Abstract
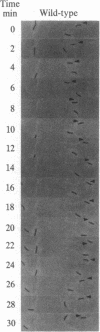
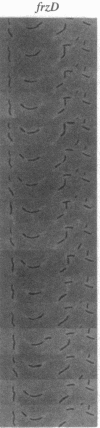
Myxococcus xanthus, a Gram-negative bacterium, has a complex life cycle that includes fruiting body formation. Frizzy (frz) mutants are unable to aggregate normally, instead forming frizzy filamentous aggregates. We have found that these mutants are defective in the control of cell reversal during gliding motility. Wild-type cells reverse their direction of gliding about every 6.8 min; net movement occurs since the interval between reversals can vary widely. The frzA-C, -E and -F mutants reverse their direction of movement very rarely, about once every 2 hr. These mutants cannot aggregate normally and give rise to frizzy filamentous colonies on fruiting agar or motility agar. In contrast, frzD mutants reverse their direction of movement very frequently, about once every 2.2 min; individual cells show little net movement and form smooth-edged "nonmotile" type colonies. Genetic analysis of the frzD locus shows that mutations in this locus can be dominant to the wild-type allele and that its gene product(s) must interact with the other frz gene products. Our results suggest that the frz genes are part of a system responsible for directed movement of this organism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blackhart B. D., Zusman D. R. Cloning and complementation analysis of the "Frizzy" genes of Myxococcus xanthus. Mol Gen Genet. 1985;198(2):243–254. doi: 10.1007/BF00383002. [DOI] [PubMed] [Google Scholar]
- Boyd A., Simon M. Bacterial chemotaxis. Annu Rev Physiol. 1982;44:501–517. doi: 10.1146/annurev.ph.44.030182.002441. [DOI] [PubMed] [Google Scholar]
- Bryan R., Purucker M., Gomes S. L., Alexander W., Shapiro L. Analysis of the pleiotropic regulation of flagellar and chemotaxis gene expression in Caulobacter crescentus by using plasmid complementation. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1341–1345. doi: 10.1073/pnas.81.5.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard R. P. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Dworkin M., Eide D. Myxococcus xanthus does not respond chemotactically to moderate concentration gradients. J Bacteriol. 1983 Apr;154(1):437–442. doi: 10.1128/jb.154.1.437-442.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller K. H., Grady M., Dworkin M. Surface tension gradients: feasible model for gliding motility of Myxococcus xanthus. J Bacteriol. 1983 Sep;155(3):1358–1366. doi: 10.1128/jb.155.3.1358-1366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner J. M., Kaiser D. Introduction of transposon Tn5 into Myxococcus for analysis of developmental and other nonselectable mutants. Proc Natl Acad Sci U S A. 1981 Jan;78(1):425–429. doi: 10.1073/pnas.78.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus I. R., Berg H. C. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982 Jul;151(1):384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C. E., Zusman D. R. Myxococcus xanthus mutants with temperature-sensitive, stage-specific defects: evidence for independent pathways in development. J Bacteriol. 1979 Dec;140(3):1036–1042. doi: 10.1128/jb.140.3.1036-1042.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. J Bacteriol. 1983 Jul;155(1):317–329. doi: 10.1128/jb.155.1.317-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko S. M. Protein and lipid methylation by methionine and S-adenosylmethionine in Myxococcus xanthus. Can J Microbiol. 1983 Sep;29(9):1224–1228. doi: 10.1139/m83-188. [DOI] [PubMed] [Google Scholar]
- Zusman D. R. "Frizzy" mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982 Jun;150(3):1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]