Abstract
Background:
microRNA-9 is a key regulator of neuronal development aberrantly expressed in brain malignancies, including medulloblastoma. The mechanisms by which microRNA-9 contributes to medulloblastoma pathogenesis remain unclear, and factors that regulate this process have not been delineated.
Methods:
Expression and methylation status of microRNA-9 in medulloblastoma cell lines and primary samples were analysed. The association of microRNA-9 expression with medulloblastoma patients' clinical outcome was assessed, and the impact of microRNA-9 restoration was functionally validated in medulloblastoma cells.
Results:
microRNA-9 expression is repressed in a large subset of MB samples compared with normal fetal cerebellum. Low microRNA-9 expression correlates significantly with the diagnosis of unfavourable histopathological variants and with poor clinical outcome. microRNA-9 silencing occurs via cancer-specific CpG island hypermethylation. HES1 was identified as a direct target of microRNA-9 in medulloblastoma, and restoration of microRNA-9 was shown to trigger cell cycle arrest, to inhibit clonal growth and to promote medulloblastoma cell differentiation.
Conclusions:
microRNA-9 is a methylation-silenced tumour suppressor that could be a potential candidate predictive marker for poor prognosis of medulloblastoma. Loss of microRNA-9 may confer a proliferative advantage to tumour cells, and it could possibly contribute to disease pathogenesis. Thus, re-expression of microRNA-9 may constitute a novel epigenetic regulation strategy against medulloblastoma.
Keywords: medulloblastoma, miRNA-9, epigenetic silencing, paediatric cancer
Medulloblastoma (MB) is the most common malignant brain tumour in children. Despite recent treatment advances, approximately 30% of children with MB will die from their disease (Northcott et al, 2012a). Those who survive often have a significantly reduced quality of life as result of therapy-related sequelae (Mulhern et al, 2005). Novel and targeted therapeutic strategies are thus needed to reduce mortality and morbidity. Recently, molecular profiling studies identified four core subgroups of MB tumours with potential targets for specific therapies based on varying gene expression (Kool et al, 2008; Northcott et al, 2011). WNT tumours are characterised by activated wingless pathway and carry a favourable prognosis under current treatment regimens. SHH tumours possess active hedgehog signalling and have an intermediate prognosis. Group 3, characterised by high MYC levels, and Group 4 tumours, molecularly less well characterised, are usually associated with poor prognosis (Taylor et al, 2012). Despite the great advance in the fight against MB resulting from molecular-based sub-grouping, additional efforts are needed to identify key biological alterations that could be targeted by tailored therapies.
MicroRNAs (miRNAs) are small RNA molecules that have emerged as key regulators during the development of both normal human brain and brain tumours, including MB (Hummel et al, 2011; Dubuc et al, 2012). Upon binding to the 3′ UTR, miRNAs downregulate the target genes expression by blocking the translation and/or by inducing mRNA degradation. Recently, specific miRNA expression signatures helped improving the molecular classification of brain tumours and facilitated the diagnosis and prediction of therapy response and prognosis (Fernandez et al, 2009). miRNAs are suggested to have a critical role in MB tumour pathogenesis because of their altered expression observed in a large number of MB primary samples (Pang et al, 2009; Turner et al, 2010). A high throughput expression profile of miRNAs expressed either in neuronal tissues or associated with tumour development revealed a tumour-specific pattern of microRNAs expression in human primary MB samples (Ferretti et al, 2009). In particular, miR-9 was found to be one of the key miRNAs expressed at low levels in MB tumours compared with normal fetal cerebellum.
MiR-9 is a regulator of neuronal progenitor cell fate during neurogenesis (Wienholds et al, 2005; Shibata et al, 2011), which has recently been implicated in cancer (Delaloy et al, 2010). Although most studies indicate a tumour-suppressor activity for miR-9 in cancer cells (Laios et al, 2008), conflicting data exist, and the outcome of miR-9 function appears to be tumour specific (Khew-Goodall and Goodall, 2010). miR-9 is downregulated in breast cancer, renal cell carcinoma, and gastric cancer due to promoter methylation (Lehmann et al, 2008; Hildebrandt et al, 2010; Tsai et al, 2011). In contrast, miR-9 has been found to be upregulated in gliomas and in colorectal cancer (Malzkorn et al, 2010; Zhu et al, 2012). Recent studies also report that miR-9 is heterogeneously expressed within a given tissue (Bonev et al, 2011). To date, it remains unclear how aberrant miR-9 expression contributes to MB pathogenesis. Therefore, we investigated the expression of miR-9 in MB, its functional role, and its association with the clinical outcome of MB patients.
Materials and methods
Human MB primary samples and cell lines
The tumour material used in this study originates from two sets of archival MB samples from patients treated at the University Children's Hospital of Zürich, Zürich, Switzerland (n=78; 63 formalin-fixed paraffin-embedded+15 fresh frozen). DNA from 64 MB samples (49 formalin-fixed paraffin-embedded+15 fresh frozen) was available for the methylation study. mRNA was available from 14 out of the 63 formalin-fixed paraffin-embedded samples and from 15 additional fresh frozen MB samples. All tissue specimens used in the study were obtained from the Swiss Pediatric Oncology Group (SPOG) Tumour Bank. Written informed consent was obtained from each patient by the hospital that provided the tissue samples. The use of SPOG Tumour Bank tissue samples for cancer research purposes was approved by the Ethical Review Board of Zurich (Ref. No. StV-18/02). Gene expression profile of 88 primary MB tumour samples was performed in Dr. Taylor's Lab (The Hospital for Sick Children, Division of Neurosurgery, Toronto, ON, Canada) and used for gene expression and survival analysis. Normal fetal cerebella were used as control for the mRNA expression studies (n=5; gestational age: 16–26 weeks). Publicly available microarray data from 285 primary MB samples was used to determine the correlation between the expression of selected genes (Northcott et al, 2012b). MB expression profiles were generated on Affymetrix Human Gene 1.1 ST Array (Santa Clara, CA, USA). Data are accessible through the open access database R2 for visualisation and analysis of microarray data (http://r2.amc.nl).
MB cell lines were cultured as previously published (von Bueren et al, 2007) and maintained at 37 °C in a humidified atmosphere with 5% CO2. DAOY human MB cells were purchased from American Type Culture Collection (Manassas, VA, USA). D341, D425, UW-228-2 and Med-1 human MB cells were the kind gift of Dr Henry Friedman (Duke University, Durham, UK). PFSK cells were the kind gift of Dr Peter Phillips (Children's Hospital of Philadelphia, Philadelphia, PA, USA).
MicroRNA and RNA analysis and expression profiling of human primary MBs
Total RNA for microRNA analysis was isolated using the mirVana miRNA Isolation Kit (Ambion, Life Technology, NY, USA) according to the manufacturer's instructions. First-strand synthesis of mature microRNAs was followed by qRT–PCR using microRNA-specific TaqMan MGB probes for miR-9 (ID 000583) and, for control RNA, U6 small nuclear 2 (RNU6B) (ID 001093) (Applied Biosystems, Life Technology, Carlsbad, CA, USA). Total RNA for mRNA analysis was extracted using the RNeasy Mini Kit (Qiagen, Basel, Switzerland) following the manufacturer's instructions. For the qRT–PCR reaction, the Gene Expression Master Mix was used, and the protocol was optimised for the ABI7900HT reader (Applied Biosystems, Life Technology). Probe-primer solutions specific for the following genes were used: HES1 (Hs00172878_m1), p21 (Hs00355782_m1), MASH1/ASCL1 (Hs00269932_m1), NEUROD1 (Hs00159598_m1), MAP2 (Hs00258900_m1), NEFH (Hs00606024_m1), GFAP (Hs00909236_m1), NES (Hs00707120_s1), and TUBB3 (Hs00964962_g1). Normal human cerebellum was used as a reference (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France). The relative gene expression was calculated for each gene of interest by using the ΔΔCT method, where cycle threshold (CT) values were normalised to the housekeeping genes RNA, U6 small nuclear 2 (RNU6B), succinate dehydrogenase complex subunit A (SDHA) (Hs00188166_m1), and 18S (Hs99999901_s1) (Applied Biosystems, Life Technology).
