The validation of breast cancer risk models is important, and that by MacInnis et al (2013) of the BOADICEA model, which is based solely on family history, is very welcome. A recent development has been the identification of 67 breast cancer risk SNPs (Michailidou et al, 2013), whose main use will be together as a panel to identify women at increased risk of breast cancer. We investigated how a polygenic SNP score based on these SNPs would compare with classical risk factors including family history, and how much information it might add to risk assessment.
Our analysis was based on simulated SNP scores from 100 000 women with population allele frequencies for the 67 SNPs, and treating them as independent so that a combined risk score can be obtained by multiplying their relative risks; and the Tyrer-Cuzick (TC) risk model (Tyrer et al, 2004) predictions from the first 10 000 women enrolled to the PROCAS study (predicting risk of breast cancer at screening) in Manchester, UK (Evans et al, 2012). The TC model is based on classical phenotypic factors including age, family history, age at menopause and menarche, and parity. The outcome measure was the 10-year relative risk of developing breast cancer.
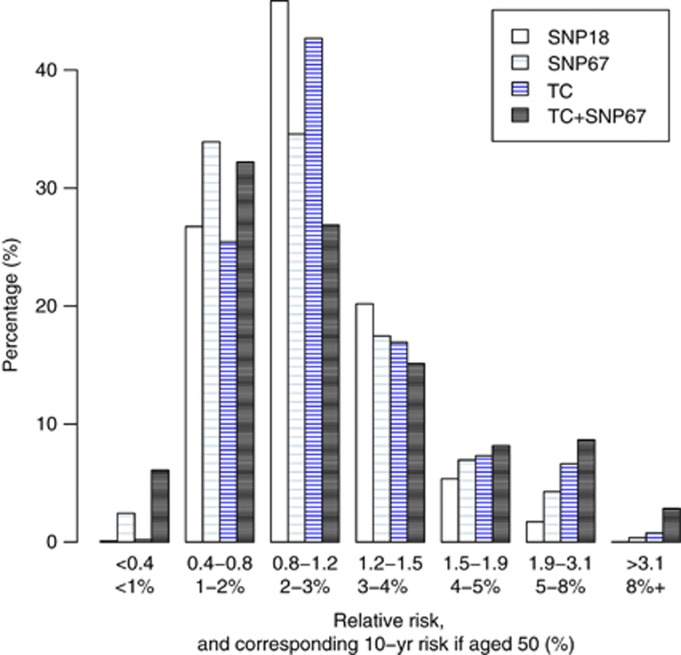
Histograms are shown in Figure 1 for the TC model, the SNP score using 18 genes previously published (Turnbull et al, 2010) but with risks updated from the COGS analysis in Michailidou et al (2013), the full set of 67 SNPs, and a combined TC+SNP67 distribution assuming independence. Initial evaluations have shown the TC and SNP18 scores appear to be independent (Evans et al, 2012).
Figure 1.
Predicted 10-year relative risk and corresponding absolute 10-year risk for a woman aged 50 using classical factors (TC), 18 SNPs from Turnbull et al (2010), the 67 COGS SNPs (SNP67) and combined (TC+SNP67). The phenotypic markers are from 10 000 women of routine screening age (46–70 years) in the UK.
Of particular interest is the >8% 10-year risk group, where NICE (2013) guidelines in the UK advise offering the preventive use of tamoxifen. The SNP score was less able to identify women at high risk than the TC model (0.02% for SNP18, 0.37% for SNP67 and 0.77% for the TC model). However, adding SNP67 to TC gave a substantial increment to 2.85%, and similar effects were seen in the 5–8% 10-year risk group (1.72%, 4.28%, 6.64% and 8.17%, respectively), which is equivalent to the NICE moderate-risk category where tamoxifen may be ‘considered'. It is also noticeable that the SNPs identified more low-risk women than the TC model, which mainly uses uncommon high-risk phenotypes.
These data suggest that although the spread towards high-risk currently achieved by SNP67 is not as large as that obtained from classical phenotypic markers, SNPs may add substantially to classic factors when used together.
References
- Evans DGR, Warwick J, Astley SM, Stavrinos P, Sahin S, Ingham S, McBurney H, Eckersley B, Harvie M, Wilson M, Beetles U, Warren R, Hufton A, Sergeant JC, Newman WG, Buchan I, Cuzick J, Howell A. Assessing individual breast cancer risk within the U.K. National Health Service Breast Screening Program: a new paradigm for cancer prevention. Cancer Prev Res. 2012;5:943–951. doi: 10.1158/1940-6207.CAPR-11-0458. [DOI] [PubMed] [Google Scholar]
- MacInnis RJ, Bickerstaffe A, Apicella C, Dite GS, Dowty JG, Aujard K, Phillips KA, Weideman P, Lee A, Terry MB, Giles GG, Southey MC, Antoniou AC, Hopper JL. Prospective validation of the breast cancer risk prediction model BOADICEA and a batch-mode version BOADICEACentre. Br J Cancer. 2013;109:1296–1301. doi: 10.1038/bjc.2013.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Fletcher O, Peto J, Gibson L, Dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomäki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waisfisz Q, Meijers-Heijboer H, Adank M, van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Müller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LF, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJ, van der Schoot CE, Guénel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JI, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MW, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE, Schumacher F, Le Marchand L, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Lindblom A, Margolin S, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Bui QM, Stone J, Dite GS, Apicella C, Tsimiklis H, Giles GG, Severi G, Baglietto L, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Brenner H, Müller H, Arndt V, Stegmaier C, Swerdlow A, Ashworth A, Orr N, Jones M, Figueroa J, Lissowska J, Brinton L, Goldberg MS, Labrèche F, Dumont M, Winqvist R, Pylkäs K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Brüning T, Radice P, Peterlongo P, Manoukian S, Bonanni B, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Durda K, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Bogdanova NV, Antonenkova NN, Dörk T, Kristensen VN, Anton-Culver H, Slager S, Toland AE, Edge S, Fostira F, Kang D, Yoo KY, Noh DY, Matsuo K, Ito H, Iwata H, Sueta A, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Shu XO, Lu W, Gao YT, Cai H, Teo SH, Yip CH, Phuah SY, Cornes BK, Hartman M, Miao H, Lim WY, Sng JH, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Shen CY, Hsiung CN, Wu PE, Ding SL, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Blot WJ, Signorello LB, Cai Q, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Simard J, Garcia-Closas M, Pharoah PD, Chenevix-Trench G, Dunning AM, Benitez J, Easton DF. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICE 2013. Clinical Guideline 164, , http://publications.nice.org.uk/familial-breast-cancer-cg164 . Accessed 19 October 2013.
- Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, Hughes D, Warren-Perry M, Tapper W, Eccles D, Evans DG, Hooning M, Schutte M, van den Ouweland A, Houlston R, Ross G, Langford C, Pharoah PDP, Stratton MR, Dunning AM, Rahman N, Easton DF. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Stat Med. 2004;23:1111–1130. doi: 10.1002/sim.1668. [DOI] [PubMed] [Google Scholar]