoparticles (c-AgNPs)
Silver nanoparticles were also synthesized by chemical methods in order to compare the biological activity with biosynthesized silver nanoparticles. In a typical experiment, c-AgNPs were prepared by the reduction of 150 µL of AgNO3 (10-2 M) using 2.8 mL of NaBH4 (0.05 mg/mL). The loose dark ash pellet was collected from chemically synthesized c-AgNPs by centrifugation at 14,000 rpm at 100 C for 1 h in Thermo scientific, Sorvall-WX ultra 100. This chemically synthesized c-AgNPs were used for antibacterial and anticancer studies and compared with biosynthesized b-AgNPs Supplementary Material: SI.Fig.4.a-c, SI-Table 1).
2.5. Antibacterial activity of b-AgNPs towards E. coli (DH5α strain)
The bacterial growth was maintained at 37 ºC in shaker (rotating at 80 rpm) in LB (Luria bertani) broth media. The inhibition growth of bacteria treated with biosynthesized silver nanoparticles was monitored by multimode spectrophotometer (Varioskan) at 600 nm up to 7 hours. Spread plate technique was used to measure the antibacterial activity of b-Ag-NPs in a dose dependent manner (3-30 µM) by counting of CFU (colony forming units). Agar disc diffusion technique was used to evaluate antibacterial activity of Gram-negative E. coli against b-Ag-NPs by measuring the inhibitory zone. SEM (Hitachi S-3000 N, Japan) technique was used to measure the interaction of silver nanoparticles with E. coli. The experimental procedures for bacteria culture have been discussed in detail to the Supplementary Material.
2.6. Bacterial cell lysate preparation for biochemical assays
Initially, bacterial cell pellet was obtained by centrifugation of bacteria cells at 4°C temperature for 30 minutes at 5000 rpm. The pellet was suspended in bacterial lysis buffer followed by the addition of lysozyme and the mixture was allowed to incubate at 4 oC for 4h followed by probe sonication for 5 minutes. Cell debris was removed by centrifugation at 10,000 rpm and the supernatant was collected and used for enzyme assays.
2.7. Assay of Catalase
The catalase enzyme activity has been measured according to published report(32. Initially, a reaction mixer was prepared by mixing 500 µl of 0.1 (M) phosphate buffer (pH = 7.5) and 500 µl of freshly prepared 0.9 % H2O2 solution (say RM solution). 100 µl of bacterial cell lysate was added to RM solution and the mixture was incubated for 3 minutes. 2(N) H2SO4 was added drop wise to stop the reaction. The unreacted H2O2 was titrated with 0.1(N) KMnO4. Similarly, for blank experiment, 100 µl of bacterial cell lysate was heated to inactivate the catalytic activity of catalase enzyme. The boiled lysate was added to RM solution and it was titrated with 0.1(N) KMnO4.
Enzyme activity (EA) was calculated by using the following formula:
| EA = (B-T/3) x 2 |
where B is blank, T: is test value/experimental value; 3 indicates incubation time of enzyme, 2 is constant value). Specific enzyme activity was calculated using enzyme activity and total protein concentration in the bacterial cell lysate calculated by Bradford method 33.
2.8. SDS-PAGE analysis of proteins of bacterial cells
Protein was estimated from bacterial cell lysate using Bradford method. Equal amount of proteins obtained from different treatment groups were loaded into 12% SDS-PAGE according to Laemmli method 34. Molecular mass marker was used as reference sample. Finally, the gel was stained with 100 mL of coomassie brilliant blue R-250 (Hi-media; Cat. # 42660) solution [0.25gms of dye, 45 ml methanol, 45 ml water and 10 ml of acetic acid] for 3 hours of incubation. The gel was distained with 100 mL of similar above solution without dye for overnight.
2.9. Cell culture experiments
All cancer cell lines [A549: lung cancer cells; B16: mouse melanoma cell line; H9C2: rat cardiomyoblast cell line; CHO: Chinese hamster ovary] purchased from ATCC were cultured in DMEM (Dulbecco's Modified Eagle Medium) media supplemented with 10% fetal bovine serum (FBS), 5% L-glutamine, and 1% antibiotics (penicillin-streptomycin) in a humidified 5% CO2 incubator at 37 0C for all in vitro experiments and fluorescence imaging. On the other hand, human umbilical vein endothelial cells (HUVEC) were cultured in EBM complete media at 37 0C incubator with 5% CO2. The cells were incubated with various concentrations of Olax extract and silver nanobioconjugates, which were kept under UV irradiation inside the cell culture hood for 15 mins before use.
2.10. Cell viability test using MTT reagent
Viability of H9C2, CHO, A549, B16 and HUVEC cells were tested by cell viability assay using MTT reagents by incubation of cells with (i) b-AgNPs, (ii) c-AgNPs in a dose dependent manner for 24 h. The detailed procedure has been discussed in the Supplementary Material.
2.11. Cell cycle assay using Fluorescence activated cell sorting (FACS)
Cell cycle analysis was performed according to a standard procedure by staining B16 cells with propidium iodide (PI). B16 cells were first treated with (i) Ox-100 µg/ml, (ii) b-Ag-30 µM, (iii) c-Ag-30 µM for 24 hours and after that the cells were washed in DPBS and fixed in 70% ethanol for 24 h. Cells were then re-hydrated, washed in DPBS, and treated with RNase mixture (50 μg/ml) followed by staining with PI (100 μg/mL). Similar experiments were done with untreated control cells. Quantification of DNA content was analysed with a FACScan flow cytometer. (BD bioscience) Extent of propidium iodide staining of the gated population was displayed in a histogram and the regions are defined as: sub-G1, G0/G1, S, and G2/M.
2.12. Determination of superoxide anion radicals and hydrogen peroxide (ROS)
Initially, the A549 & B16 cells were cultured in DMEM complete media. In order to determine and compare the presence of superoxide radicals in the A549 cells treated with and without b-AgNPs, the cells (2 X 104 cells per 1 mL) were incubated on cover slips in 24-well plates in DMEM starved media for 24 hours. The media replaced with fresh DMEM media and the cells were incubated with 10 µM of DHE (Sigma Aldrich: D7008) for 15 min. Finally, after extensive washing the cells with DPBS, the fluorescence images of A549 cells in HBSS buffer were examined by fluorescence microscopy (Nikon Eclipse TE2000-E). The red fluorescence emission (λEx = 605 nm), indicating the presence of superoxide anion, was collected with a 10 X microscope objective after excitation at λEx = 510 and 560 nanometers nm (green). Similarly, after treatment the cells were incubated with DCFDA (30 µM) for 30 mins and the cells were extensively washed with DPBS. Finally, the fluorescence images of A549 (treated and untreated) in HBSS buffer were examined by Nikon Eclipse TE2000-E). The green fluorescence emission (λEx = 525 nm), indicating the presence of especially hydrogen peroxide, was collected with a 10 X microscope objective after excitation at λEx = 420 and 495 nanometers (blue).
2.13. Immunoblot analysis (Western Blot Analysis)
Total protein extraction and immunoblotting was performed as described previously [63]. Protein concentration was determined by Bradford method (Bio-Rad, Hercules, CA, USA). An equal amount (30 μg) of protein of each of the samples under study was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, protein was transferred to PVDF membranes (GE Healthcare, USA). The membranes were then blocked in Tris-buffered saline Tween-20 (TBS-T; 10 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween-20) and 5% non-fat dry milk for 1 h, and subsequently washed and incubated with primary antibodies in TBST with 2.5% non-fat dry milk at 4°C for overnight. The following polyclonal antibodies and titers were used: P53 (1:500, ABBIOTEC #200143), Pro-caspase3 (1-5 µg/ml , IMGENEX # IMG-144A), Active-caspase3 (3 µg/ml , IMGENEX # IMG-144A) and β-Actin (1:1000, ABCAM #ab8224). After washing with TBS-T, membranes were incubated with Goat Anti-Rabbit IgG-HRP (1:5000 dilution, SANTA CRUZ, # SC 2004) or Goat Anti-Mouse IgG-HRP (1:5000 dilution, SANTA CRUZ, #2005) horseradish peroxidase conjugated secondary antibody with 2.5% non-fat dry milk at room temperature for 1 h. After washing with TBS-T immunoreactions were visualized with a chemiluminescence detection kit (EMD MILLIPORE, Prod No- WBLUF0500). Then the blots were developed in Bio Spectrum 810 Imaging System (UVP). Quantification of band intensity was performed using the Image J Software (NIH, Bethesda, MD, USA). For quantitative analysis, the signal intensity of each of the protein was normalized with the corresponding β-Actin signal.
2.14. Quantification of released silver ions (AAS and UV-Visible studies)
The as synthesized b-AgNPs was centrifuged at 14,000 r.p.m and the as collected pellets have incubated with acidic (pH-5.0) and neutral buffer solutions (pH-7.4) for 24 hours [10 ppm of b-Ag has been incubated with 10 mL of pH-5.0 and pH-7.4 buffers]. After 24 hours of incubation the solutions have been centrifuged at 25,000 rpm to eliminate all the AgNPs and to keep only the silver ions. The supernatant is submitted for AAS to determine the concentration of silver ions. Also, the b-Ag incubated with acidic and neutral buffers for 24 hours is monitored in the UV-Visible spectroscopy.
3. CHARACTERIZATION TECHNIQUES
The as synthesized b-AgNPs and c-AgNPs were thoroughly characterized by several physico-chemical techniques that are elaborately described in Supplementary Material. Before characterization, 100 mL of as-synthesized b-AgNPs & c-AgNPs were centrifuged for 1h at 14,000 rpm at 10 0C using Thermo scientific, Sorvall-WX ultra 100. The intense loose ash colored AgNPs pellet was used for further characterizations and in-vitro experiments. The absorption and crystallinity of as-synthesized AgNPs were measured by UV-VIS spectroscopy and X-ray diffraction (XRD) analysis, respectively. The morphology, shape and size of nanoparticles were examined by transmission electron microscope (TEM). Fourier transformed infrared spectroscopy (FTIR) was used to detect the possible functional groups in biomolecules present in the plant extract. The presence of proteins in the leaf extract was confirmed by gel electrophoresis using 12% SDS-PAGE 35. The surface charge of as-synthesized AgNPs was measured by zeta potential analyzer (DLS). Inductively coupled plasma optical emission spectrometer (ICP-OES) was carried out for the detection of AgNPs at very low concentration. Fluorescence images of A549 cancer cells were collected by fluorescence microscopy (Olympus IX71, Olympus U-CMAD3, T2 Tokyo Japan) through excitation of 518 nm (green) and emission of 605 nm (Red). The detailed characterization procedures are described in Supplementary Material.
4. RESULTS AND DISCUSSION
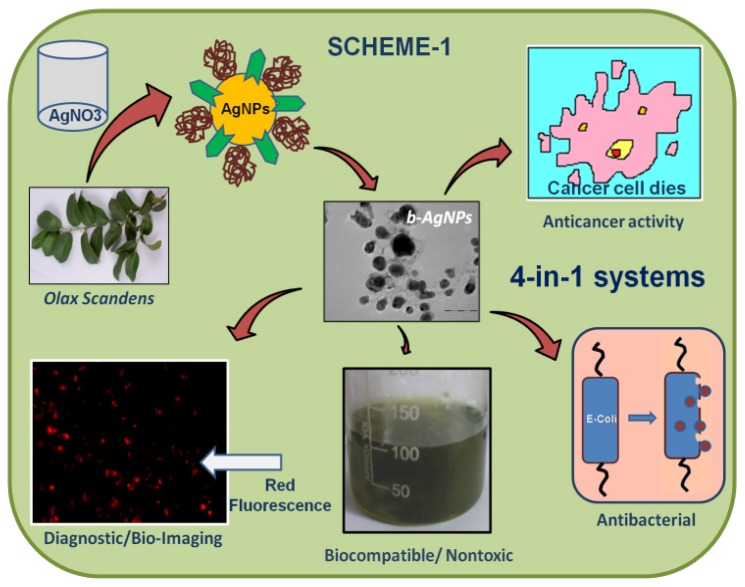
Biosynthesized silver nanoparticles (b-AgNPs) have been synthesized using Olax scandens leaf extract, which acts as reducing as well as stabilizing/capping agent. The one step synthesis reaction was carried out at ambient conditions (room temperature and atmospheric pressure) in water solvent, a universally accepted solvent. Our biosynthesis method for b-AgNPs follows almost all principles of green chemistry approach 36. In order to get the optimized and stable b-AgNPs, we have carried out a series of reactions using the various concentrations of Olax leaf extract (Table 1). According to experimental conditions described in Table 1 and characterization data (discussed later), the experiment number # 3 is the optimized reaction conditions for the biosynthesis of nanoparticles (b-AgNPs-500) base on color, stability, size & charge of the nanoparticles. Therefore the optimized silver nanoparticles (b-AgNPs-500) have been utilized for detailed characterizations and further in vitro assays for several biomedical applications (biocompatibility, anti-bacterial & anti-cancer activity, and imaging study). The overall synthesis and their applications have been schematically presented in SCHEME 1 (Figure A).
Figure A.
(Scheme 1) Over all presentation for synthesis, characterization and biomedical applications (diagnostic, anticancer antibacterial applications) of biosynthesized silver nanoparticles (b-AgNPs) using Olax Scandens leaf extract.
4.1. UV visible spectroscopy
A series of reactions were performed for the synthesis of silver nanoparticles using various concentrations of Olax leaf extract (150-750 µL) that is presented in Table 1. Appearance of ash yellow color indicates the formation of silver nanoparticles, which was further confirmed, by the appearance of absorption peak around 410-430 nm, measured by UV visible spectroscopy 37, 38. The reaction time for the formation of silver nanoparticles decreases from 7h to 1.5h with increasing concentration of extract from 150 µL to 750 µL.