Promoter methylation analysis of the miR-9genes
Promoter methylation status of the miR-9-1, -2, and -3 genes was analysed by methylation-specific PCR (MSP) in MB cell lines and 64 primary samples of MB tissues. Bisulfite conversion of DNA was carried out using the EZ DNA Methylation Kit (Zymo Research Co., Irvine, CA, USA) following the manufacturer's instructions. The primers and the protocol used for MS-PCR have been previously described (Lujambio et al, 2008; Bandres et al, 2009). For each MS-PCR reaction, universal methylated DNA (Chemicon International, Temecula, CA, USA) was included as positive control and normal lymphocyte DNA as negative control. Samples of normal tissue on paraffin sections were used as negative controls. PCR products were run on a 3% agarose gel and were visualised with UV trans-illumination (Bio-Rad, Hercules, CA, USA). Methylated bands were detectable if >25% of the DNA was methylated. Similarly, methylated bands were not detectable when all DNA was unmethylated and, vice versa, confirming the sensitivity and specificity of the method.
In silico search for miR-9 target genes
Predictions on miR-9-HES1 interactions were extracted either from databases that predict miRNA target sequences, for example, TargetScan (Lewis et al, 2003), Pictar (Krek et al, 2005), DIANA-microT 3.0 algorithm (Maragkakis et al, 2009) and microRNA.org (Betel et al, 2008) or from databases that can predict miRNA binding sites on potential target genes, such as StarBase v2.0 (Yang et al, 2011), microPIR (Piriyapongsa et al, 2012), and miRWalk (Dweep et al, 2011).
Western blotting and immunofluorescence analysis
Western blotting analyses were performed as previously published (Fiaschetti et al, 2011). The membranes were incubated with the following first antibodies: HES1 (H-140) (sc-25392, Santa Cruz Biotechnology, Santa Cruz, CA, USA), β-tubulin Class III (TUBB3) (2G10) (Ab78078, Abcam, Cambridge, UK), MASH1/Achaete-scute homolog 1 (ab38557, Abcam), and Cip1/WAF-1/p21 (CP74) (P1484, Sigma-Aldrich, Buchs, Switzerland). Protein detection was performed with Supersignal Western Bio Chemo-luminescent Substrate (Pierce, Thermo Fisher Scientific, Rockford, IL, USA). β-Actin (AC-74) (A5316, Sigma-Aldrich) was used as a loading control. For immunofluorescence staining, cells were fixed in 4% PFA and blocked in medium containing 10% FCS and 0.5% Triton. Cells were stained over night at 4 °C for NES (196908) (MAB1259, R&D Systems, Minneapolis, MN, USA) and TUBB3 (Ab78078, Abcam). Alexa Fluor 488 or 594 (1/200) (Invitrogen, Life Technology) antibodies were used as secondary antibodies. All stainings were analysed with an Axioskop2 mot plus fluorescence microscope (Zeiss, Jena, Germany).
Cell cycle and apoptosis analysis
MB cells were harvested, PBS washed, and immediately fixed in 5 ml ice-cold 70% ethanol. Before flow cytometry analysis, cells were resuspended in 300 μl propidium iodide solution, 2 μl of RNase (100 mg ml−1) was added, and the cells were incubated at 37 °C for 45 min. Flow cytometry analysis was performed on a BD FACS Canto II instrument (BD Biosciences, Franklin Lakes, NJ, USA) following the manufacturer's recommended protocol. Data were collected with DIVA software (BD Biosciences) and analysed with FlowJo software (TreeStar Inc., Ashland, OR, USA) using appropriate controls and gates.
Activation of caspases 3 and 7 was detected using the Caspase-Glo 3/7 Assay (Promega Corporation, Madison, WI, USA). Annexin V–APC staining (550475, BD Biosciences) was measured by flow cytometry. Apoptosis induction was confirmed by quantifying hystone-associated DNA fragments by Cell Death Detection ELISAPLUS assay (Roche Diagnostics, Rotkreuz, Switzerland).
Cell transfection and 5-Aza-2′-deoxycytidine (5-Aza) treatment
Cells were seeded in six-well plates at a cell density of 4 × 104 (DAOY, PFSK) and 2 × 105 (D341, D425), respectively. Transfection of pre-miR-9 (PM10022, Ambion) or negative control oligonucleotide referred to as scrambled (AM17110, Ambion) in a final concentration of 30 nmol l−1 was performed using siPORT NeoFX transfection reagent (Ambion) according to the manufacturer's recommendations. Transfection efficiency was assessed with a FAM-labeled pre-miR-9 negative control (AM17121, Ambion) 24 h post transfection. FAM-positive cells were counted under a fluorescence microscope and expressed as a percentage of total cells. Six random fields were counted for each sample. DAOY and PFSK cells (70–80% confluent) were transfected using HES1 siRNA (ID 6922, Ambion, Silencer select pre-designed siRNA) at 20 nM final concentration. As control, Silencer Negative Control siRNA no. 1 (AM4611, Ambion) was used under the same conditions. Cells were harvested 48 and 72 h after transfection for subsequent quantitative qRT–PCR and western blotting analysis. Cells were plated in six-well plates and treated with 0.1 mM 5-Aza (A3656; Sigma-Aldrich) or control (DMSO) for 24–48 h before further analysis.
Construction of 3′UTR reporter plasmids and luciferase assays
A 459-bp fragment of the 3′UTR of HES1 containing the putative miR-9 binding site wasPCR-amplified from PFSK cell's genomic DNA (FW: 5′-AAAACTCGAGAACGCAGTGTCACCTTCC-3′, RE: 5′-AAAACTCGAGCAGTTCGAAGACATAAAAGCC-3′). The 459-base pair segment was designed to contain XhoI and NotI sites and was cloned into the psiCHECK-2 vector (Promega Corporation), between the XhoI and NotI site, immediately 3′ downstream of the renilla luciferase gene. All constructs were verified by DNA sequencing. Ten nanograms of psiCHECK-2 construct was co-transfected with 10 nM miR-9 duplex or scrambled duplex into HEK-293T cells in a 96-well plate using lipofectamin-2000 (Invitrogen). After 48 h, the cell extract was obtained; firefly and renilla luciferase activities were measured with the Dual-Luciferase reporter system (Promega Corporation) according to the manufacturer's instructions.
Statistical analysis
All experiments were performed at least in triplicates. Data are represented as mean±s.d. For in vitro experiments Student's t-test was used. P-values of <0.05 were considered significant. Pearson's correlation test was used for gene correlation in patient's samples.