Synthesis of silver nanoparticles using different concentrations of extract is presented in Supplementary Material: SI.Fig.1.a-d that has been monitored by UV visible spectroscopy in a time dependent manner. Appearance of absorption peak around 410-430 nm in UV visible spectra suggests the formation of spherical silver nanoparticles (SI.Fig.1.a-d). Increasing the absorption intensity with increasing the reaction time supports the gradual formation of AgNPs towards the completion of the reaction. In Experiment #1 (Table 1), there is no further increase in the absorption intensity after 120 h. It indicates the completion of the reaction. Similarly for Experiment numbers # 2 takes 1h to complete the reaction and beyond that time the intensity decreases because of instability of the nanoparticles. For Experiment number # 3, the maximum intensity is observed at 24 h and intensity is slightly decreased around 120 hours. Also, we have compared the absorption intensity of the four different sets at 24 hours (Fig.1.a). It has been clearly visible that for b-AgNPs-500 (Experiment number #3) the absorption intensity is maximum. That is why, Experiment number #3 where we have used 500 µL of extract for the synthesis of AgNPs is the optimized reaction conditions considering the color, stability, size and charge of the nanoparticles. Therefore, b-AgNPs-500 (written as b-AgNPs later) has been used for further detailed characterizations and in vitro experiments.
Figure 1.
Physico-chemical characterization and stability studies of b-AgNPs. (a) absorbance of b-AgNPs obtained after 24 hours with different concentration of Olax extract has been measured by UV visible spectroscopy (150, 400, 500 & 750 values indicate the volume of Olax extract in µL), (b) X-ray diffraction pattern (XRD) of b-AgNPs-500 demonstrates the crystalline nature of silver nanoparticles obtained with 500 µL of extract, (c) TEM indicates the morphology of b-AgNPs-500 and (d) in-vitro stability studies of as synthesized b-AgNPs-500 (24 h to 14 days) has been carried out to several physiological buffer and salts solution at different pH. The results indicate the high stability of b-AgNPs.
4.2. X-ray diffraction (XRD) spectroscopy
The crystal structure of the as-synthesized b-AgNPs-500 has been investigated by X-ray diffraction analysis (XRD). The Bragg reflections indicate that b-AgNPs-500 is specifically indexed to a face centered cubic (FCC) crystal structure (Fig.1.b). Extra peaks in XRD pattern suggest the presence of silver oxide as impurity 39. The diffraction peaks are consistent with the standard JCPDS database (No. 04-0783 for AgNPs and No. 42-0874 for silver oxide) for all reflections.
4.3. Transmission electron microscopy (TEM)
In order to investigate the size, shapes of biosynthesized silver nanoparticles at different reaction conditions, TEM has been carried out. Fig.1.c indicates the TEM images of b-AgNPs-500. TEM images of b-AgNPs-150; b-AgNPs-400, b-AgNPs-500 and b-AgNPs-750 obtained after 24 hours of reactions, are shown in the Supplementary Material (SI-Fig.2-a-d). The silver nanoparticles consist of almost monodispersed spherical nanoparticles (20-60 nm) along with very few bigger particles (~90 nm). The as-synthesized silver nanoparticles are highly stable for several days. In order to prove that we have carried out the stability studies of b-AgNPs in different physiological buffers (pH -6 to pH -8) and solutions (water, 5% and 10% NaCl solution, FBS & DPBS etc) (Fig.1.d) and observed the stability up to 14 days.
4.4. Dynamic light scattering (DLS) technique
The stability of biosynthesized silver nanoparticles (b-AgNPs) depends on both size and surface charge of the nanoparticles. The data obtained from DLS measurements indicates that the size of the particles range is varied from 55-85 nm and negative charge is from -10.1 ± 0.7 to -15.2 ± 0.5. The high negative ξ value supports long-term stability, good colloidal nature and high dispersity of biosynthesized silver nanoparticles due to negative-negative repulsion. The size and zeta potential (ξ) values for biosynthesized nanoparticles are tabulated in Table 1. The size distribution of the nanoparticles obtained by DLS analysis is presented in S.I.Fig.3.a-d.
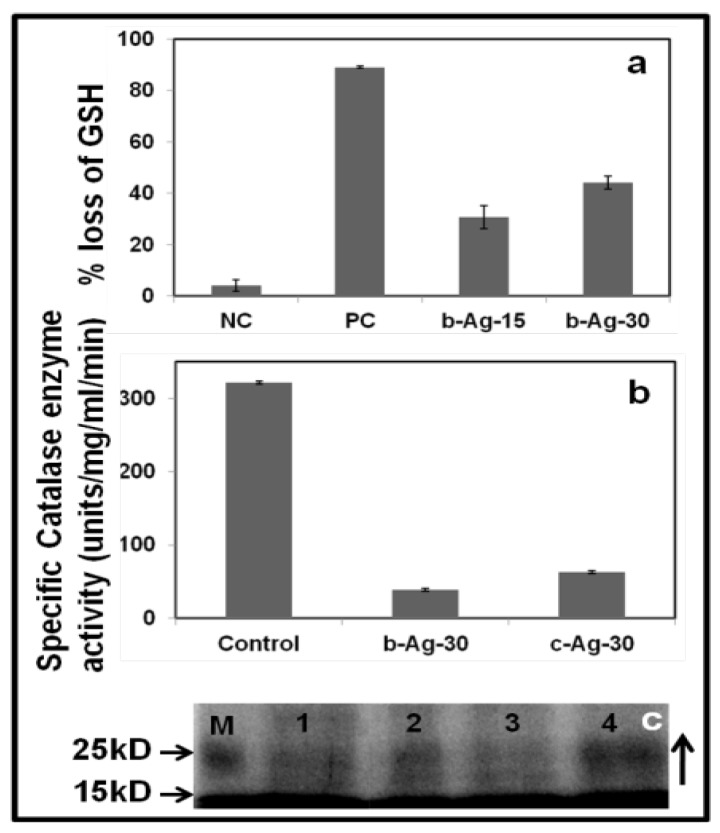
Figure 3.
Study of oxidative stress and SDS PAGE. (a) In-vitro GSH oxidation by Ellman's assay: PC: positive control (1 mM H2O2), and b-AgNPs-15 and b-AgNPs-30: biosynthesized silver nanoparticles synthesized by Olax leaf extract (15 & 30 indicates µM). Results show that b-AgNPs (in both the concentrations) shows considerable amount of oxidation of glutathione, which generates oxidative stress inside bacteria cell. (b) In vitro catalase activity: Control, biosynthesized b-AgNPs (30 µM) treated E. coli and chemically synthesized c-AgNPs (30 µM) treated E. coli. Biosinthesized AgNPs show maximum inhibition effect. (c) Coomassie blue staining of SDS page of bacterial proteins: 1D gel electrophoresis study of E. coli proteins treated with (2) Ampicillin (100μg/ml), (3) c-AgNPs (30 µM) and 4 b-AgNPs (30 µM). M: standard protein marker and (1) untreated E. coli. Bacteria cells treated with b-AgNPs show the up regulation of stress proteins (10 kD-25 kD) in b-AgNPs (30 µM).
4.5. Fourier transformed infrared (FTIR) spectroscopy
The bio-synthesis of silver nanoparticles has been carried out using Olax scandens leaf extract that contains some phytochemicals which are responsible for synthesis and stabilization of the nanomaterials. In order to find out the exact role of phytochemicals present in the leaf extract, FTIR spectroscopy has been carried out for both Olax leaf extract and b-AgNPs-500 that are presented in Supplementary Material: SI.Fig.5.a-b. The major stretching frequencies appeared at ν = 1512.18 cm-1 and ν = 1258.79 cm-1 due to the presence as amide II and amide III in proteins of Olax scandens, have been almost disappeared for b-AgNPs-500 (SI.Fig.5.b) confirming the role of proteins for the synthesis and stablization of b-AgNPs-500 (SI.Fig.5.b). These results are supported by our previous literature 22, 23. The detailed discussions are incorporated in to Supplementary Material. The role of proteins for the synthesis and stablization of b-AgNPs-500 has been further confirmed by SDS gel electrophoresis (discussed later in section 4.6).
4.6. SDS-PAGE analysis of proteins
According to earlier literature, proteins present in the plant leaf have an important role for the formation and stabilization of b-AgNPs 19, 21. In order to find out the role of proteins present in the Olax Scandens leaf extract, SDS-PAGE has been carried out where we have loaded concentrated Olax leaf extract and concentrated supernatant of b-AgNPs-500 (obtained after centrifugation), in Lane-1 and Lane-2, respectively. The protein around 15-17 kDa (in Lane-1) is almost disappeared in Lane-2 suggesting the role of some low molecular weight proteins (~15-17 kDa) present in Olax leaf extract responsible for formation and stabilization of b-AgNPs. Supplementary Material: SI.Fig.6)
4.7. Inductively coupled plasma optical emission spectrometer (ICP-OES)
The concentration of silver in the loose pellets (obtained after centrifugation) of bio-synthesized (b-AgNPs) and chemically synthesized (c-AgNPs) nanoparticles has been quantitatively determined by ICP-OES analysis (22, 23. The results show the concentration of b-AgNPs and c-AgNPs are 332.8 mg/mL and 383.7 mg/mL, respectively.
4.8. Antibacterial activity: Growth Inhibition and Colony counting assay
The time dependent growth of Gram Negative bacteria E. coli in LB-broth has been studied by measuring the OD of E. coli treated with b-AgNPs. Graphical representation of inhibition kinetics for E. coli treated with different concentrations of biosynthesized silver nanoparticles (b-AgNPs) is presented in Fig. 2.a. The inhibition of growth is observed with b-AgNPs in a dose dependent manner compared to control untreated bacteria. There is no growth at all (~100% inhibition) when the bacteria are treated with 30 µM of b-AgNPs up to 24 hours (data shown up to 7 hours). The 100% inhibition of E. coli growth is termed as minimum inhibitory concentration (MIC), which is observed at 30 µM of biosynthesized silver nanoparticles. However, chemically synthesized silver nanoparticles show MIC at concentration of 60 µM according to published literature 40. These results clearly indicate that biosynthesized AgNPs have almost double anti-bacterial effect than chemically synthesized AgNPs. The inhibition effect is comparable with positive control experiment where the bacteria are induced with ampicillin (100μg/ml). The anti-bacterial effect of b-AgNPs has been confirmed by further in vitro experiments (described below).
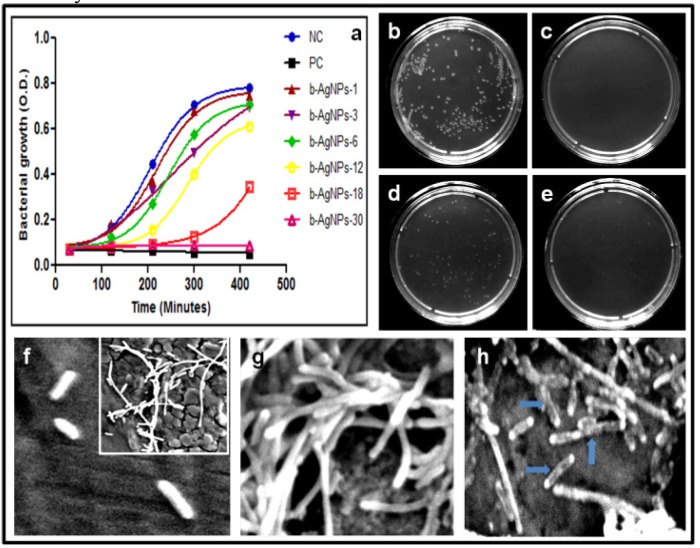
Figure 2.
Study of anti-bacterial activities: (a) liquid growth inhibition kinetics of E. coli using different concentrations of b-AgNPs. b-AgNP-30 (at 30 µM) shows almost 100% growth inhibition. Ampicillin has been used as a positive control (PC) & NC: negative control or untreated E. coli. The numerical number indicates the concentration of b-AgNPs in µM, (b-e) optical images of bacterial colonies formed by E. coli cells i.e. colony counting assay (after 24 h): b: Control, c:Ampicillin (100μg/ml), d: b-AgNPs (18 μM), e: b-AgNPs (30μM) and (f-h) SEM images of E. coli cells (f) without being treated (control), (g) treated with Olax for 1 hour, (h) treated with b-AgNPs (30 µM) for 1 hour. The SEM images show the silver nanoparticles damages the bacterial cell membrane (marked by blue arrow), whereas, the bacterial membranes of untreated and treated E. coli with Olax is intact.
Colony forming unit (CFU) is an indirect method for the determination of antibacterial activity. The anti-bacterial effect of E. coli treated with Olax extract, ampicillin and different concentrations of bio-synthesized AgNPs has been measured using CFU of viable E. coli through spread plate method 38, 41, 42(Fig-2.b-e). The antibacterial efficacy (ABE) of b-AgNPs, presented in Table 2 can be calculated according to the following equation (Equ.-1)
Table 2.
Antibacterial activity of biosynthesized silver nanoparticles (b-AgNPs): CFU, ABE and Inhibition Zone experimental results.
| Sample Name | Concentration | No. of colonies | ABE | Inhibition Zone (mm) |
|---|---|---|---|---|
| Control | - | 300 +/- 20 | - | 0 |
| Ampicillin | 100μg/ml | 0 | 100% | 19 |
| Olax | 100μg/ml | Mat | 0% | 9 |
| b-Ag-Ox-18 | 18µM | 75 +/- 5 | 75 % | - |
| b-Ag-Ox-30 | 30µM | 10 +/- 2 | 96.67% | 20 |
| c-Ag-30 | 30µM | - | - | 10 |
| ABE = % (Vc-Vt)/Vc × 100 | (1) |
Here, Vc =Number of viable bacterial colonies on control (blank) plate.
And, Vt = Number of viable bacterial colonies on test specimen.
Table 2 indicates that 97% of ABE is observed when E. coli is treated with 30µM of b-AgNPs. However, Raffi et al observed 100 % of ABE to E. coli at 353 µM of chemically synthesized silver nanoparticles 42. These results clearly indicate that our bio-synthesized AgNPs shows almost 10 times more anti-bacterial effect compared to chemically synthesized nanoparticles.
Agar diffusion disc method has been utilized in order to investigate the antibacterial activity of Ag-NPs at different concentration on E. coli. The antibacterial activity of b-AgNPs is compared with Olax, c-AgNPs, AgNO3 & ampicillin (100 μg/ml) by measuring the zone of inhibition. The results are tabulated in Table 2. Interestingly, the b-AgNPs at 30 μM concentration shows almost similar inhibition zone (~20 mm) with respect to ampicillin at 100 μg/ml Supplementary Material: SI.Fig.7.a-b). Again, same concentration of chemically synthesized AgNPs shows only 10 mm of inhibition zone. These results also clearly demonstrate that our biosynthesized AgNPs are more effective than chemically synthesized AgNPs.
4.9. Scanning electron microscopy (SEM)
In order to visualize the interaction of cell wall of E. coli & b-AgNPs and morphological changes of E. coli treated with b-AgNPs, the scanning electron microscopic analysis has been performed. We have not observed any morphological changes of control untreated bacteria (Fig.2.f), bacteria treated with Olax extract (Fig.2.g). However, biosynthesized AgNPs at 30 µM of concentration rupture the E. coli bacterial cell wall by making strong interactions (Shown by blue arrow in Fig.2.h). The results clearly indicate that the b-AgNPs is making direct interactions with bacterial cell wall which accelerates the breakdown of cell wall. Due to cell wall rupturing cellular cytoplasm has been released and consequently b-AgNPs kills the E. coli. Our result corroborates with published literature (38, 40. The probable mechanism for the antibacterial activity of b-AgNPs is schematically presented in Scheme-2 (discussed later).
4.10. Oxidative Stress: In-vitro GSH Oxidation: Possible mechanism for antibacterial effect
GSH prevent from cellular damages caused by oxidative stress 43. To examine the reactive oxygen species (ROS) independent oxidative stress the GSH oxidation has been measured through Ellman's assay 43. GSH can be oxidized to glutathione disulfide (GSSG). Hence the loss of GSH is the measure of glutathione disulfide formation. Accordingly, 1 mM of H2O2 (considered as positive control experiment) induces almost 89.3% of GSH loss. Similarly, bio-synthesized AgNPs induced almost 30% & 44% of GSH loss at the concentrations of 15 µM and 30 µM, respectively (Fig.3.a). This result indicates that either bio-synthesized AgNPs directly converts GSH to GSSG or induces the formation of ROS which converts the GSH to GSSG. Ultimately, the depletion of GSH is responsible for bacterial cell damage.
4.11. Catalase enzyme levels in bacteria
Catalase, an enzymatic anti-oxidant in prokaryotic and eukaryotic cells, mainly removes hydroxyl radicals from the cells 44. The oxidative stress in the cells reduces the level of antioxidants that indirectly indicates the cytotoxicity in bacteria cell. Hence measurement of catalase enzyme levels is required to understand the mechanism of oxidative stress. The bacterial cell lysate treated with b-AgNPs (30 µM) shows the lower level of catalase (38.20 units/min/mg proteins) compared to control untreated bacterial cell lysate (321.3 units/min/mg proteins). Even it is lower than that of bacterial cell lysate (62 units/min/mg proteins) treated with c-AgNPs (30 µM) (Fig.3.b). The results altogether indicate that biosynthesized AgNPs reduces additional catalase level compared to other experiments may be due to formation of hydroxyl radical or silver ions 45.
4.12. Protein expression profile on SDS-PAGE
According to SDS-PAGE, we have observed the up-regulation of some low molecular weight protein (15-25 kD) in the bacterial cell lysate treated with b-AgNPs (Ln:4) compared to control untreated (Ln:1) , ampicillin (Ln: 2) and c-AgNPs (Ln:3) treated samples (Fig. 3.c). We assume that the up-regulation of proteins may be bacterial heat shock proteins and stress responsive proteins that are expressed in the bacteria during oxidative stress according to the published literature 46.
4.13. Reactive oxygen species and anti-bacterial effect (discussion)
The exact mechanism of anti-bacterial activity using colloidal biosynthesized silver nanoparticles (b-AgNPs) is poorly understood. However, combination of our results and published literature entirely give us an idea about the formation of ROS which is responsible for the cytotoxicity of bacteria cells in presence of b-AgNPs. The bio-synthesized silver nanoparticles may interact with the cell wall of the E. coli which may destabilize the plasma-membrane potential (Fig.2.f-h) and reduced the levels of intracellular adenosine triphosphate (ATP) resulting in bacterial cell death 38, 40. We assume that one, or combination of the following reasons may help to explain the bacterial cell death using b-AgNPs.
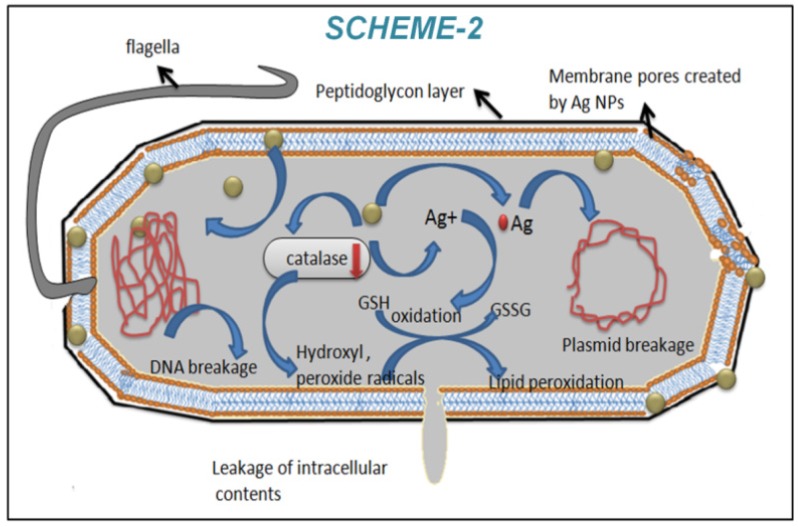
(i) Glutathione contributes the role in cellular protective system from oxidative stress 43. Moreover cellular protein thiol groups are protected by glutathione 47. The internalized bio-synthesized silver nanoparticles inside the cells may generate silver ions that interact with cellular glutathione and oxidize it. This oxidized glutathione results in formation of excess ROS that may help bacterial cellular toxicity and consequently inhibition of bacterial growth. (ii) Secondly, glutathione oxidation by silver ions may increase the lipid peroxidation in cellular membrane that may cause leakage of cellular constituents through membrane damage 48. (iii) Thirdly, silver nanoparticles may interact with sulphur and phosphorous groups of the DNA due to their soft acidic nature 49, 50. This type of interaction may help in the damage of chromosomal DNA and plasmid DNA 49, 50, (iv) we have observed the lower level of catalase (anti-oxidant enzyme) in bacterial cell lysate when bacteria cells were treated with b-AgNPs. Lower level of catalase is responsible for oxidative stress, which help in the up regulation of several stress proteins (presented in Fig.3.c) including heat shock proteins. Results together support stress related antibacterial mechanisms in presence of b-AgNPs. In order to know the exact mechanism for silver nanoparticles induced cellular toxicity, rigorous research needs to be done systematically. Most recently, Zhang et al demonstrated that ROS generation using magnetic nanoparticles is responsible for both anti-bacterial and anti-cancer activity 51. The probable mechanism of antibacterial effect of b-AgNPs is shown in the Scheme 2 (Figure B).
Figure B.
(Scheme 2) Schematic presentation of antibacterial activity of b-AgNPs towards E. coli using biosynthesized silver nanoparticles.
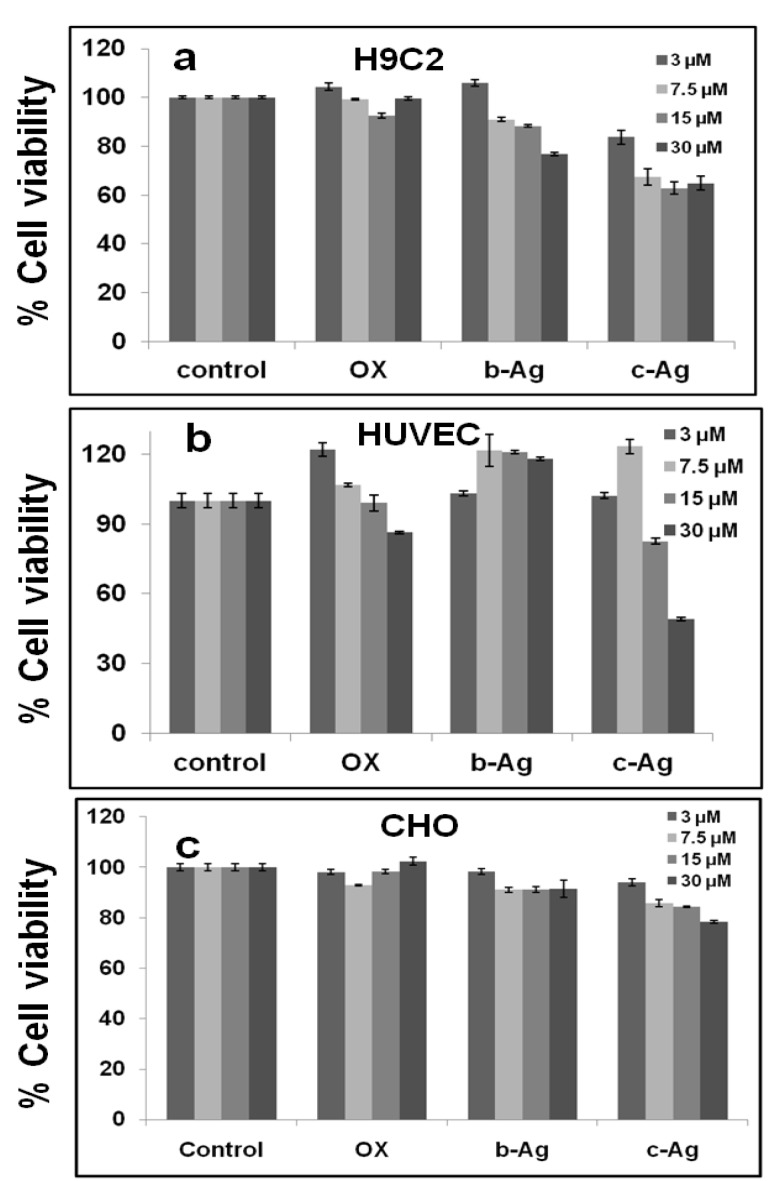
4.14. Biocompatibility and cytotoxicity study
Recently, nanoparticles alone and nanoparticles based drug delivery systems have been extensively used for the diagnostic and therapeutic purpose in biomedical technology. Hence, safety is the main concern before applying the nanomaterials to in vivo system especially animal model. Mild toxicity of nanomaterials can ultimately limit the use of nanomaterials for therapeutic efficacy. Toxicity of a nanomaterial is highly dependent of its physical, chemical and structural properties. Therefore, in vitro biocompatibility study of b-AgNPs has been evaluated to different normal cells (HUVEC: human umbilical vein endothelial cells, H9C2: rat cardiomyoblast cell line & CHO: Chinese hamster overy cells) using MTT assay. Initially, HUVEC, CHO and H9C2 cells are incubated with both biosynthesized silver nanoparticles (b-AgNPs) and chemically synthesized nanoparticles (c-AgNPs) in a dose dependent manner (3-30 µM) (Fig.4.a-b). Surprisingly, we have observed that b-AgNPs are not toxic to normal cells up to 15 µM compared to c-AgNPs. However, it shows mild cytotoxicity towards H9C2 cells at high concentration (30 µM). But HUVEC & CHO cells are not toxic to b-AgNPs even at 30µM concentration (Fig.4.b). Biocompatibility of H9C2 cells towards b-AgNPs has been further verified by microscopic images (using inverted microscope) of H9C2 cells treated with Olax extract and b-AgNPs Supplementary Material: SI.fig.8.a-d). On the other hand, c-AgNPs shows cytotoxic effect to normal cells even at low doses may be due to the anti-angiogenic properties of silver nanoparticles (13Supplementary Material: SI.Fig.7.a-d).
Figure 4.
Cell viability assay using MTT reagents. In vitro anti proliferative assay by MTT reagent in (a) H9C2, (b) HUVEC & (c) CHO. The numerical value represents the concentrations in µM for respective samples. Biosynthesized silver nanoparticles, biocompatible towards H9C2, HUVEC & CHO. *All values are significant, i.e. P < 0.05.