Results
Tumour-specific CpG island hypermethylation downregulates the expression of miR-9 in MB cells
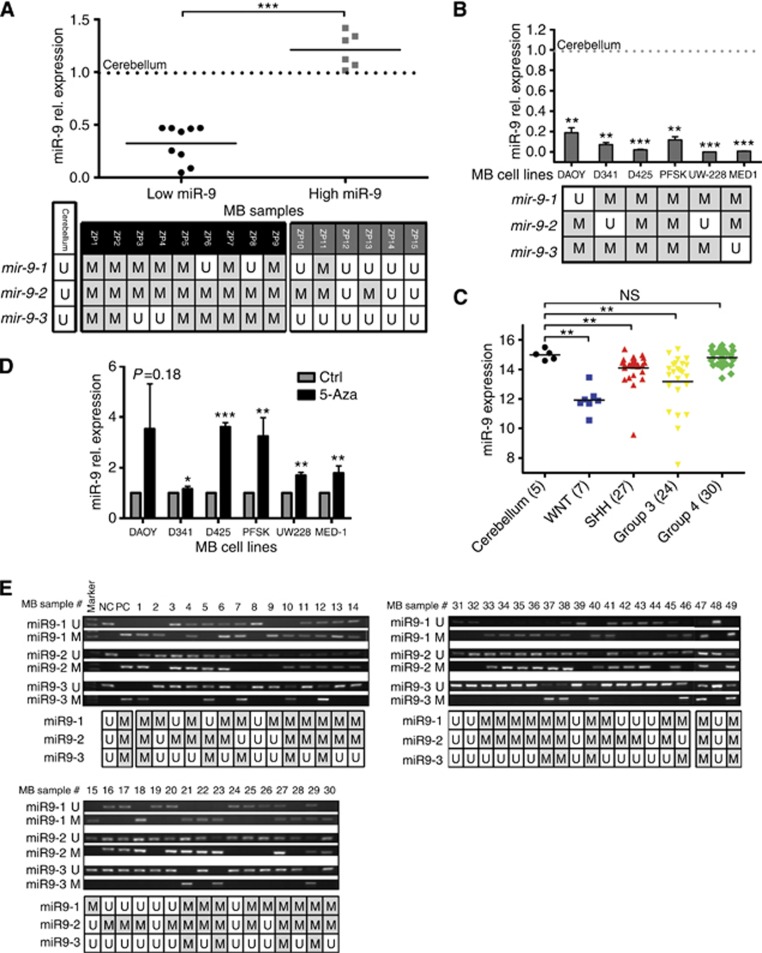
To gain insight into the expression of miR-9 in MB, we analysed a cohort of MB primary samples (n=29) and six MB-derived cell lines. Compared with normal human fetal cerebellum, a decreased expression level of miR-9 was observed in the majority of MB tumour tissues (Figure 1A, and Supplementary Figure S1A) and in all the six MB cell lines tested (Figure 1B, upper panel). To confirm that miR-9 downregulation is a common feature in MB, we validated our findings in an independent, non-overlapping cohort of primary MBs (n=88) with molecular subgroup information. miR-9 expression was found to be lower in a large subset of MB tumour samples (91%, 80/88) than in normal fetal cerebellum, regardless of their genetic backgrounds (Figure 1C). Across the four MB molecular subtypes, miR-9 downregulation was most pronounced in WNT and Group 3 subgroups, which are characterised by high expression of the MYC proto-oncogene (Figure 1C and Supplementary Figures S1B and C). In contrast, Group 4 tumours, which are characterised by the expression of high levels of genes involved in neuronal differentiation, displayed higher miR-9 levels than other subgroups that are comparable to normal cerebellar samples.
Figure 1.
Expression of miR-9 is epigenetically repressed in MB cells. Relative miR-9 expression as determined by qRT–PCR (A) in 15 fresh, frozen MB primary samples and (B) in the indicated MB cell lines. Values represent the fold decrease of miR-9-2 mRNA expression relative to normal cerebellum. Dotted line indicates cerebellum level at arbitrary value=1. (A and B, lower panels) Methylation status of the three miR-9 genes in the correspondent MB samples and cell lines. (C) Box plots showing relative expression of miR-9-2 across MB subgroups compared with fetal cerebellum. Relative expression is a measure of the luminosity of the gene probe signal, corrected for the background luminosity of each array and normalised using control probes across different arrays. (D) qRT–PCR analysis of miR-9 in the indicated human MB cell lines treated with 5-Aza or control (DMSO). (*P<0.05, **P<0.01, ***P<0.001 according to Student's t-test.) (E) Agarose gel showing MS-PCR products of the three miR-9 genes' CpG island regions in 49 FFPE MB primary samples. Lower panel represents methylation status of the three miR-9 genes in the correspondent MB samples. Abbreviations: M=methylated; NC=negative control; NS=not significant; PC=positive control; U=unmethylated.
Tumour-suppressor genes can be inactivated in various ways, including CpG promoter hypermethylation (Esteller, 2002). Our preliminary investigation revealed no deletions in miR-9 genes in MB cells (Supplementary Figure S1D). To determine whether hypermethylation caused reduced miR-9 expression in MB cells, we examined methylation of the promoter regions of hsa-miR-9 by MSP. Hsa-miR-9 is represented by three genomic loci hsa-miR-9-1 (1q22), hsa-miR-9-2 (5q14.3), and hsa-miR-9-3 (15q26.1) (Bandres et al, 2009). All three loci could independently contribute to the expression of miR-9. In contrast to the normal cerebellum, the three miR-9 loci were independently methylated in the subset of primary samples with low miR-9 expression and in all the tested MB cell lines (Figure 1A and B, lower panels).
To establish the causal relationship between methylation and the low miR-9 expression level, we treated MB cells with the demethylating agent 5-Aza. After treatment with 5-Aza, miR-9 expression markedly increased in all MB cell lines except in D341 cells, where only a small increase was observed (Figure 1D), confirming the role of promoter methylation in miR-9 silencing. To determine the role of epigenetic regulation of hsa-miR-9 in a clinically more relevant model, the promoter methylation status of the three has-miR-9 genes was analysed in a panel of 64 MB samples using MSP analysis. In 78% (50/64) of MB patients, at least one of the three miR-9 loci showed promoter methylation (Figure 1A, lower panel, and 1E). In particular, the promoter regions of miR-9-1 were methylated in 64% (41/64) and miR-9-2 was methylated in 73% (47/64), whereas miR-9-3 promoter was methylated in 34% (23/64) of MB cases. These results indicate that the reduced expression of miR-9 found in MB tissue samples and cell lines is likely caused by aberrant methylation of one, or more, of the three pre-miR-9 paralogous loci.
MYC is involved in the regulation of miR-9 levels in MB
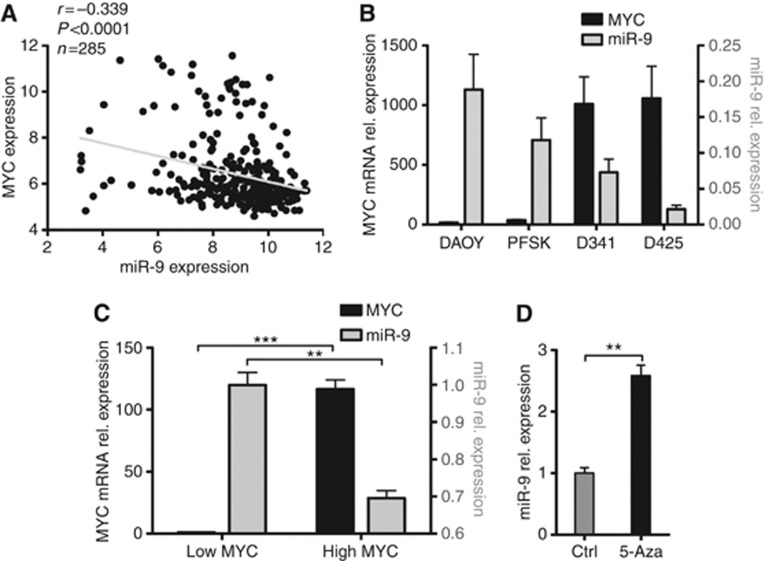
Given the prominent role of aberrant MYC in MB pathogenesis (Northcott et al, 2012b) and its capability to directly bind to the promoter region of miR-9 (Ma et al, 2010), we investigated whether MYC is involved in the regulation of miR-9 expression in MB cells. The analysis of gene expression profiles of 285 MBs revealed a statistically significant negative correlation between miR-9 level and MYC expression in primary tumour samples (Figure 2A). A similar inverse correlation was observed when the expression levels of MYC and miR-9 were analysed in a representative panel of MB cell lines. The levels of miR-9 were higher in cell lines bearing low level of MYC (DAOY and PFSK), whereas high-MYC cells (D341 and D425) showed a relatively low expression of miR-9 (Figure 2B). To verify this finding, we examined the level of miR-9 in MB cells ectopically overexpressing MYC (DAOY-MYC) and compared it with empty-vector-transfected cells (low MYC-DAOY cells) (Stearns et al, 2006). MYC overexpression resulted in a 30% decrease in miR-9 expression (Figure 2C), suggesting the involvement of MYC in the regulation of miR-9 expression in MB cells.