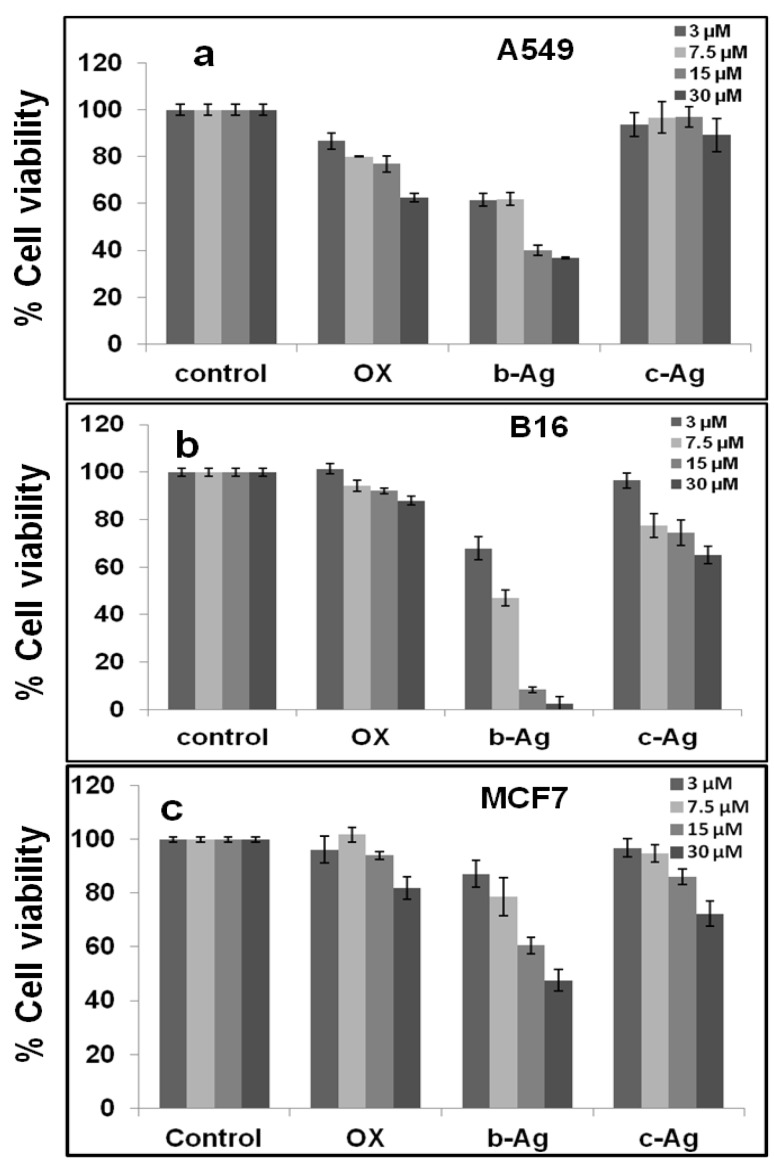
Similarly, we have performed the MTT assays to different cancer cells (A549: human lung cancer cell lines, B16: mouse melanoma cell line & MCF7: human breast cancer cell lines) in a dose dependent manner (3 µM-30 µM) where we have found reverse effect of bio-synthesized silver nanoparticles compared to chemically synthesized silver nanoparticles (Fig.5.a-c). Biosynthesized silver nanoparticles shows significant inhibition of proliferation to both cancer cells (A549 & B16) in a dose dependent manner compared to chemically synthesized AgNPs. For example, 40-70% inhibition of A549 cell proliferation is observed with increasing the concentration of b-AgNPs from 3 µM-30 µM. Similarly, 35-90% inhibition of B16 cell proliferation and 25-55% inhibition of MCF7 cell proliferation are observed with increasing the concentration of b-AgNPs from 3 µM-30 µM. However, there is almost none inhibition of cell proliferation is observed when A549 cells are incubated with c-AgNPs in dose dependent manner. However, c-AgNPs shows mild cytotoxicity towards B16 cells and MCF7 cells. The results altogether demonstrate that b-AgNPs are biocompatible towards normal cells whereas it highly toxic towards cancer cells indicating future use of these b-AgNPs as drug delivery vehicle as well as anti-cancer agent.
Figure 5.
Cell viability assay using MTT reagents. In vitro anti proliferative assay by MTT reagent in (a) A549, (d) B16 and (c) MCF7 cell lines for 24 hours in dose dependent manner. Biosynthesized silver nanoparticles (b-AgNPs) and chemically synthesized AgNPs (c-AgNPs) have been given as treatment as different doses. The numerical value represents the concentrations in µM for respective samples. Biosynthesized silver nanoparticles show anti-cancer activity towards A549 and B16 cancer cell lines. *All values are significant, i.e. P < 0.05.
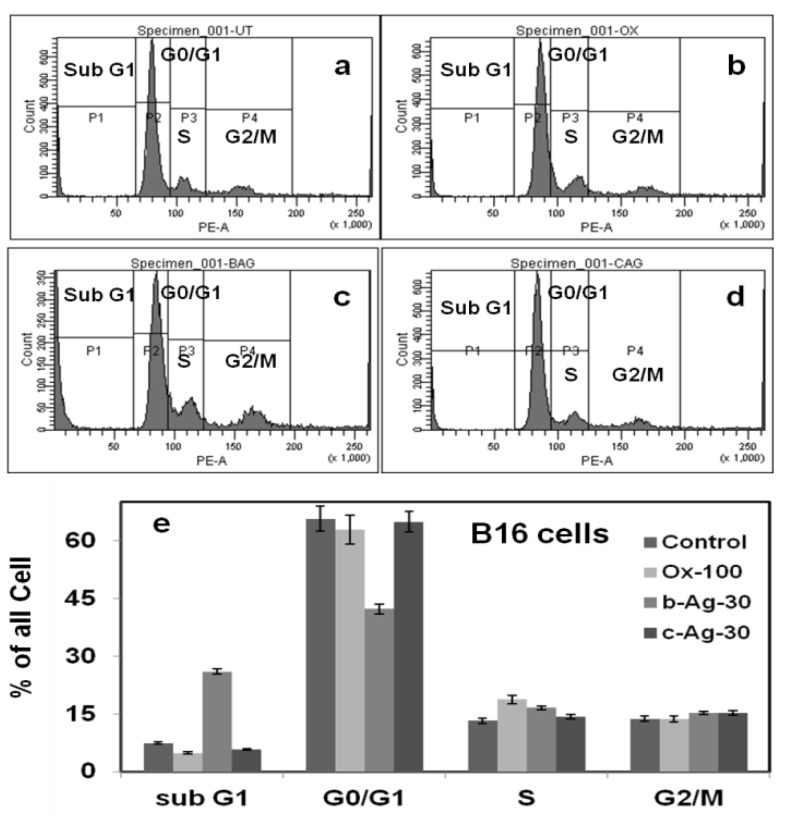
4.15. Cell cycle assay using Fluorescence activated cell sorting (FACS)
Cell cycle assay of B16 melanoma cancer cells treated with Olax extract, b-Ag-30 (30 µM) and c-Ag-30 (30 µM) has been carried out using propidium iodide staining. The extent of staining of the gated population is presented as sub-G1, G0/G1, S, and G2/M in Fig.6.a-e. Olax extract treated cells show almost no change in the cell populations except slight increase the population of S-phase. Also there is no change of cell population in G0/G1, S and G2/M phase when cells were treated with c-Ag-30. However, interestingly, we have observed that there is a significant increase of sub-G1 phase (~ 20%) and consequently decrease of G0/G1 phase cell population. This results indicate that b-Ag-30 treated cancer cells are gone through the sub-G1 phase arrest which may be one of the reasons of induction of apoptosis according to published reports. Beach et al demonstrated that DEX treated cancer cells shows accumulation of cells with sub-G1 DNA content leads to the induction of apoptosis (52. On the other hand, Mao et al, showed that the cell population increase in the sub-G1 phase is connected with the activation of apoptotic protease caspase-3 which is inducing the apoptosis of cells 53. Similarly, Chang et al, also showed that the curcumin treated cancer cells are gone through the sub-G1 phase arrest which is directly related to the apoptotic effect of curcumin towards the cancer cells 54. According to published reports, we assume that killing of B16 cancer cells treated with biosynthesized silver (b-Ag-30) may be due to the increase of sub-G1 phase population that directly correlates with the induction of apoptosis. The exact mechanism for the anti-cancer activity of b-AgNPs needs further investigation which is beyond the scope of our present study.
Figure 6.
Cell cycle assay using PI-RNase reagents in B16 cells. Extent of propidium iodide staining of the gated population was displayed in a histogram and the regions are defined as: sub-G1, G0/G1, S, and G2/M. Result shows the sub-G1 cell accumulation for b-Ag-30 treated B16 melanoma cancer cells indicates the apoptotic cell arrests or death.
4.16. Reactive oxygen species (ROS) and anti-cancer activity
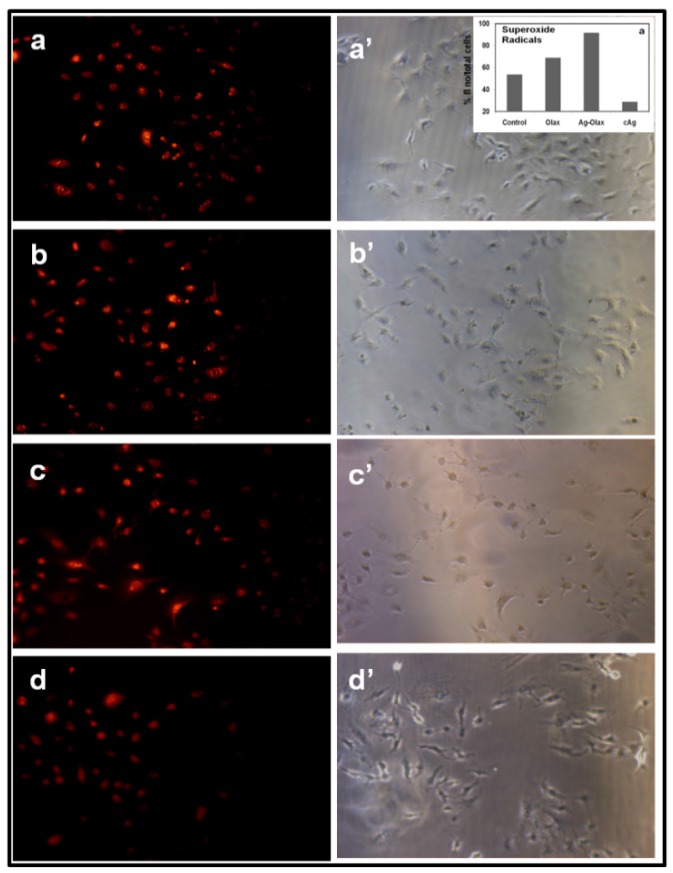
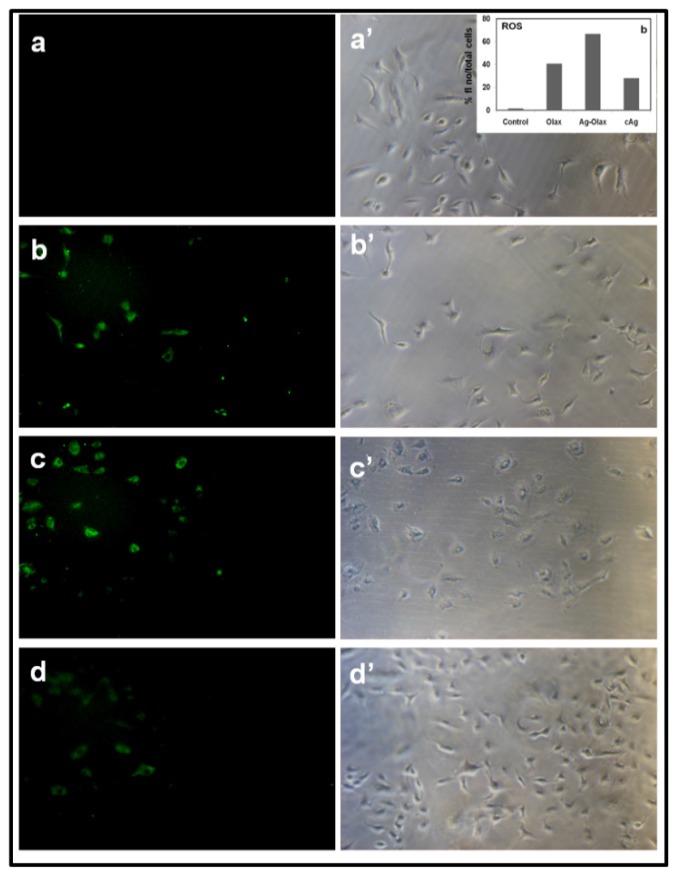
Recent reports suggest that formation of reactive oxygen species (ROS) are involved in anticancer activities 55-57. Hence, anti-cancer activity of bio-synthesized AgNPs towards A549 and B16 cells may be attributed due to the formation of ROS especially super oxide anion (O•-) and hydrogen peroxide (H2O2). In this context, 2',7'-dichlorofluorescin diacetate (DCFH-DA) and dihydroethidium (DHE), redox sensitive probes have been used in cell culture experiments to evaluate reactive oxygen species (ROS) production. In presence of H2O2, DCFH is oxidized to the fluorescent compound 2',7'-dichlorofluorescin (DCF). Similarly, DHE (a blue fluorescent in cell cytoplasm) on oxidation with superoxide anion turns red fluorescent product (oxyethidium) which intercalates with DNA.