Figure 2.
The oncogene MYC is involved in the regulation of miR-9 expression. (A) Correlation between miR-9 and MYC mRNA expression in 285 MB patients' samples data sets as determined by microarray-based expression profiles (Northcott et al, 2012b; r=Pearson's correlation coefficient). (B) Relative mRNA expression of MYC (left y axis) and expression of miR-9 (right y axis) in the indicated cell lines as determined by qRT–PCR. (C) Relative mRNA expression of MYC (left y axis) and expression of miR-9 (right y axis) in empty-vector-transfected DAOY cells (low MYC) and in MYC-transfected DAOY cells (high MYC) as determined by qRT–PCR. (D) Relative expression of miR-9 in MYC-transfected DAOY cells upon 24 h of 5-Aza treatment as determined by qRT–PCR. (*P<0.05, **P<0.01, ***P<0.001 according to Student s t-test.)
MYC-mediated repression of several miRNAs has been reported (Chang et al, 2008; Bui and Mendell, 2010), although the underlying mechanisms are not fully clarified (Frenzel et al, 2010). To confirm the capability of MYC to mediate a downregulation of miR-9 in MB and to shed light on the mechanisms behind it, we investigated whether DNA methylation was required for MYC-mediated miR-9 repression. Indeed, MYC-induced miR-9 repression was counteracted by 5-Aza treatment, suggesting that the oncogene MYC is involved in the reduction of miR-9 level likely by regulating the DNA methylation status of miR-9 promoter.
miR-9 regulates the expression of human HES1 gene in MB
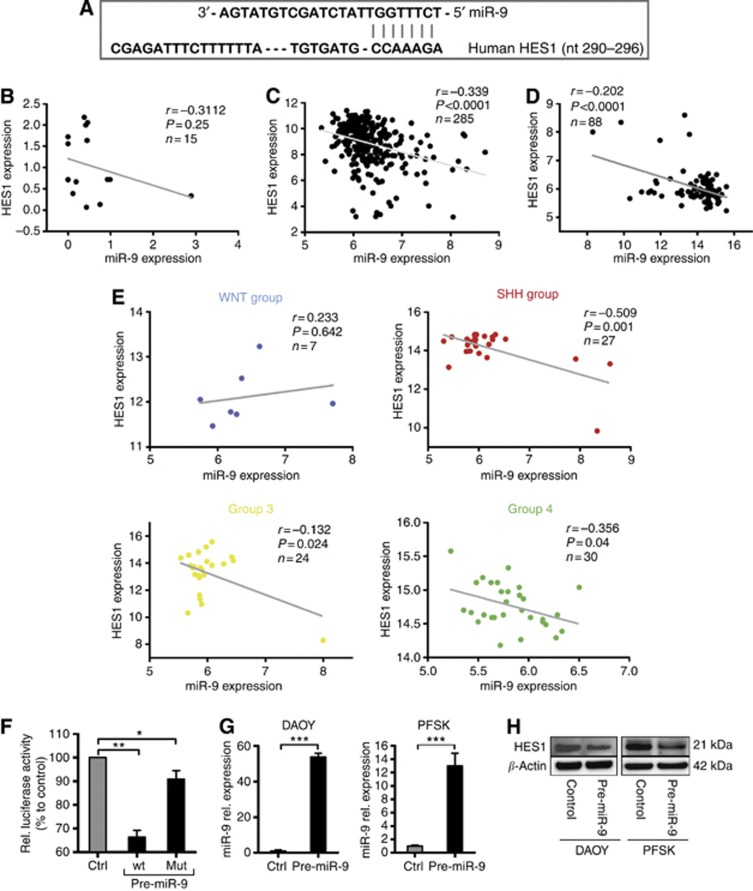
To identify putative targets for miR-9 with potential implications in MB tumour biology, we conducted an in silico search using several independent databases. Among potential miR-9 targets, based on target site conservation in humans, distinct programs identified HES1 (hairy and enhancer of split 1) gene as a potential target for miR-9 (Figure 3A and Supplementary Figure S2A). As HES1 has a central role in normal brain development and in MB pathogenesis (Hallahan et al, 2004; Dakubo et al, 2006; de Bont et al, 2008; Ingram et al, 2008), we decided to focus our further investigation on miR-9-mediated regulation of HES1.
Figure 3.
miR-9 regulates HES1 expression in MB cells. (A) Sequence alignment of predicted miR-9 binding site in the 3′UTR of HES1 promoter (seed-complementary region). (B–D) Correlation study of miR-9 and HES1 expression in three independent sets of MB patients' samples as determined by qRT–PCR and by microarray-based expression profiles. (r=Pearson's correlation coefficient). (E) Correlation study of HES1 and miR-9 levels according to MB subgroups (WNT, SHH, Group 3, Group 4) (n=88). (F) Luciferase reporter activity in 293T cells transfected with pre-miR-9 or negative control and co-transfected with the pisCheck2 vector containing the 3′UTR of HES1 gene downstream of the firefly luciferase reporter gene. (G) Relative miR-9 expression as determined by qRT–PCR 48 h following transfection with either pre-miR-9 or control in the indicated MB cell lines. Values represent fold increase of miR-9 mRNA relative to control. (*P <0.05, **P <0.01, ***P <0.001 according to Student's t-test.) (H) Pre-miR-9 mediated downregulation of HES1 protein expression in a representative immunoblotting experiment.
To gain insight into the miR-9/HES1 interaction in MB, we compared the expression of HES1 and miR-9 transcripts in MB primary samples. A pilot analysis of 15 MBs revealed a slight inverse correlation between the expression of HES1 and miR-9 levels (Figure 3B). To further corroborate these findings, we analysed the relationship between HES1 and miR-9 transcripts in two independent sets of 285 and 88 primary MB tumours (Northcott et al, 2012b). The analysis confirmed that miR-9 and HES1 expression levels are inversely correlated in both data sets (Figure 3C and D; Supplementary Figure S2B and C). The analysis of MB sugroups revealed a negative correlation between miR-9 and HES1 across SHH, Group 3, and Group 4 MBs in both sets of MB patient samples (Figure 3E and Supplementary Figure S2D). In contrast to the other subgroups, WNT tumours demonstrated a positive correlation between miR-9 and HES1 expression. Although the number of WNT samples in the analysed cohort is not large enough to validate this observation, the unexpected results obtained analyzing WNT cases might reflect a distinctive microRNA signature existing within the WNT subgroup (Gokhale et al, 2010).
To functionally validate miR-9 regulation of HES1 expression, we designed a luciferase reporter fused to either the wild-type HES1 3′UTR sequence or to a sequence that carries a mutation in the miR-9 seed-complementary region. The expression of the wild-type luciferase reporter in 293T cells was significantly repressed by miR-9 precursor mimics (pre-miR-9) compared with scrambled precursors, whereas the expression of the mutated luciferase reporter was not significantly affected, confirming the direct repression of HES1 by miR-9 (Figure 3F). We then examined the effect of miR-9 overexpression on HES1 protein level in MB cell lines. Ectopic restoration of low endogenous miR-9 expression (Figure 3G) effectively triggered a 30% decrease of HES1 protein level (Figure 3H and Supplementary Figure S2E). This result was confirmed in two additional MB cell lines (Supplementary Figure S2E). Overall, these findings show that miR-9 regulates HES1 expression and suggest HES1 as functional target of miR-9 in MB cells.