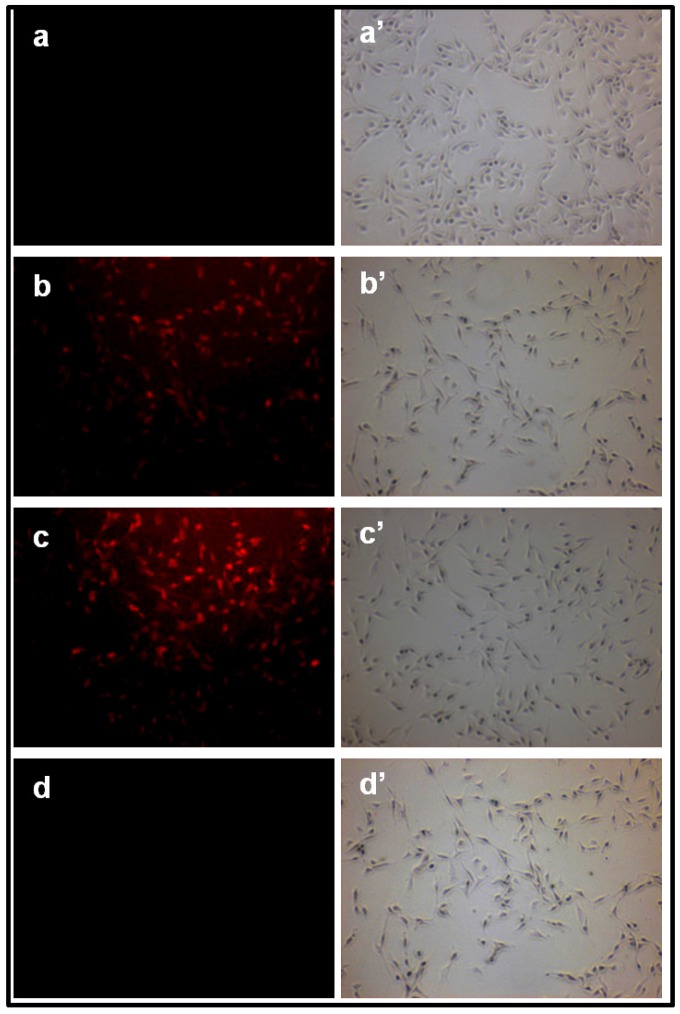
The Fig.7. (a-d; a'-d') indicates the fluorescence images of (a) untreated A549 cells; (b-d) cells treated with Olax extract, b-AgNPs and c-AgNPs, respectively and a'-d' indicate their corresponding phase images. The control sample, untreated A549, show much weaker red fluorescence because of the formation of endogenous superoxide anion (Fig.7.a-a'). The bright red fluorescence as seen in Figure 7.b-d indicates that the production of superoxide anion inside the A549 cells treated with Olax extract, b-AgNPs and c-AgNPs, respectively. The maximum red fluorescence intensity (inset Figure 7.a') is observed inside the A549 cells while treated with biosynthesized silver nanoparticles indicates the formation of maximum superoxide anion.
Figure 7.
Formation of superoxide anion inside A549 cancer cell measured by fluorescence microscopy: Fluorescence images of A549 cells treated with (a) nothing; negative control, (b) Olax extract (100 µg/ ml), (c) b-AgNPs at 30 µM and (d) c-AgNPs at 30 µM, respectively. Images of a'-d' correspond to bright-filed phase images. Biosynthesized AgNPs support the formation of super oxide anion inside the A549 cells. Insets of figure.5.a'. shows the quantitative analysis of % (total number of fluorescence/ cell numbers).
Similarly, the Fig.8. (a-d; a'-d') indicates the green fluorescence images of (a) untreated A549 cells; (b-d) cells treated with Olax extract, b-AgNPs and c-AgNPs, respectively and a'-d' indicate their corresponding phase images. The control sample, untreated A549, show much weaker green fluorescence because of the formation of endogenous hydrogen peroxide (Fig.8.a-a'). The bright green fluorescence as seen in Figure 8.b-d indicates that the production of hydrogen peroxide inside the A549 cells treated with Olax extract, b-AgNPs and c-AgNPs, respectively. The maximum green fluorescence intensity (inset Figure 8.a') is observed inside the A549 cells while treated with biosynthesized silver nanoparticles indicates the formation of maximum hydrogen peroxide. Our results altogether (Fig.7 & Fig.8) support that formation of both hydrogen peroxide and superoxide anion inside the cancer cells in presence of b-AgNPs. Numerous reports suggest the anti-tumoral activity of colloidal silver and gold nanoparticles to cancer cells is observed due to the formation of ROS inside the cells 51, 58, 59. Therefore, we hypothesized that biosynthesized colloidal silver nanoparticles (b-AgNPs) induce for the formation of ROS, which is ultimately responsible for cellular damage. Our results also support the published reports.
Figure 8.
Formation of hydrogen peroxide inside A549 cancer cell measured by fluorescence microscopy: Fluorescence images of A549 cells treated with (a) nothing; negative control, (b) Olax extract (100 µg/ ml), (c) b-AgNPs at 30 µM and (d) c-AgNPs at 30 µM, respectively. Images of a'-d' correspond to bright-filed phase images. Biosynthesized AgNPs support the formation of hydrogen peroxide inside the A549 cells. Insets of figure.6.a'. shows the quantitative analysis of % (total number of fluorescence/ cell numbers).
Signal transduction pathways which are involved in activation of apoptosis mechanism are influenced by increased cellular ROS especially superoxide anion and hydrogen peroxide 60. Mitochondrial transmembrane potential and uncoupling of respiration are associated with increased generation of superoxide radicals 61. It is well established that several drugs show anti-cancer activities by generating the superoxide radicals 62. Fig.7 and Fig.8 together indicates the formation of both these ROS in cancer cells. Our results and reported literature together demonstrate that biosynthesized silver nanoparticles may be one of the plausible mechanisms for anti-cancer activity by generating ROS.
Also, similar experiments we have performed in B16 Supplementary Material: SI.Fig.9-10) & CHO cells (SI.Fig.11). Similar kind of result is observed in B16 cancer cells where b-AgNPs treated B16 cells show more amount of H2O2 & SOD signals than untreated or c-AgNPs treated B16 cells. Whereas, in case of CHO cells the result is reversed i.e. c-AgNPs treated CHO normal overy cells have shown more ROS production than b-AgNPs treated CHO cells. It proves that b-AgNPs is biocompatible towards normal cells due to less production of ROS than c-AgNPs.
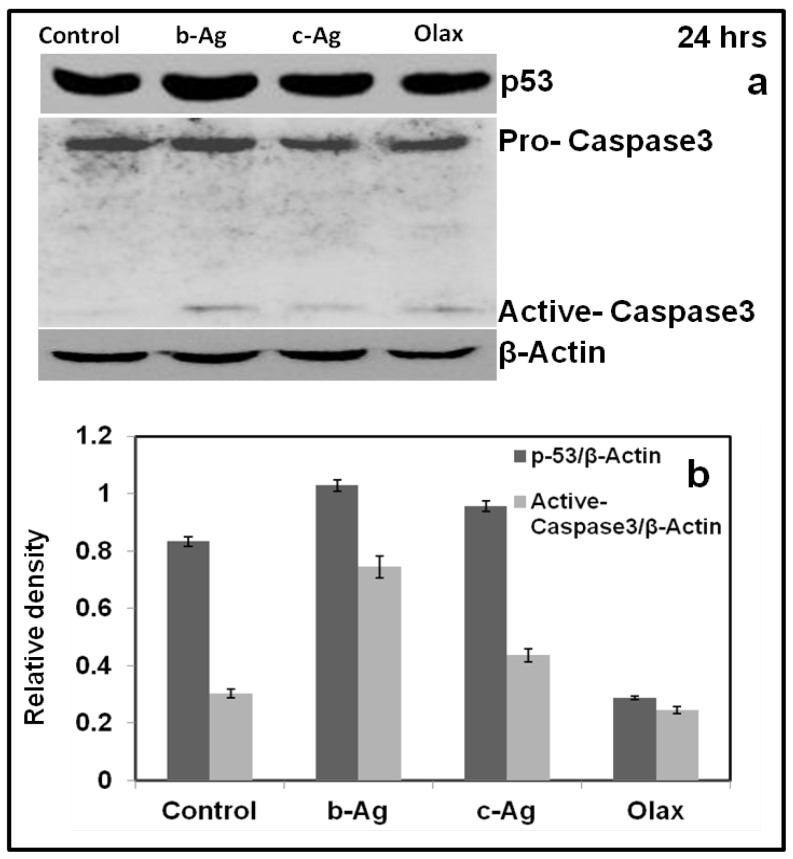
4.17. Immunoblot analysis (Western Blot Analysis & Cancer mechanistic overview
Additionally we have carried out western blot analysis in order to understand the mechanism of anticancer activity of b-AgNPs (fig.9.a-b)(63. Accordingly, our western blot analysis clearly indicates the upregulation of p53 protein in the lysate of B16 cells treated with b-AgNPs (for 24 h) compared to c-AgNPs and olax extract. Similarly, we have observed the upregulation of cleaved caspase or active caspase-3 expression for B16 cells treated with b-AgNPs than c-AgNPs and olax (Fig.9.a). These results have been further supported by the quantification of band intensity performed using the Image J Software (NIH, Bethesda, MD, USA) where the signal intensity of each of the protein was normalized with the corresponding β-Actin signal (Fig.9.b).
Figure 9.
a-b: Western blot analysis of b-AgNPs treated B16 cells show upregulation of p53 and active Caspase-3; (b) Quantification of signal intensity of each of the protein was normalized with the corresponding β-Actin signal shows increase in p53 & active caspase-3.
It is well established that silver nanoparticles induce the oxidative stress in the cancer cells that helps in the induction of cancer cell death through the activation of apoptosis pathwayvia the triggering the p53 upregulation. 64-66 We have investigated the formation of ROS in A549 & B16 cell lines while cancer cell was treated with b-AgNPs. Gurunathan et al, demonstrated the generation of silver ions from AgNPs, the main culprit for the formation oxidative stress that activates the caspase-3. Therefore we hypothesized that b-AgNPs can affect in the mitochondrial respiratory chain, which directly correlates with production of ATP, finally leads to DNA damage 67. Finally we assumed that acidic tumor environment helped to release of anticancer phytoconstituents from biosynthesized silver nanoconjugates that helps further increase in anticancer activity of b-AgNPs. However, it was not happened in normal cell lines and therefore b-AgNPs are biocompatible to normal cell lines. The detailed mechanistic study is beyond the scope of our present communication, however it is progress in our laboratory.
4.18. Cancer cell specific cytoxicty: pH dependent release of Silver ions (AAS & UV)
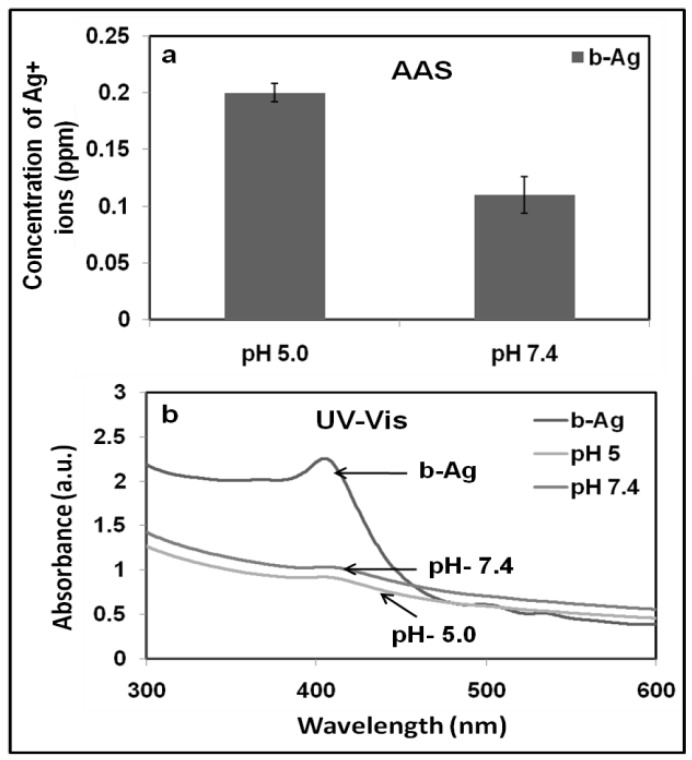
According to published literature, one of the reasons for the death of cancer cells using silver nanoparticles is the release of silver ions from nanoparticles. Hence death of cells is concentration dependent of silver ions and it varies from normal to cancer cells 68. Again, release of silver ions from silver nanoparticles is pH dependent (lower pH, higher release of ions). It is well established that pH of tumor cells are slightly acidic than normal cells 69. Therefore, we hypothesized release of silver ions from AgNPs in tumor cells are more than that of normal cells and consequently it kills more cancer cells. Accordingly we have observed the more killing of cancer cells compared to normal cells while cells were treated with biosynthesized silver nanoparticles. In order to proof our hypothesis, in a typical experiment, a certain amount of b-AgNPs was added to acidic (pH = 5) and physiological buffer solution (pH ~7.4) and incubates overnight. Finally, we have observed that release of silver ions are higher (around double) in acidic environment compared to neutral buffer observed by atomic absorption spectroscopy (AAS) (quantitative analysis) and UV visible spectroscopy (qualitative analysis) [Fig.10.a-b]. The higher concentration of release silver ions in cancer cells compared to normal cells is responsible for selective cancer cell death. However, we have not observed the similar fashion while c-AgNPs was used for the same experiment (data not shown here).
Figure 10.
a-b: (a) atomic absorption spectroscopy to quanitify the amount of silver ion released in acidic and physiological pH, (b) UV-Visible studies show that the comparitive UV spectrum of b-AgNPs in acidic and physiological pH solution.
Again, we have already observed the formation of H2O2 (Fig.8) and superoxide ions (Fig.7) in cancer cells (Fig.7-8, SI-Fig. 9-10) but not in normal cells Supplementary Material: SI-Fig. 11) in presence of b-AgNPs. These results suggest thatin acidic tumor environment biosynthesized silver nanoparticles release more silver ions that facilitate the formation of ROS which ultimately kills the cancer cells. According to Asharani et al. b-AgNPs initially generate ROS that helps to produce silver ions from b-AgNPs through the oxidative dissolution process in acidic pH (= 5.0 -6.4):
| 2Ag + H2O2 + 2H+ → 2Ag+ + 2H2O, E0 = 0.17 V |
The initial formation of ROS thatgenerates of silver ions may be another reason to toxicity for selective cancer cells.
The detailed experimental procedure for AAS & UV has been included in Supplementary Material.
Multifunctional activities of our nanoparticles have been compared with published literature (using chemical and biological approach) and commercially available nanoparticles with different sizes (Table 3. The comparison indicates that b-AgNPs show excellent anti-bacterial and anti-cancer activity compared to published and commercially available Ag nanoparticles.
Table 3.