miR-9 restoration in MB cells induces cell cycle arrest and impairs clonal growth
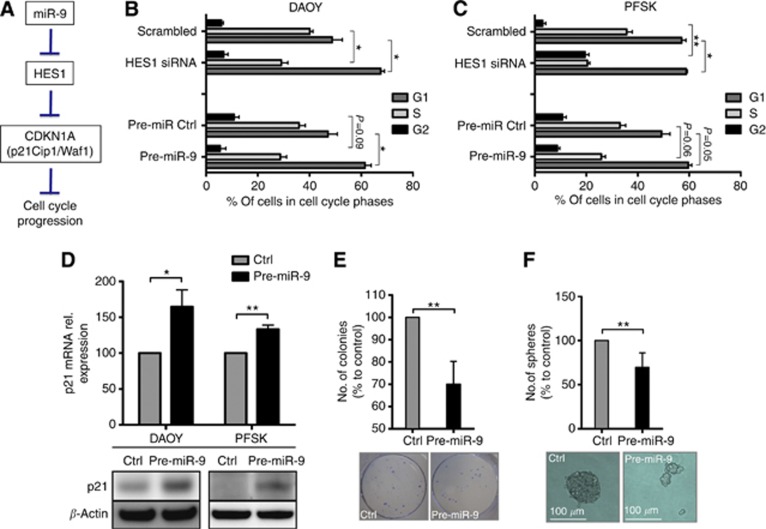
HES1 controls tumour cell growth through the transcriptional repression of cell cycle inhibitors, such as p21 (waf1/Cip1), which is central to cell cycle regulation and is altered in the vast majority of human cancers, including MB (Figure 4A; Kabos et al, 2002; Murata et al, 2005; Monahan et al, 2009). We hypothesised that by repressing HES1 miR-9 restoration could restrict the oncogenic induction of cell cycle progression in MB cells by promoting upregulation of p21. To test this possibility, we measured cell cycle distribution, changes in p21 expression, and the capability of MB cells to grow clonally following the restoration of miR-9. MB cells transfected with pre-miR-9 showed consistent changes in cell cycle distribution compared with control-transfected cells. We observed a decrease in the number of cells in S-phase and an increase of cells in G1 phase, indicative of cell cycle arrest. Analogously siRNA-mediated silencing of HES1 decreased the number of cells in S-phase, thereby phenocopying the effects of miR-9 overexpression (Figure 4B and C). The restoration of miR-9 resulted in an increase of p21, both at mRNA and protein levels, when compared with control-transfected MB cells (Figure 4D). Transfection of two additional MB cell lines with pre-miR-9 confirmed this result (Supplementary Figure S3A). Although re-establishment of miR-9 did not affect cell viability, cell senescence (data not shown), or cell death (Supplementary Figure S3B and C), it decreased the cell capability to form colonies (Figure 4E), and it reduced the size and number of neuronal spheres (Figure 4F). Taken together, these results highlight the potential benefit of miR-9 restoration against MB cell growth.
Figure 4.
Restoration of miR-9 upregulates HES1 responsive gene P21Cip1 and impairs clonal growth of MB cells. (A) Cartoon depicting an overview of cell cycle-related miR-9 downstream responsive genes and their functions. (B, C) Flow cytometric analysis of cell cycle distribution for the indicated MB cell lines 72 h after transfection with either pre-miR-9 or control and with HES1 siRNA or siRNA scrambled control. (D) Pre-miR9 mediated upregulation of p21 mRNA (upper panel) and protein (lower panels) expression in the indicated MB cell lines. (E) Colony formation and (F) sphere formation analysis of human MB DAOY cells treated with pre-miR-9 or control for 6 days. Lower panels: representative pictures. (*P<0.05, **P<0.01 according to Student‘s t-test.)
Re-establishment of miR-9 in MB cells promotes neural differentiation
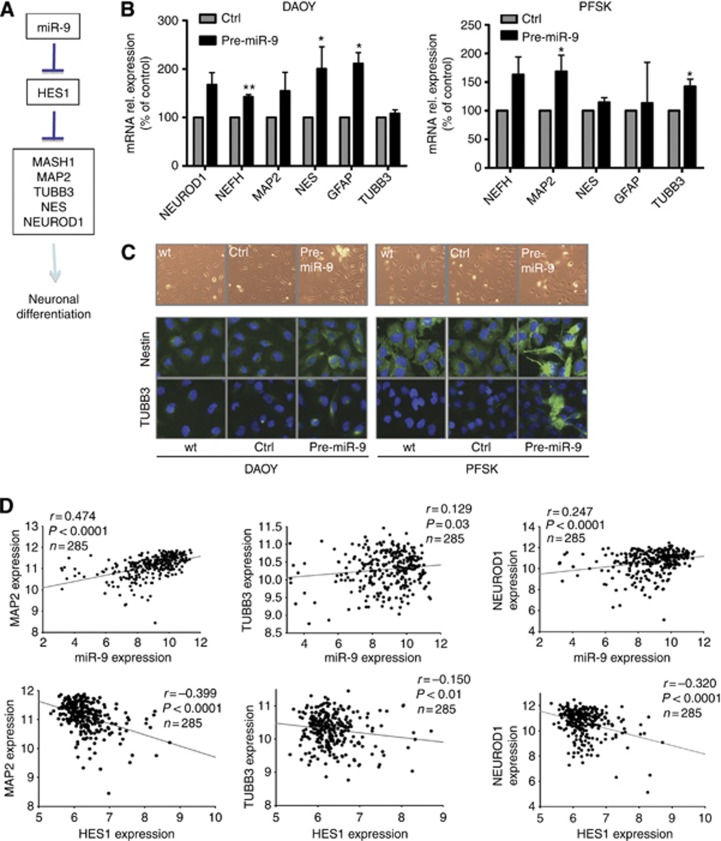
The induction of endogenous HES1 inhibits neuronal differentiation (Kageyama and Ohtsuka, 1999; Ohtsuka et al, 1999), while HES1 knockout or mutation lead to neuronal differentiation in vivo (Figure 5A) (Ishibashi et al, 1995; Tomita et al, 1996). Given the important regulatory function of HES1 in neuronal differentiation (Kabos et al, 2002; Sang et al, 2008), we hypothesised that miR-9-mediated repression of HES1 could trigger the differentiation of MB cells. To test this possibility, we evaluated the effect of miR-9 restoration on MB cell differentiation. mRNA expression level of a panel of canonical neural differentiation markers were analysed by qRT–PCR: neurogenic differentiation 1 (NEUROD1) (Lee et al, 1995), neurofilament H (NEFH) (Lee and Cleveland, 1996), neuron-specific microtubule-associated protein (MAP2) (Dinsmore and Solomon, 1991), nestin (NES) (Wiese et al, 2004), β-tubulin 3 class III (TUBB3) (Katsetos et al, 2003), and glial fibrillary acidic protein (GFAP) (Eng et al, 2000). Transfection of MB cells with pre-miR-9 resulted in the upregulation of the mRNA expression of a subset of neural differentiation marker in MB cells compared with controls (Figure 5B). Among pre-miR-9-treated DAOY cells, some contacted with each other through their branching protrusions, suggesting a primary stage of differentiation (Figure 5C, upper panel), while control-treated cells retained flat morphology. Immunoflorescence analysis confirmed at the protein level that upon miR-9 re-expression both cell lines exhibited upregulation of TUBB3 and Nestin (Figure 5C and Supplementary Figure S4A) (Bhoopathi et al, 2011). To determine the contribution of HES1 repression to miR-9-mediated effects on neuronal differentiation, we examined the effects of HES1 silencing on mRNA expression of neuronal differentiation markers. As observed in MB cells treated with pre-miR-9, siRNA-mediated knockdown of HES1 resulted in an upregulation of a subset of neuronal differentiation genes (Supplementary Figure S4B). Similarly to the effects caused by the restoration of miR-9, the expression of NeuroD1, Nestin, and GFAP genes were upregulated by HES1 silencing. Thus, miR-9-mediated regulation of HES1 appears to be an essential aspect of miR-9 function on MB cell differentiation.
Figure 5.
miR-9 restoration mediates increase of the expression of a panel of pro-neural differentiation genes in MB cells. (A) Cartoon depicting an overview of neuronal differentiation-related miR-9/HES1 downstream responsive genes. (B) Relative mRNA expression of the indicated pro-neural differentiation genes as determined by qRT–PCR 72 h following transfection with either pre-miR-9 or control. Values represent fold change relative to control (*P<0.1, **P<0.01 according to Student's t-test). (C) Morphological changes of MB cells following miR-9 restoration and immunofluorescence analysis of β-tubulin class III (TUBB3) and nestin (NES) 72 h following transfection with either pre-miR-9 or control. (D) Correlation study of expression of miR-9/HES1 and selected pro-neural differentiation genes in 285 MB patients' samples as determined by microarray-based expression profiles (r=Pearson's correlation coefficient).
To validate our results in a clinically relevant model, microarray-based expression profiling data obtained from a set of 285 MB tumours (Northcott et al, 2012b) were analysed searching for correlations between mRNA levels of miR-9, HES1, and previously selected neuronal differentiation markers. A trend to positively correlate was found between mRNA expression of MAP2, TUBB3 and NEUROD1 and miR-9-2 (Figure 5D, upper panels), parallel to an inverse correlation with HES1 mRNA expression (Figure 5D, lower panels). The analysis of the expression of miR-9-1 and miR-9-3 in the same MB samples also showed similar results, although less statistically significant (Supplementary Figure S4C and D).
HES1 regulatory function on cell differentiation status was previously shown to be mediated by the pro-neural differentiation gene MASH1 (Kageyama et al, 1997). Therefore, we determined whether miR-9 restoration affected the expression of MASH1. Indeed, besides the downregulation of HES1, we observed a moderate but measurable upregulation of MASH1 protein level when compared with control-transfected cells (Supplementary Figure S4E). These results suggest that miR-9 has a critical role in MB cells by decreasing MB cell differentiation and highlight the potential use of miR-9 restoration for reactivating the differentiation programme in cancer cells.
miR-9 and HES1 expression associates with clinicopathological characteristics of paediatric MB
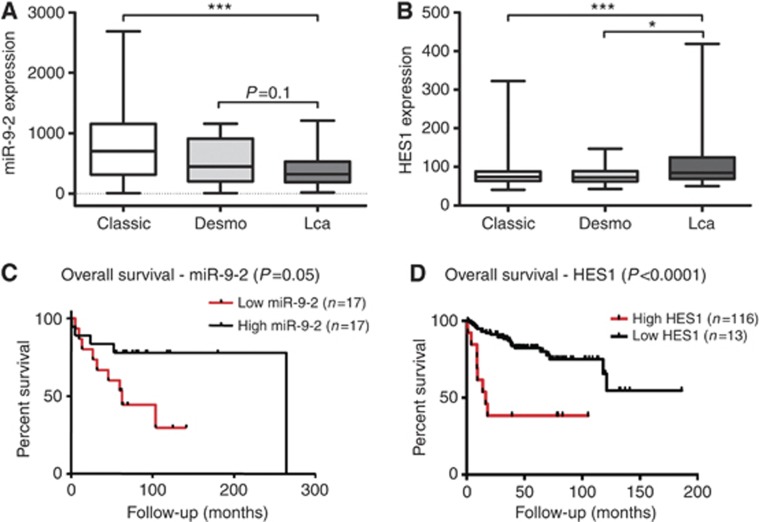
Finally, we investigated the association of miR-9 and HES1 expression with clinicopathological characteristics of paediatric MB. Three main histological subtypes of MB are included in the current WHO classification (Sarkar et al, 2006) among which large-cell anaplastic (LC/A) MBs represents a distinct and more aggressive variant compared with classic and desmoplastic subtypes (Leonard et al, 2001; von Hoff et al, 2010). To determine the relation of miR-9 expression with MB histopathological subtypes, we analysed a set of MB primary samples with available information about histological features. Strikingly, LC/A MB samples possess lower miR-9-2 expression compared with the other variants. In particular, a statistically significant differential expression was observed on comparing LC/A MBs with tumours of classic subtype (Figure 6A). Analysis of miR-9-1 and miR-9-3 expression pattern in the same MB samples showed a similar trend to inversely correlate with anaplastic MB subtypes (Supplementary Figure S5A and B), corroborating the hypothesis that MB tissues with decreased expression of miR-9 tend to have a more severe pathological grade. Notably, LC/A MBs also showed a slightly higher HES1 expression (Figure 6B). Furthermore, to determine the potential impact of miR-9 on clinical outcome, miR-9 expression level was correlated with survival. Patient survival information was available for 34 MB samples. The lower overall survival probability of patients with low miR-9-2 expression revealed by Kaplan–Meier analysis suggests a strong trend towards prognostic significance (Figure 6C). Although it did not reach statistical significance, the analyses of miR-9-1 and miR-9-3 expression pattern in the same MB samples showed a similar inverse correlation between miR-9 expression and patient's survival (Supplementary Figure S5C and D). Additionally, in line with previous reports (Cho et al, 2011), high HES1 expression correlated significantly with lower overall survival in a distinct cohort of 129 MB samples (Figure 6D). We also investigated the relation between miR-9 expression and metastatic stage, age, and gender in the same cohort of MB samples but found no significant correlation (data not shown). Together, these data indicate that low expression of miR-9 (in particular miR-9-2) and the consequently increased expression of HES1 could be associated with worse MB clinical outcome. Hence, miR-9 might be considered as a candidate marker predicting unfavourable prognosis for patients with paediatric MB that deserves further validation studies.
Figure 6.
Expression of miR-9 and HES1 are associated with clinicopatholigical characteristics of paediatric MB. Box plots showing (A) miR-9-2 and (B) HES1 expression according to MB histological variants: classic (n=200), (desmo) desmoplastic (n=21) and (Lca) large cells/anaplastic (n=30); centre line=median. (*P<0.05, ***P<0.001 according to Student's t-test.) (C) Kaplan–Meier survival curves for miR-9-2 expression in paediatric MBs. Patients were sub-divided into high (n=17) and low (n=17) miR-9 expression groups based on the median expression value in each population. (D) Kaplan–Meier survival curves for HES1 expression in paediatric MBs. Patients were sub-divided into high (n=13) and low (n=116) HES1 expression groups based on the median expression value in each population. Optimal cutoff selection was combined with Bonferroni correction because of the low number of HES1-expressing tumours.
Discussion
The current study provides evidence that in paediatric MB miR-9 is significantly downregulated when compared with normal fetal cerebellum and that miR-9 has a tumour-suppressor role in MB tumours. We evaluated the prognostic significance of these findings, because neither the functional role nor the diagnostic implications of miR-9 in MB have been previously defined. Our investigation revealed that the reduced expression of miR-9 in MB cells is largely due to tumour-specific CpG promoter hypermethylation, a finding previously confirmed in other cancer types (Lehmann et al, 2008; Lujambio et al, 2008; Bandres et al, 2009). Epigenetic deregulation of miRNA genes highlights a potential mechanism implicated in the development of various malignancies. Considering the high frequency of methylation in miR-9 promoter in paediatric MB we revealed in this study, the degree of promoter methylation of the miR-9 genes could be considered as a possible useful tumour marker. Furthermore, the use of DNA-demethylating agents could have significant consequences for cancer patients, opening the possibility for new therapeutic strategies for selected patients with MB.
In MB, as in several human cancers, deregulation of the oncogene MYC is associated with tumour pathogenesis. Although in breast cancer MYC-mediated induction of miR-9 has been observed (Khew-Goodall and Goodall, 2010), our data rather suggest a potential pathological function of MYC in MB involving the repression of miR-9 expression. This miR-9 repression likely involves epigenetic silencing, thus underlining the distinct, context-dependent regulation of miRNAs networks in different tumour backgrounds. Moreover, this observation emphasises the notion that MYC-mediated repression of tumour-suppressing miRNAs, such as miR-9, may be a fundamental component of the MYC tumourigenic programmes in MB, in particular in MB subgroups expressing high levels of MYC. In fact, miR-9 downregulation was most pronounced in WNT and Group 3 tumours, which highly express the oncogene MYC. Conversely, Group 4 cases rarely express high levels of MYC (Northcott et al, 2011) and are characterised by an over-representation of genes involved in neuronal differentiation (Cho et al, 2011; Taylor et al, 2012). This is in accordance with our findings correlating high miR-9 levels with induction of cell differentiation. Interestingly, Group 4 tumours are characterised by high NMYC levels, thus suggesting distinct functional significances of MYC and NMYC for miR-9 expression in MB.