Comparative studies between the antibacterial and anticancer activity of b-AgNPs synthesized by Olax scandens leaf extract with published & commercially available silver nanoparticles.
| Synthesis of silver nanoparticles (AgNPs) | Shape with size (nm) | Bacterial activity (E. coli) (MIC, MBC in µg/mL & ZOI in mm) | Cellular toxicity IC50 (µg/mL) |
|---|---|---|---|
| Bacterium, B. Licheniformis13 | Spherical; (2-100) |
- | 500.0 nM, Antiangiogenic toxicity in Bovine retinal endothelial cells (BRECs) |
| Olax scandens leaf extract (Green chemistry methods)* | Spherical; (5-60) |
MIC & MBC: 5.0 ZOI: 20 |
A549: 3.5, MCF7: 5.0 and B16: 2.5 |
| Na-hydrazine and Citrate70 | Spherical; (5-50) |
MIC = 14-29 | - |
| SDS capped71 | Spherical; (45 ± 10) |
MBC = 5.0 ZOI = 7.3 |
- |
| Citrullus colocynthis leaves72 | Spherical; (31 |
- | Human epidermoid larynx carcinoma (HEp-2) cell line: 60.0 |
| Chemical reducing agents (Nanotechnologies, Inc)73 | Spherical; (15-21) | MBC = 75.0 | - |
| Inert gas condensation (IGC) process42 | Spherical; (16) | MBC = 60.0 | - |
| NaBH474 | Spherical; (10-20) | ZOI = 2.0-3.5 | - |
| D-glucose and hydrazine38 | Spherical; (10-15) | MIC, MBC = 25.0 | - |
| Syzygium cumini fruit extract75 | Spherical; (10-15) | - | Dalton lymphoma cells:100.0 |
| Chitosan76 | Spherical; (10-12) | MIC, MBC = 10.0 | HepG2: 6.09, Lu: 5.20, MCF-7: 5.71, KB: 3.74 |
| (Shanghai Huzheng Nanotechnology Company limited)40 | Spherical; (2-5) | MIC = 10.0 | Not available |
* Present article, MIC (Minimum Inhibitory concentration), MBC (Minimum bactericidal concentration) & ZOI (Zone of Inhibition).
4.19. Fluorescence Imaging (Diagnostic approach)
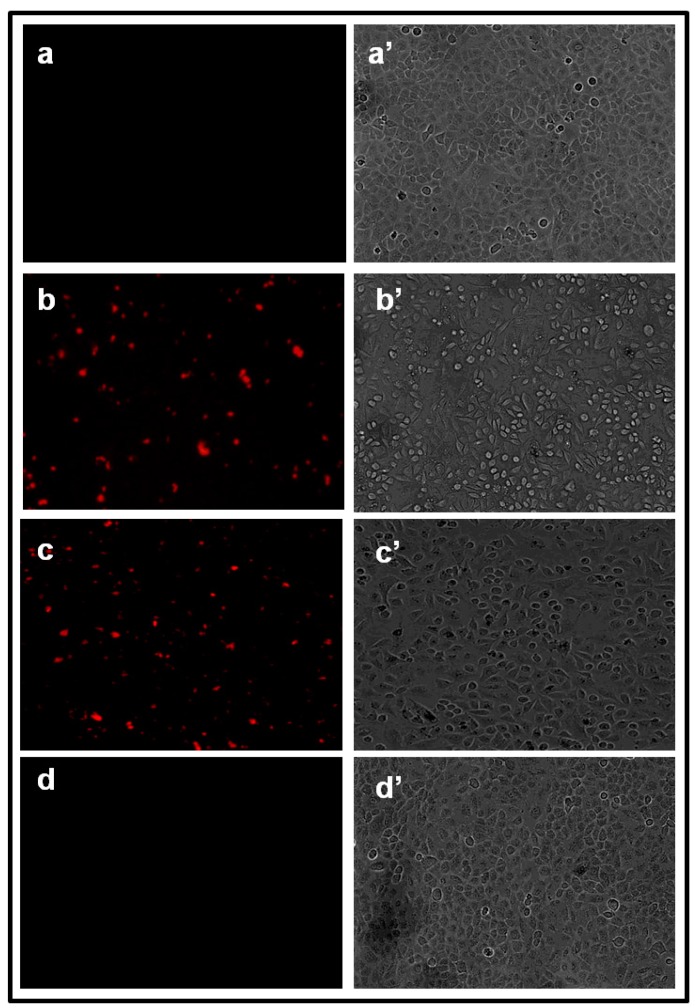
We have already demonstrated the presence of some phytochemicals which exhibit strong fluorescence properties in Olax scandens methanolic leaf extract 22. During the synthesis of AgNPs, these fluorescent molecules can attach with those nanoparticles that enter the cell when cells are incubated with these b-AgNPs. The biosynthesized silver nanoconjugates (b-AgNPs) could be used as diagnostic tool in cancer disease due to presence of fluorescence properties. In order to observe the fluorescence image of b-AgNPs, we have taken the fluorescence image of A549 & B16 cells treated with Olax and b-AgNPs using a fluorescence microscopy. The cells were extensively washed with DPBS before taking the fluorescence image. The control untreated A549 & B16 cells (Fig.11.a & Fig.12.a) and A549 & B16 cells treated with c-AgNPs do not show any red fluorescence (Fig.11.d & Fig.12.d), which indicates the absence of fluorescent molecules. The red fluorescence is observed inside the A549 & B16 cells when the cells are incubated with Olax extract indicating the presence of fluorescent molecules in Olax extract (Fig.11.b & Fig.12.b). However, very strong red fluorescence is observed inside the A549 & B16 cells that confirms the internalization of AgNPs inside A549 cells & B16 cells (Fig.11.c & Fig.12.c). We have taken the fluorescence images of cancer and normal cells treated with different concentration of nanoparticles (10 - 75 µM) and we have observed the fluorescence image of the cells treated with low concentration of nanoparticles (30 µM) for 16-20 h of incubation where cell death is almost negligible. [Fig. 11-12 & Supplementary Material: SI.Fig. 12] Fig.11.a'-d' & Fig.12.a'-d' are the corresponding phase images of untreated & treated cells respectively.
Figure 11.
Fluorescence and the corresponding phase images of untreated A549 cells and cells treated with Olax, b-AgNPs c-AgNPs, observed by an Olympus Fluorescence Microscope. Fluoresence images of A549 cells treated with (a) untreated or control, (b) Olax (100 µg/ ml) leaf extract ,(c) b-AgNPs (at 30 µM) and (d) c-AgNPs (at 30 µM). Images of a', b', c' & d' correspond to phase images. All the treated A549 cells were extensively washed with DPBS (6 times) before taking the fluorescence images. It is to be noted that there is no significant cell killing is observed at 30 µM.
Figure 12.
Fluorescence and the corresponding phase images of untreated B16 cells and cells treated with Olax, b-AgNPs c-AgNPs, observed by an Olympus Fluorescence Microscope. Fluoresence images of B16 cells treated with (a) untreated or control, (b) Olax (100 µg/ ml) leaf extract ,(c) b-AgNPs (at 30 µM) and (d) c-AgNPs (at 30 µM). Images of a', b', c' & d' correspond to phase images. All the treated B16 cells were extensively washed with DPBS (6 times) before taking the fluorescence images. It is to be noted that there is no significant cell killing is observed at 30 µM.
Additionally, we have carried out similar experiments in CHO cells and observed similar results. Supplementary Material: Fig.S12)
4.20. Mechanism of biosynthesized AgNPs
The exact mechanism for the formation of silver nanoparticles using plant extract is still under investigation. However, according to our published literature and other reports published by other groups we have hypothesized that the phytochemicals like proteins, polyphenols present (octacosanol, β-sitosterol, the glucosides of β -sitosterol), in the Olax scandens leaf extract are responsible for the synthesis of silver nanoparticles 22, 77. The reduction process of AgNO3 for the formation of silver nanoparticle might go through a two-step redox or free radical process.
| AgI + R´-OH → Ag0 + R´= O | (2) |
| AgNO3 + 3NR3 → Ag0 + 3NR3+. | (3) |
As the redox potential of alcohol/ aldehyde system (E0RCO/RCHO = 1.80 V) is higher than that of AgI/Ag0 system (E0Ag+/Ag0 = 0.80 V), reduction of AgNO3 by polyphenols or other alcoholic compounds is favorable. Again, free radical mechanism using the different kinds of amines coming from proteins present in methanolic extract of Olax scandens is another pathway for the synthesis of silver nanoparticles.
5. CONCLUSIONS
We have developed a simple green chemistry approach for the synthesis of silver nanoparticle (b-AgNPs) by Olax scandens leaf extract that demonstrate the multifunctional activities of bio-synthesized AgNPs (4-in-1 system). Surprisingly, we have observed the biocompatible nature of b-AgNPs towards normal cells and the results indicates the future application as drug delivery vehicle for cancer therapeutics. Again, the nanobioconjugates shows anticancer activity towards different cancer cells that indicates the future application as anti-cancer agent. Apart from that, bio-synthesized silver nanoparticles (b-AgNPs) shows enhanced antibacterial activity compared to chemically synthesize silver nanoparticles (c-AgNPs). Red fluorescence of b-AgNPs inside the cancer cells could be used as diagnostic tool for cancer disease in near future.All results together demonstrate the multifunctional activities of bio synthesized AgNPs (4-in-1 system) towards biomedical applications that could be applied as (i) anti-bacterial & (ii) anti-cancer agent, (iii) drug delivery vehicle, and (iv) imaging facilitator. A plausible mechanistic approach has been investigated for anti-bacterial & anti-cancer activity of b-AgNPs. Formation of reactive oxygen species induced by b-AgNPs is one of the plausible mechanisms for the anti-cancer and anti-bacterial activity. We strongly believe that bio-synthesized AgNPs will open a new direction towards various biomedical applications in near future.
Supplementary Material
Supplementary Figures and Tables.
Acknowledgments
This research was supported by the 'Ramanujan Fellowship grant' (SR/S2/RJN-04/2010; GAP0305), DST-New Delhi to CRP and generous financial support by Council of Scientific and Industrial Research (ADD: CSC0302; SMiLE: CSC0111), Government of India, New Delhi. SM is thankful to CSIR, New Delhi for a Junior Research Fellowship. Publication cost for this article has been defrayed from 'Ramanujan Fellowship grant' (SR/S2/RJN-04/2010). CRP is grateful to Dr. (Ms.) M. Lakshmi Kantam, Director of CSIR-IICT, for her constant help, support and motivation towards nanomedicine research at CSIR-IICT, Hyderabad.
Biographies
 Mr. Sudip Mukherjee earned his M.Sc in Chemistry from Ramakrishna Mission Residential College, Narendrapur, University of Calcutta in 2011. Currently, he is working as CSIR-JRF at Biomaterials Group, LST Division, CSIR-Indian Institute of Chemical Technology, Hyderabad under the guidance of Dr.Chittaranjan Patra. His research is involved in the design & development of advanced nanomaterials for drug delivery in cancer theranostics and angiogenesis.
Mr. Sudip Mukherjee earned his M.Sc in Chemistry from Ramakrishna Mission Residential College, Narendrapur, University of Calcutta in 2011. Currently, he is working as CSIR-JRF at Biomaterials Group, LST Division, CSIR-Indian Institute of Chemical Technology, Hyderabad under the guidance of Dr.Chittaranjan Patra. His research is involved in the design & development of advanced nanomaterials for drug delivery in cancer theranostics and angiogenesis.
 Debabrata Chowdhury is a research scholar presently pursuing his Ph.D degree in Biotechnology from Centre for Chemical Biology, CSIR-IICT, Hyderabad (Registered with JNTU, Hyderabad). He has done his M.Sc in Microbiology from Vidyasagar university, West Bengal. His research interests lie in understanding molecular mechanism and diagnostic markers of cardiac hypertrophy by applying molecular biology techniques coupled with proteomics.
Debabrata Chowdhury is a research scholar presently pursuing his Ph.D degree in Biotechnology from Centre for Chemical Biology, CSIR-IICT, Hyderabad (Registered with JNTU, Hyderabad). He has done his M.Sc in Microbiology from Vidyasagar university, West Bengal. His research interests lie in understanding molecular mechanism and diagnostic markers of cardiac hypertrophy by applying molecular biology techniques coupled with proteomics.
 Mr. Rajesh Kotcherlakota earned his M.Sc biotechnology from D.N.R College, Bhimavaram, Andhra University in 2012. Currently he is working as CSIR-Project Assistant-II under the guidance of Dr.Chittaranjan Patra, at Biomaterials Group, CSIR-Indian Institute of Chemical Technology, Hyderabad, India. His research involves the development of metal nanoparticles based targeted drug delivery systems containing peptide, antibody, DNA for the treatment of cancers.
Mr. Rajesh Kotcherlakota earned his M.Sc biotechnology from D.N.R College, Bhimavaram, Andhra University in 2012. Currently he is working as CSIR-Project Assistant-II under the guidance of Dr.Chittaranjan Patra, at Biomaterials Group, CSIR-Indian Institute of Chemical Technology, Hyderabad, India. His research involves the development of metal nanoparticles based targeted drug delivery systems containing peptide, antibody, DNA for the treatment of cancers.
 Sujata Patra obtained her M.Sc. In Ecology and Environmental Sciences from Sikkim Manipal University, India in 2003. She worked as Research Assistant for almost 7 years in the field of Molecular Biology, primary cell culture & isolation, DNA isolation & nanobiotechnology from various Institutes (Bar-Ilan University, Israel; Mayo Clinic Cancer Center, USA and CSIR-Indian Institute of Chemical Technology, Hyderabad. She has published 14 research articles in high impact International journals as co-authors and submitted one paper as first author.
Sujata Patra obtained her M.Sc. In Ecology and Environmental Sciences from Sikkim Manipal University, India in 2003. She worked as Research Assistant for almost 7 years in the field of Molecular Biology, primary cell culture & isolation, DNA isolation & nanobiotechnology from various Institutes (Bar-Ilan University, Israel; Mayo Clinic Cancer Center, USA and CSIR-Indian Institute of Chemical Technology, Hyderabad. She has published 14 research articles in high impact International journals as co-authors and submitted one paper as first author.
 Vinothkumar B earned his M.Pharm degree in Pharmacology from Mother Theresa Post Graduate and Research Institute of Health Sciences, Pondicherry university, India, and completed his M.Pharm project work at CSIR-IICT-Hyderabad, India where he was involved in design, development of inorganic nanoparticles using green chemistry approach and the study of antibacterial activity of silver nanoparticles under the supervision Dr. Chittaranjan Patra. Currently, he is assistant professor in the Department of Pharmacology, Bharat Institute of Technology, India.
Vinothkumar B earned his M.Pharm degree in Pharmacology from Mother Theresa Post Graduate and Research Institute of Health Sciences, Pondicherry university, India, and completed his M.Pharm project work at CSIR-IICT-Hyderabad, India where he was involved in design, development of inorganic nanoparticles using green chemistry approach and the study of antibacterial activity of silver nanoparticles under the supervision Dr. Chittaranjan Patra. Currently, he is assistant professor in the Department of Pharmacology, Bharat Institute of Technology, India.
 Manika Pal Bhadra is presently heading the Division of Chemical Biology at the Indian Institute Of Chemical Technology Hyderabad India. She has done her Ph.D from University of Calcutta and postdoctoral research from University of Missouri Columbia USA. Her research interest includes understanding the mechanism of RNA interference in heterochromatin formation and cell division as well as finding small molecules and understanding the role in controlling cell division with major emphasis to epigenetics.
Manika Pal Bhadra is presently heading the Division of Chemical Biology at the Indian Institute Of Chemical Technology Hyderabad India. She has done her Ph.D from University of Calcutta and postdoctoral research from University of Missouri Columbia USA. Her research interest includes understanding the mechanism of RNA interference in heterochromatin formation and cell division as well as finding small molecules and understanding the role in controlling cell division with major emphasis to epigenetics.
 Bojja Sreedhar received his M.Phil and Ph. D degrees in Physics from Sri Venkateswara University, Tirupati, Andhra Pradesh (India) in 1990. In 1993 he worked as a post-doctoral fellow in the Department of Chemistry, Ritsuemeikan University, Kyoto (Japan) and between 1994 to 1997 as faculty in Department of Chemistry, Ritsuemeikan University, Kusatsu, Shiga (Japan). He is currently working as Principal Scientist at CSIR-Indian Institute of Chemical Technology, Hyderabad, India and his research interests include, nanomaterials - synthesis, characterization and applications as catalysts for various organic transformations, glass science, and homogeneous catalysis.
Bojja Sreedhar received his M.Phil and Ph. D degrees in Physics from Sri Venkateswara University, Tirupati, Andhra Pradesh (India) in 1990. In 1993 he worked as a post-doctoral fellow in the Department of Chemistry, Ritsuemeikan University, Kyoto (Japan) and between 1994 to 1997 as faculty in Department of Chemistry, Ritsuemeikan University, Kusatsu, Shiga (Japan). He is currently working as Principal Scientist at CSIR-Indian Institute of Chemical Technology, Hyderabad, India and his research interests include, nanomaterials - synthesis, characterization and applications as catalysts for various organic transformations, glass science, and homogeneous catalysis.
 Chitta Ranjan Patra received his B.Sc (1995) and M.Sc (1997) in Chemistry from the University of Burdwan, India. He was awarded the “Dr. Mrigendranath Ghosh Gold Medal” for obtaining first class and first rank in B.Sc Chemistry (Hons) Examination. He completed his Ph.D. in Chemistry in 2003 (as CSIR Fellow: April 1998- March 2003) from National Chemical Laboratory (NCL), Pune. Thereafter, he received his postdoctoral experience (March 2003-October 2004) as 'Nanotechnologist' from Bar-Ilan University, Ramat-Gan, Israel. After that he moved to Mayo Clinic in October 2004 and explored the field of nanobiotechnology at Department of Biochemistry and Molecular Biology, Mayo Clinic College of Medicine, Rochester, MN, USA. Finally, he received his academic rank as Assistant Professor in the same Department. Dr. Patra moved to CSIR-IICT-Hyderabad after receiving prestigious Ramnujan Fellowship, from DST-New Delhi. Currently, Dr. Patra is Scientist at Biomaterials Group, LST Division, CSIR-IICT. He has 61 publications including 54 peer-reviewed research articles in high impact International journals, 4 book chapters, and 3 patents. Dr. Patra serves as repeated invited reviewer of more 20 journals such as Advanced Drug Delivery Review, Biomaterials, ACS Applied Materials & Interfaces, ChemComm, Nanomedicine etc. Dr. Patra is the member of ACS, AACR, AAAS, CRSI, ISCA. He serves as Editorial Board Member of 'Nanoscience and Nanotechnology', 'Gene Therapy and Molecular Biology (GTMB)' and 'Journal of Nanomedicine & Nanotechnology'. Apart from that, he has received 'Young Investigator Award' and Michael A. O'Connor Travel Award' from Mayo Clinic angiogenesis symposium. Dr. Patra's current research involves design and developments of advanced nanoparticles based drug delivery system for cancer diseases and cardiovascular diseases. His research focus includes, nanotechnology, nanomedicine nanobiotechnology, cancer and angiogenesis.
Chitta Ranjan Patra received his B.Sc (1995) and M.Sc (1997) in Chemistry from the University of Burdwan, India. He was awarded the “Dr. Mrigendranath Ghosh Gold Medal” for obtaining first class and first rank in B.Sc Chemistry (Hons) Examination. He completed his Ph.D. in Chemistry in 2003 (as CSIR Fellow: April 1998- March 2003) from National Chemical Laboratory (NCL), Pune. Thereafter, he received his postdoctoral experience (March 2003-October 2004) as 'Nanotechnologist' from Bar-Ilan University, Ramat-Gan, Israel. After that he moved to Mayo Clinic in October 2004 and explored the field of nanobiotechnology at Department of Biochemistry and Molecular Biology, Mayo Clinic College of Medicine, Rochester, MN, USA. Finally, he received his academic rank as Assistant Professor in the same Department. Dr. Patra moved to CSIR-IICT-Hyderabad after receiving prestigious Ramnujan Fellowship, from DST-New Delhi. Currently, Dr. Patra is Scientist at Biomaterials Group, LST Division, CSIR-IICT. He has 61 publications including 54 peer-reviewed research articles in high impact International journals, 4 book chapters, and 3 patents. Dr. Patra serves as repeated invited reviewer of more 20 journals such as Advanced Drug Delivery Review, Biomaterials, ACS Applied Materials & Interfaces, ChemComm, Nanomedicine etc. Dr. Patra is the member of ACS, AACR, AAAS, CRSI, ISCA. He serves as Editorial Board Member of 'Nanoscience and Nanotechnology', 'Gene Therapy and Molecular Biology (GTMB)' and 'Journal of Nanomedicine & Nanotechnology'. Apart from that, he has received 'Young Investigator Award' and Michael A. O'Connor Travel Award' from Mayo Clinic angiogenesis symposium. Dr. Patra's current research involves design and developments of advanced nanoparticles based drug delivery system for cancer diseases and cardiovascular diseases. His research focus includes, nanotechnology, nanomedicine nanobiotechnology, cancer and angiogenesis.
References
- 1.Alivisatos P. The use of nanocrystals in biological detection. Nature Biotechnology. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 2.Salata O. Applications of nanoparticles in biology and medicine. J Nanobiotechnology. 2004;2(1):3.. doi: 10.1186/1477-3155-2-3. Epub 2004/05/04. doi: 10.1186/1477-3155-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nature Biotechnology. 2004;22:969–76. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 4.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Gold Nanoparticles for Biology and Medicine. Angew Chem Int Ed. 2010;49:3280–94. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patra CR, Bhattacharya R, Patra S, Vlahakis NE, Gabashvili A, Koltypin Y. et al. Pro-angiogenic properties of europium(III) hydroxide nanorods. Advanced Materials. 2008;20(4):753–6. [Google Scholar]
- 6.Patra CR, Bhattacharya R, Wang E, Katarya A, Lau JS, Dutta S. et al. Targeted delivery of gemcitabine to pancreatic adenocarcinoma using cetuximab as a targeting agent. Cancer Res. 2008;68(6):1970–8. doi: 10.1158/0008-5472.CAN-07-6102. [DOI] [PubMed] [Google Scholar]
- 7.Patra CR, Kim J-H, Pramanik K, d'Uscio LV, Patra S, Katusic ZS. et al. Reactive Oxygen Species Driven Angiogenesis by Inorganic Nanorods. Nano Letters. 2011;11:4932–38. doi: 10.1021/nl2028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barui AK, Veeriah V, Mukherjee S, Manna J, Patel AK, Patra S. et al. Zinc oxide nanoflowers make new blood vessels. Nanoscale. 2012;4:7861–9. doi: 10.1039/c2nr32369a. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Gu FX, Chan JM, Wang AZ, Langer RS, Farokhzad OC. Nanoparticles in Medicine: Therapeutic Applications and Developments. Clinical Pharmacology & Therapeutics. 2008;83(5):761–9. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 10.Ahamed M, Alsalhi MS, Siddiqui MK. Silver nanoparticle applications and human health. Clin Chim Acta. 2010;411(23-24):1841–8. doi: 10.1016/j.cca.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Modak A, Barui AK, Ranjan Patra C, Bhaumik A. A luminescent nanoporous hybrid material based drug delivery system showing excellent theranostics potential for cancer. Chem Commun (Camb) 2013;49(69):7644–6. doi: 10.1039/c3cc43487g. [DOI] [PubMed] [Google Scholar]
- 12.Sironmani A, Daniel K. Silver Nanoparticles - Universal Multifunctional Nanoparticles for Bio Sensing, Imaging for Diagnostics and Targeted Drug Delivery for Therapeutic Applications. Kapetanovic I, editor. Europe and China: In Tech. 2011;463:pp.1–27. [Google Scholar]
- 13.Gurunathan S, Lee KJ, Kalishwaralal K, Sheikpranbabu S, Vaidyanathan R, Eom SH. Antiangiogenic properties of silver nanoparticles. Biomaterials. 2009;30(31):6341–50. doi: 10.1016/j.biomaterials.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Prucek R, Tuček J, Kilianová M, Panáček A, Kvítek L, Filip J. et al. The targeted antibacterial and antifungal properties of magnetic nanocomposite of iron oxide and silver nanoparticles. Biomaterials. 2011;32(21):4704–13. doi: 10.1016/j.biomaterials.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 15.Lee K-S, El-Sayed MA. Gold and Silver Nanoparticles in Sensing and Imaging: Sensitivity of Plasmon Response to Size, Shape, and Metal Composition. J Phys Chem B. 2006;110:19220–5. doi: 10.1021/jp062536y. [DOI] [PubMed] [Google Scholar]
- 16.Im AR, Kim JY, Kim HS, Cho S, Park Y, Kim YS. Wound healing and antibacterial activities of chondroitin sulfate- and acharan sulfate-reduced silver nanoparticles. Nanotechnology. 2013;24(39):395102. doi: 10.1088/0957-4484/24/39/395102. [DOI] [PubMed] [Google Scholar]
- 17.Centi G. "Greening Chemistry"--in Turin and the world. ChemSusChem. 2008;1(8-9):663. doi: 10.1002/cssc.200800149. [DOI] [PubMed] [Google Scholar]
- 18.Kou J, Varma RS. Beet juice-induced green fabrication of plasmonic AgCl/Ag nanoparticles. ChemSusChem. 2012;5(12):2435–41. doi: 10.1002/cssc.201200477. [DOI] [PubMed] [Google Scholar]
- 19.Shukla R, Nune KS, Chanda N, Katti K, Mekapothula S, Kulkarni RR. et al. Soybeans as a Phytochemical Reservoir for the Production and Stabilization of Biocompatible Gold Nanoparticles. Small. 2008;4(9):1425–36. doi: 10.1002/smll.200800525. [DOI] [PubMed] [Google Scholar]
- 20.Shankar SS, Rai A, Ahmad A, Sastry M. Controlling the Optical Properties of Lemongrass Extract Synthesized Gold Nanotriangles and Potential Application in Infrared-Absorbing Optical Coatings. Chem Mater. 2005;17:566–72. [Google Scholar]
- 21.Xie J, Lee YJ, Wang ICD, Ting PY. Identification of Active Biomolecules in the High-Yield Synthesis of Single-Crystalline Gold Nanoplates in Algal Solutions. Small. 2007;3(4):672– 82. doi: 10.1002/smll.200600612. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee S, Sushma V, Patra S, Barui AK, Pal Bhadra M, Sreedhar B, Patra CR. Green chemistry approach for the synthesis and stabilization of biocompatible gold nanoparticles and their potential applications in cancer therapy. Nanotechnology. 2012;23(45):455103. doi: 10.1088/0957-4484/23/45/455103. [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee S, Vinothkumar B, Prashanthi S, Bangal PR, Sreedhar B, Patra CR. Potential therapeutic and diagnostic applications of one-step in situ biosynthesized gold nanoconjugates (2-in-1 system) in cancer treatment. RSC Adv. 2013;3:2318–29. [Google Scholar]