Our experimental evidence shows that HES1 is a direct target of miR-9-mediated translational inhibition in MB cells, corroborating a recent report about miR-9-mediated effects on the homolog of mammalian HES1 (Hairy1) in low vertebrates (Bonev et al, 2012). This is highly significant because HES1, one of the main NOTCH target genes, controls cell fate decisions of the mammalian neural progenitor cells from which MBs arise (Solecki et al, 2001; Pomeroy et al, 2002). Moreover, the inverse correlation between miR-9 and HES1 expression across distinct independent sets of MB patients' samples is clinically relevant. A subgroup-specific analysis of miR-9 and HES1 expression revealed negative correlation in the non-WNT MBs. On the other hand, WNT-type MBs showed rather an increase of miR-9 expression. This might indicate that miR-9 has a subgroup-specific effect on translational inhibition and regulates different targets in WNT MBs, analogous to the distinctive microRNA signatures proposed within the WNT subgroup (Gokhale et al, 2010). Nevertheless, an analysis of a bigger cohort of WNT tumours is needed to confirm this prediction.
Our investigation showed that restoration of miR-9 expression led to G1/S cell cycle phase arrest in MB cells and that the tumour-suppressive role of miR-9 is mediated primarily through HES1 regulation. Thus miR-9 restoration may reduce MB cell growth by stalling the cells in G1/S phase and preventing cell cycle progression via HES1 inhibition and p21 induction. miR-9-mediated inhibition of HES1 also promoted the expression of distinct neuronal differentiation markers, including early neural differentiation marker TUBB3, neural-progenitor marker Nestin, and astrocytic differentiation marker GFAP, which has been associated with less aggressive MBs and higher sensitivity to anticancer drugs (Sadatomo et al, 1996; Son et al, 2003). Indeed, our results show that miR-9 is downregulated in poorly differentiated MB cells, whereas it is normally expressed in normal fetal cerebellum from which MB tumours arise. Therefore, miR-9 restoration may re-establish the neurogenic differentiation programme and reduce MB tumourigenic phenotype through the repression of HES1.
The role of miR-9 as a prognostic factor has been recently described in different tumours, such as acute lymphocytic leukemia (Agirre et al, 2009), acute myeloid leukemia (Marcucci et al, 2008), and colon cancer (Schetter et al, 2008). Based on the analysis of primary tumour data sets, the present study suggests that miR-9 might also be useful as a predictor of MB patient outcome. Indeed, MB patients with tumours expressing low levels of miR-9 (and concomitantly high HES1) show poorer survival rates. This is in agreement with recently published reports demonstrating that high HES1 expression levels are associated with poor MB prognosis (Hallahan et al, 2004; Ingram et al, 2008). The significant association between high HES1/low miR-9 expressions with established high risk indicators for MB suggests a diagnostic significance of miR-9 expression in MB histology.
In conclusion, we identified MB-specific CpG island hypermethylation of miR-9, a critical regulator of MB cell differentiation and cell cycle progression. Given the dynamic and, importantly, reversible nature of DNA methylation, our results suggest that miR-9 may serve as an important prognostic molecular marker. Furthermore, restoration of miR-9 expression may represent a novel therapeutic strategy to contribute to improvements in the treatment of MB patients, and future studies in animal models of MB are required to evaluate miR-9-mediated regulation of MB tumour growth in vivo.
Acknowledgments
We thank the Swiss Pediatric Oncology Group (SPOG) for providing medulloblastoma samples. This project was supported by the Swiss Research Foundation Child and Cancer and by Krebsliga Zürich.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, Martin-Subero JI, Cordeu L, Garate L, San Jose-Eneriz E, Abizanda G, Rodriguez-Otero P, Fortes P, Rifon J, Bandres E, Calasanz MJ, Martin V, Heiniger A, Torres A, Siebert R, Roman-Gomez J, Prosper F. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoopathi P, Chetty C, Dontula R, Gujrati M, Dinh DH, Rao JS, Lakka SS. SPARC stimulates neuronal differentiation of medulloblastoma cells via the Notch1/STAT3 pathway. Cancer Res. 2011;71:4908–4919. doi: 10.1158/0008-5472.CAN-10-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Stanley P, Papalopulu N. MicroRNA-9 modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012;2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui TV, Mendell JT. Myc: maestro of microRNAs. Genes Cancer. 2010;1:568–575. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, Lau CC, Olson JM, Gilbertson RJ, Gajjar A, Delattre O, Kool M, Ligon K, Meyerson M, Mesirov JP, Pomeroy SL. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakubo GD, Mazerolle CJ, Wallace VA. Expression of Notch and Wnt pathway components and activation of Notch signaling in medulloblastomas from heterozygous patched mice. J Neurooncol. 2006;79:221–227. doi: 10.1007/s11060-006-9132-2. [DOI] [PubMed] [Google Scholar]
- de Bont JM, Packer RJ, Michiels EM, den Boer ML, Pieters R. Biological background of pediatric medulloblastoma and ependymoma: a review from a translational research perspective. Neuro-oncol. 2008;10:1040–1060. doi: 10.1215/15228517-2008-059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsmore JH, Solomon F. Inhibition of MAP2 expression affects both morphological and cell division phenotypes of neuronal differentiation. Cell. 1991;64:817–826. doi: 10.1016/0092-8674(91)90510-6. [DOI] [PubMed] [Google Scholar]
- Dubuc AM, Mack S, Unterberger A, Northcott PA, Taylor MD. The epigenetics of brain tumors. Methods Mol Biol. 2012;863:139–153. doi: 10.1007/978-1-61779-612-8_8. [DOI] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by ‘walking' the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP thirty-one years (1969–2000) Neurochem Res. 2000;25:1439–1451. doi: 10.1023/a:1007677003387. [DOI] [PubMed] [Google Scholar]
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Northcott PA, Taylor MD, Kenney AM. Normal and oncogenic roles for microRNAs in the developing brain. Cell Cycle. 2009;8:4049–4054. doi: 10.4161/cc.8.24.10243. [DOI] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R, Giangaspero F, Farcomeni A, Nofroni I, Laneve P, Gioia U, Caffarelli E, Bozzoni I, Screpanti I, Gulino A. MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124:568–577. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- Fiaschetti G, Castelletti D, Zoller S, Schramm A, Schroeder C, Nagaishi M, Stearns D, Mittelbronn M, Eggert A, Westermann F, Ohgaki H, Shalaby T, Pruschy M, Arcaro A, Grotzer MA. Bone morphogenetic protein-7 is a MYC target with prosurvival functions in childhood medulloblastoma. Oncogene. 2011;30:2823–2835. doi: 10.1038/onc.2011.10. [DOI] [PubMed] [Google Scholar]
- Frenzel A, Loven J, Henriksson MA. Targeting MYC-regulated miRNAs to combat cancer. Genes Cancer. 2010;1:660–667. doi: 10.1177/1947601910377488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, Kunder R, Goel A, Sarin R, Moiyadi A, Shenoy A, Mamidipally C, Noronha S, Kannan S, Shirsat NV. Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J Cancer Res Ther. 2010;6:521–529. doi: 10.4103/0973-1482.77072. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, Olson JM. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, Wood CG, Wu X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29:5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- Hummel R, Maurer J, Haier J. MicroRNAs in brain tumors: a new diagnostic and therapeutic perspective. Mol Neurobiol. 2011;44:223–234. doi: 10.1007/s12035-011-8197-x. [DOI] [PubMed] [Google Scholar]
- Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–1500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Kabos P, Kabosova A, Neuman T. Blocking HES1 expression initiates GABAergic differentiation and induces the expression of p21(CIP1/WAF1) in human neural stem cells. J Biol Chem. 2002;277:8763–8766. doi: 10.1074/jbc.C100758200. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ishibashi M, Takebayashi K, Tomita K. bHLH transcription factors and mammalian neuronal differentiation. Int J Biochem Cell Biol. 1997;29:1389–1399. doi: 10.1016/s1357-2725(97)89968-2. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999;9:179–188. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- Katsetos CD, Herman MM, Mork SJ. Class III beta-tubulin in human development and cancer. Cell Motil Cytoskeleton. 2003;55:77–96. doi: 10.1002/cm.10116. [DOI] [PubMed] [Google Scholar]
- Khew-Goodall Y, Goodall GJ. Myc-modulated miR-9 makes more metastases. Nat Cell Biol. 2010;12:209–211. doi: 10.1038/ncb0310-209. [DOI] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, Mrsic A, Ylstra B, Grajkowska W, Hartmann W, Pietsch T, Ellison D, Clifford SC, Versteeg R. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Laios A, O'Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, D'Arcy T, McGuinness E, Sheils O, Sheppard B, O'Leary J. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lee MK, Cleveland DW. Neuronal intermediate filaments. Annu Rev Neurosci. 1996;19:187–217. doi: 10.1146/annurev.ne.19.030196.001155. [DOI] [PubMed] [Google Scholar]
- Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- Leonard JR, Cai DX, Rivet DJ, Kaufman BA, Park TS, Levy BK, Perry A. Large cell/anaplastic medulloblastomas and medullomyoblastomas: clinicopathological and genetic features. J Neurosurg. 2001;95:82–88. doi: 10.3171/jns.2001.95.1.0082. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stuhler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA, Sethupathy P, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- Monahan P, Rybak S, Raetzman LT. The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology. 2009;150:4386–4394. doi: 10.1210/en.2009-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhern RK, Palmer SL, Merchant TE, Wallace D, Kocak M, Brouwers P, Krull K, Chintagumpala M, Stargatt R, Ashley DM, Tyc VL, Kun L, Boyett J, Gajjar A. Neurocognitive consequences of risk-adapted therapy for childhood medulloblastoma. J Clin Oncol. 2005;23:5511–5519. doi: 10.1200/JCO.2005.00.703. [DOI] [PubMed] [Google Scholar]
- Murata K, Hattori M, Hirai N, Shinozuka Y, Hirata H, Kageyama R, Sakai T, Minato N. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol Cell Biol. 2005;25:4262–4271. doi: 10.1128/MCB.25.10.4262-4271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Pfister SM, Taylor MD. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8:340–351. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, Pfister S, Taylor MD. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, Beroukhim R, Ellison DW, Marshall CR, Lionel AC, Mack S, Dubuc A, Yao Y, Ramaswamy V, Luu B, Rolider A, Cavalli FM, Wang X, Remke M, Wu X, Chiu RY, Chu A, Chuah E, Corbett RD, Hoad GR, Jackman SD, Li Y, Lo A, Mungall KL, Nip KM, Qian JQ, Raymond AG, Thiessen NT, Varhol RJ, Birol I, Moore RA, Mungall AJ, Holt R, Kawauchi D, Roussel MF, Kool M, Jones DT, Witt H, Fernandez LA, Kenney AM, Wechsler-Reya RJ, Dirks P, Aviv T, Grajkowska WA, Perek-Polnik M, Haberler CC, Delattre O, Reynaud SS, Doz FF, Pernet-Fattet SS, Cho BK, Kim SK, Wang KC, Scheurlen W, Eberhart CG, Fevre-Montange M, Jouvet A, Pollack IF, Fan X, Muraszko KM, Gillespie GY, Di Rocco C, Massimi L, Michiels EM, Kloosterhof NK, French PJ, Kros JM, Olson JM, Ellenbogen RG, Zitterbart K, Kren L, Thompson RC, Cooper MK, Lach B, McLendon RE, Bigner DD, Fontebasso A, Albrecht S, Jabado N, Lindsey JC, Bailey S, Gupta N, Weiss WA, Bognar L, Klekner A, Van Meter TE, Kumabe T, Tominaga T, Elbabaa SK, Leonard JR, Rubin JB, Liau LM, Van Meir EG, Fouladi M, Nakamura H, Cinalli G, Garami M, Hauser P, Saad AG, Iolascon A, Jung S, Carlotti CG, Vibhakar R, Ra YS, Robinson S, Zollo M, Faria CC, Chan JA, Levy ML, Sorensen PH, Meyerson M, Pomeroy SL, Cho YJ, Bader GD, Tabori U, Hawkins CE, Bouffet E, Scherer SW, Rutka JT, Malkin D, Clifford SC, Jones SJ, Korbel JO, Pfister SM, Marra MA, Taylor MD. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JC, Kwok WK, Chen Z, Ng HK. Oncogenic role of microRNAs in brain tumors. Acta Neuropathol. 2009;117:599–611. doi: 10.1007/s00401-009-0525-0. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Bootchai C, Ngamphiw C, Tongsima S. microPIR: an integrated database of microRNA target sites within human promoter sequences. PLoS One. 2012;7:e33888. doi: 10.1371/journal.pone.0033888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeck M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Sadatomo T, Yoshida J, Wakabayashi T, Mizuno M, Harada K, Kurisu K, Uozumi T, Sugita K. New approach for the treatment of medulloblastoma by transfection with glial fibrillary acidic protein gene. Surg Oncol. 1996;5:69–75. doi: 10.1016/s0960-7404(96)80003-x. [DOI] [PubMed] [Google Scholar]
- Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321:1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar C, Deb P, Sharma MC. Medulloblastomas: new directions in risk stratification. Neurol India. 2006;54:16–23. doi: 10.4103/0028-3886.24696. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Son EI, Kim IM, Kim DW, Yim MB, Kang YN, Lee SS, Kwon KY, Suh SI, Kwon TK, Lee JJ, Kim DS, Kim SP. Immunohistochemical analysis for histopathological subtypes in pediatric medulloblastomas. Pathol Int. 2003;53:67–73. doi: 10.1046/j.1440-1827.2003.01444.x. [DOI] [PubMed] [Google Scholar]
- Stearns D, Chaudhry A, Abel TW, Burger PC, Dang CV, Eberhart CG. c-Myc overexpression causes anaplasia in medulloblastoma. Cancer Res. 2006;66:673–681. doi: 10.1158/0008-5472.CAN-05-1580. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, Huang KH, Lin WC. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6:1189–1197. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JD, Williamson R, Almefty KK, Nakaji P, Porter R, Tse V, Kalani MY. The many roles of microRNAs in brain tumor biology. Neurosurg Focus. 2010;28:E3. doi: 10.3171/2009.10.FOCUS09207. [DOI] [PubMed] [Google Scholar]
- von Bueren AO, Shalaby T, Rajtarova J, Stearns D, Eberhart CG, Helson L, Arcaro A, Grotzer MA. Anti-proliferative activity of the quassinoid NBT-272 in childhood medulloblastoma cells. BMC Cancer. 2007;7:19. doi: 10.1186/1471-2407-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hoff K, Hartmann W, von Bueren AO, Gerber NU, Grotzer MA, Pietsch T, Rutkowski S. Large cell/anaplastic medulloblastoma: outcome according to myc status, histopathological, and clinical risk factors. Pediatr Blood Cancer. 2010;54:369–376. doi: 10.1002/pbc.22339. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, De Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wiese C, Rolletschek A, Kania G, Blyszczuk P, Tarasov KV, Tarasova Y, Wersto RP, Boheler KR, Wobus AM. Nestin expression—a property of multi-lineage progenitor cells. Cell Mol Life Sci. 2004;61:2510–2522. doi: 10.1007/s00018-004-4144-6. [DOI] [PubMed] [Google Scholar]
- Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng S, Ge W. MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Med Oncol. 2012;29:1037–1043. doi: 10.1007/s12032-011-9975-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.