- 24.Govindaraju K, Basha SK, Kumar VG, Singaravelu G. Silver, gold and bimetallic nanoparticles production using single-cell protein (Spirulina platensis) Geitler. J Mater Sci. 2008;43:5115–122. [Google Scholar]
- 25.Balakrishna K, Natarajan RK, Purushothaman KK. Chemical examination of Olax scandens Roxb. Bmebr. 1983;4:167–69. [Google Scholar]
- 26.Ju YH, Clausen LM, Allred KF, Almada AL, Helferich WG. beta-Sitosterol, beta-Sitosterol Glucoside, and a Mixture of beta-Sitosterol and beta-Sitosterol Glucoside Modulate the Growth of Estrogen-Responsive Breast Cancer Cells In Vitro and in Ovariectomized Athymic Mice. J. Nutr. 2004;134:1145–51. doi: 10.1093/jn/134.5.1145. [DOI] [PubMed] [Google Scholar]
- 27.Awad AB, Chan KC, Downie AC, Fink CS. Peanuts as a source of beta-sitosterol, a sterol with anticancer properties. Nutr Cancer. 2000;36(2):238–41. doi: 10.1207/S15327914NC3602_14. [DOI] [PubMed] [Google Scholar]
- 28.Thippeswamy G, Sheela ML, Salimath BP. Octacosanol isolated from Tinospora cordifolia downregulates VEGF gene expression by inhibiting nuclear translocation of NF-<kappa>B and its DNA binding activity. Eur J Pharmacol. 2008;588(2-3):141–50. doi: 10.1016/j.ejphar.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Duraipandiyan V, Ayyanar M, Ignacimuthu S. Antimicrobial activity of some ethnomedicinal plants used by Paliyar tribe from Tamil Nadu, India. BMC Complementary and Alternative Medicine. 2006;6:35. doi: 10.1186/1472-6882-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeyaprakash K, Ayyanar M, Geetha KN, Sekar T. Traditional uses of medicinal plants among the tribal people in Theni District (Western Ghats), Southern India. Asian Pacific Journal of Tropical Biomedicine. 2011:S20–S25. [Google Scholar]
- 31.Venkata Subbaiah KP, Savithramma N. Bio-prospecting and documentation of traditional medicinal plants used to treat itching, psoriasis and wounds by ethnic groups of kurnool district, Andhra Pradesh, India. Asian J Pharm Clin Res. 2012;5(2):127–31. [Google Scholar]
- 32.Radhakrishnan TM, Sarma PS. Studies on the Intracellular Localization and Incorporation of 59Fe into Catalase in Rat Liver. Biochem J. 1964;93:440–47. doi: 10.1042/bj0930440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural Proteins during the Assembly of the head of Bacteriophage t4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Kvitek L, Panacek A, Prucek R, Soukupova J, Vanickova M, Kolar M. et al. Antibacterial activity and toxicity of silver - nanosilver versus ionic silver. Journal of Physics: Conference Series. 2011;304:012029. [Google Scholar]
- 36.Tang S, Bourne R, Smith R, Poliakoff M. The 24 Principles of Green Engineering and Green Chemistry: “IMPROVEMENTS PRODUCTIVELY”. Green Chem. 2008;10:268–9. [Google Scholar]
- 37.Krpetic Z, Scarı G, Caneva C, Speranza G, Porta F. Gold Nanoparticles Prepared Using Cape Aloe Active Components. Langmuir. 2009;25(13):7217–21. doi: 10.1021/la9009674. [DOI] [PubMed] [Google Scholar]
- 38.Shrivastava S, Bera T, Roy A, Singh G, Ramachandrarao P, Dash D. Characterization of enhanced antibacterial effects of novel silver nanoparticles. Nanotechnology. 2007;18:225103. doi: 10.1088/0957-4484/18/22/225103. [DOI] [PubMed] [Google Scholar]
- 39.Inoue Y, Uota M, Torikai T, Watari T, Noda I, Hotokebuchi T. et al. Antibacterial properties of nanostructured silver titanate thin films formed on a titanium plate. J Biomed Mater Res A. 2010;92(3):1171–80. doi: 10.1002/jbm.a.32456. [DOI] [PubMed] [Google Scholar]
- 40.Li W-R, Xie X-B, Shi Q-S, Zeng H-Y, OU-Yang Y-S, Chen Y-B. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl Microbiol Biotechnol. 2010;85:1115–22. doi: 10.1007/s00253-009-2159-5. [DOI] [PubMed] [Google Scholar]
- 41.Jena P, Mohanty S, Mallick R, Jacob B, Sonawane A. Toxicity and antibacterial assessment of chitosan-coated silver nanoparticles on human pathogens and macrophage cells. International Journal of Nanomedicine. 2012;7:1805–18. doi: 10.2147/IJN.S28077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raffi M, Hussain F, Bhatti T, Akhter J, Hameed A, Hasan M. Antibacterial characterization of silver nanoparticles against E. coli ATCC-15224. J Mater Sci Technol. 2008;24:192–6. [Google Scholar]
- 43.Liu S, Helen Zeng TH, Hofmann M, Burcombe E, Wei J, Jiang R. et al. Antibacterial Activity of Graphite, Graphite Oxide, Graphene Oxide, and Reduced Graphene OXide: Membrane and Oxidative Stress. Acs Nano. 2011;5:6971–80. doi: 10.1021/nn202451x. [DOI] [PubMed] [Google Scholar]
- 44.Gaetani GF, Ferraris AM, Rolfo M, Mangerini R, Arena S, Kirkman HN. Predominant role of catalase in the disposal of hydrogen peroxide within human erythrocytes. Blood. 1996;87(4):1595. [PubMed] [Google Scholar]
- 45.Jung WK, Koo HC, Kim KW, Shin S, Kim SH, Park YH. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl Environ Microbiol. 2008;74(7):2171–8. doi: 10.1128/AEM.02001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kędzierska S, Staniszewska M, Węgrzyn A. The role of DnaK/DnaJ and GroEL/GroES systems in the removal of endogenous proteins aggregated by heat-shock from Escherichia coli cell. FEBS Lett. 1999;446:331–7. doi: 10.1016/s0014-5793(99)00154-4. [DOI] [PubMed] [Google Scholar]
- 47.Masip I, Veeravalli K, Georgiou G. The Many Faces of Glutathione in Bacteria. Antioxidants & Redox Signaling. 2006;8(5-6):753–62. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 48.Piao MJ, Kang KA, Lee IK, Kim HS, Kim S, Choi JY. et al. Silver nanoparticles induce oxidative cell damage in human liver cells through inhibition of reduced glutathione and induction of mitochondria-involved apoptosis. Toxicol Lett. 2011;201(1):92–100. doi: 10.1016/j.toxlet.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 49.Hatchett DW, Henry S. Electrochemistry of sulfur adlayers on low-index faces of silver. J Phys Chem. 1996;100:9854–9. [Google Scholar]
- 50.Prabhu S, Poulose EK. Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. International Nano Letters. 2012;2:32. [Google Scholar]
- 51.Zhang D, Zhao Y-X, Gao Y-J, Gao F-P, Fan Y-S, Li X-J. et al. Anti-bacterial and in vivo tumor treatment by reactive oxygen species generated by magnetic nanoparticles. J Mater Chem B. 2013;1:5100–7. doi: 10.1039/c3tb20907e. [DOI] [PubMed] [Google Scholar]
- 52.Beach JA, Nary LJ, Hirakawa Y, Holland E, Hovanessian R, Medh RD. E4BP4 facilitates glucocorticoid-evoked apoptosis of human leukemic CEM cells via upregulation of Bim. J Mol Signal. 2011;6(1):13. doi: 10.1186/1750-2187-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mao X, Seidlitz E, Truant R, Hitt M, Ghosh HP. Re-expression of TSLC1 in a non-small-cell lung cancer cell line induces apoptosis and inhibits tumor growth. Oncogene. 2004;23(33):5632–42. doi: 10.1038/sj.onc.1207756. [DOI] [PubMed] [Google Scholar]
- 54.Chang YJ, Tai CJ, Kuo LJ, Wei PL, Liang HH, Liu TZ. et al. Glucose-Regulated Protein 78 (GRP78) Mediated the Efficacy to Curcumin Treatment on Hepatocellular Carcinoma. Annals of Surgical Oncology. 2011;18(8):2395–403. doi: 10.1245/s10434-011-1597-3. [DOI] [PubMed] [Google Scholar]
- 55.Wan J, Liu T, Mei L, Li J, Gong K, Yu C. et al. Synergistic antitumour activity of sorafenib in combination with tetrandrine is mediated by reactive oxygen species (ROS)/Akt signaling. British Journal of Cancer. 2013;109:342–350. doi: 10.1038/bjc.2013.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whibley CE, McPhail KL, Keyzers RA, Maritz MF, Leaner VD, Birrer MJ. et al. Reactive oxygen species mediated apoptosis of esophageal cancer cells induced by marine triprenyl toluquinones and toluhydroquinones. Mol Cancer Ther. 2007;6(9):2535–43. doi: 10.1158/1535-7163.MCT-06-0760. [DOI] [PubMed] [Google Scholar]
- 57.Ravid A, Koren R. The role of reactive oxygen species in the anticancer activity of vitamin D. Recent Results Cancer Res. 2003;164:357–67. doi: 10.1007/978-3-642-55580-0_26. [DOI] [PubMed] [Google Scholar]
- 58.Minai L, Yeheskely-Hayon D, Yelin D. High levels of reactive oxygen species in gold nanoparticle-targeted cancer cells following femtosecond pulse irradiation. Sci Rep. 2013;3:2146. doi: 10.1038/srep02146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Franco-Molina MA, Mendoza-Gamboa E, Sierra-Rivera CA, Gomez-Flores RA, Zapata-Benavides P, Castillo-Tello P. et al. Antitumor activity of colloidal silver on MCF-7 human breast cancer cells. Journal of Experimental & Clinical Cancer Research. 2010;29:148. doi: 10.1186/1756-9966-29-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueda S, Masutani H, Nakamura H, Tanaka T, Ueno M, Yodoi J. Redox control of cell death. Antioxid Redox Signal. 2002;4(3):405–14. doi: 10.1089/15230860260196209. [DOI] [PubMed] [Google Scholar]
- 61.Garrido C, Galluzzi L, Brunet M, Puig PE, Didelot C, Kroemer G. Mechanisms of cytochrome c release from mitochondria. Cell Death and Differentiation. 2006;13(9):1423–33. doi: 10.1038/sj.cdd.4401950. [DOI] [PubMed] [Google Scholar]
- 62.Velayutham M, Villamena FA, Fishbein JC, Zweier JL. Cancer chemopreventive oltipraz generates superoxide anion radical. Archives of Biochemistry and Biophysics. 2005;435(1):83–8. doi: 10.1016/j.abb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 63.Chowdhury D, Tangutur AT, Khatua TN, Saxena P, Banerjee SK, Bhadra MP. A proteomic view of isoproterenol induced cardiac hypertrophy: Prohibitin identified as a potential biomarker in rats. Journal of Translational Medicine. 2013;130:1–13. doi: 10.1186/1479-5876-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mei N, Zhang Y, Chen Y, Guo X, Ding W, Ali SF, Biris AS, Rice P, Moore MM, Chen T. Silver nanoparticle-induced mutations and oxidative stress in mouse lymphoma cells. Environment and molecular mutagenesis. 2012;53:409–19. doi: 10.1002/em.21698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gurunathan S, Han JW, Eppakayala V, Jeyaraj M, Kim JH. Cytotoxicity of Biologically Synthesized Silver Nanoparticles in MDA-MB-231 Human Breast Cancer Cells. BioMed Research International. 2013;2013:535796. doi: 10.1155/2013/535796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amaral JD, Xavier JM, Steer CJ, Rodrigues CM. The Role of p53 in Apoptosis. Discov Med. 2010;45:145–52. [PubMed] [Google Scholar]
- 67.Asharani P, Wu Y, Gong Z, Valiyaveettil S. Toxicity of silver nanoparticles in zebrafish models. Nanotechnology. 2008;19:255102. doi: 10.1088/0957-4484/19/25/255102. [DOI] [PubMed] [Google Scholar]
- 68.Marambio-Jones C, Hoek EMV. A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment. J Nanopart Res. 2010;12:1531–51. [Google Scholar]
- 69.Tannock IF, Rotin D. Acid pH in Tumors and Its Potential for Therapeutic Exploitation. Cancer Research. 1989;49:4373–84. [PubMed] [Google Scholar]
- 70.Guzmán MG, Dille J, Godet S. Synthesis of silver nanoparticles by chemical reduction method and their antibacterial activity. World Academy of Science Engineering and Technology. 2008;43:357–64. [Google Scholar]
- 71.Kora AJ, Manjusha R, Arunachalam J. Superior bactericidal activity of SDS capped silver nanoparticles: Synthesis and characterization. Materials Science and Engineering: C. 2009;29:2104–09. [Google Scholar]
- 72.Satyavani K, Gurudeeban S, Ramanathan T, Balasubramanian T. Biomedical potential of silver nanoparticles synthesized from calli cells of Citrullus colocynthis (L.) Schrad. Journal of Nanobiotechnology. 2011;9:43. doi: 10.1186/1477-3155-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ram´ırez JT, Yacaman MJ. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16:2346–53. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 74.Lee SM, Song KC, Lee BS. Antibacterial activity of silver nanoparticles prepared by a chemical reduction method. Korean J. Chem. Eng. 2010;27(2):688–92. [Google Scholar]
- 75.Mittal AK, Bhaumik J, Kumar S, Banerjee UC. Biosynthesis of silver nanoparticles: elucidation of prospective mechanism and therapeutic potential. Journal of Colloid and Interface Science. 2014;415:39–47. doi: 10.1016/j.jcis.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 76.Tran HV, Tran LD, Ba CT, Vu HD, Nguyen TN, Pham DG, Nguyen PX. Synthesis, characterization, antibacterial and antiproliferative activities of monodisperse chitosan-based silver nanoparticles. Colloids and Surfaces A: Physicochemical and Engineering Aspects. 2010;360:32–40. [Google Scholar]
- 77.Newman SDJ, Blanchard JG. Formation of Gold Nanoparticles Using Amine Reducing Agents. Langmuir. 2006;22:5882–7. doi: 10.1021/la060045z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Tables.