Abstract
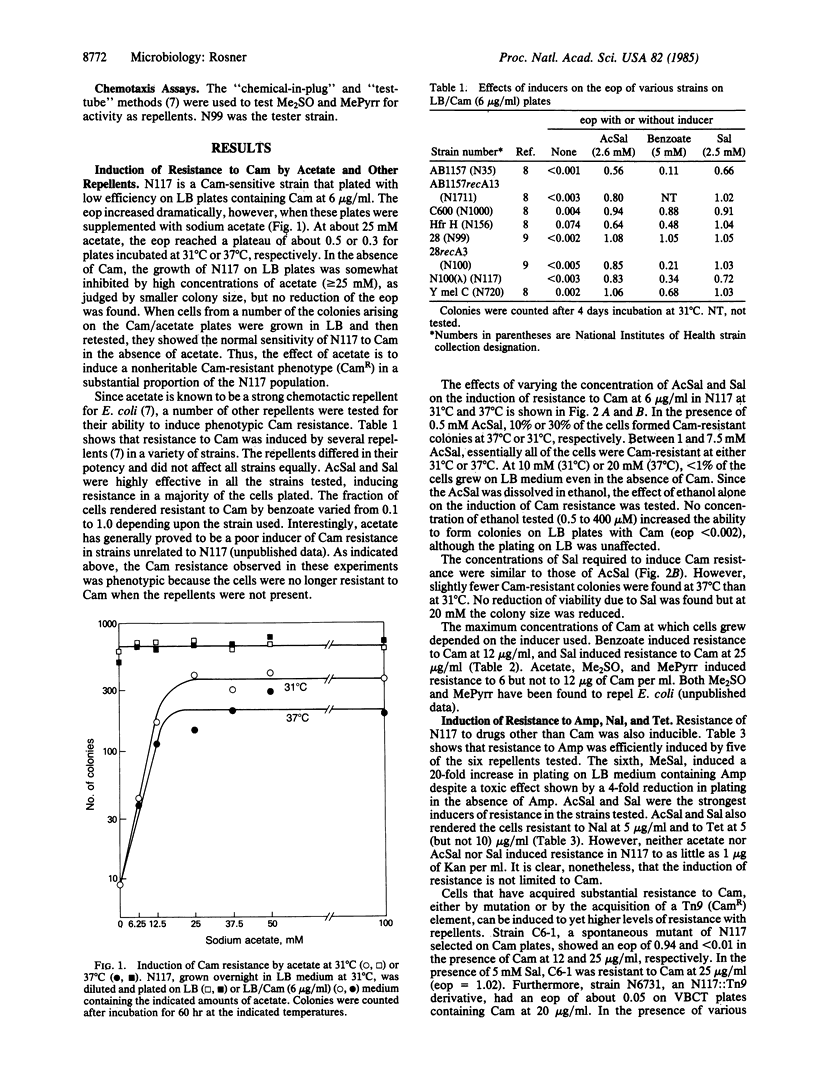
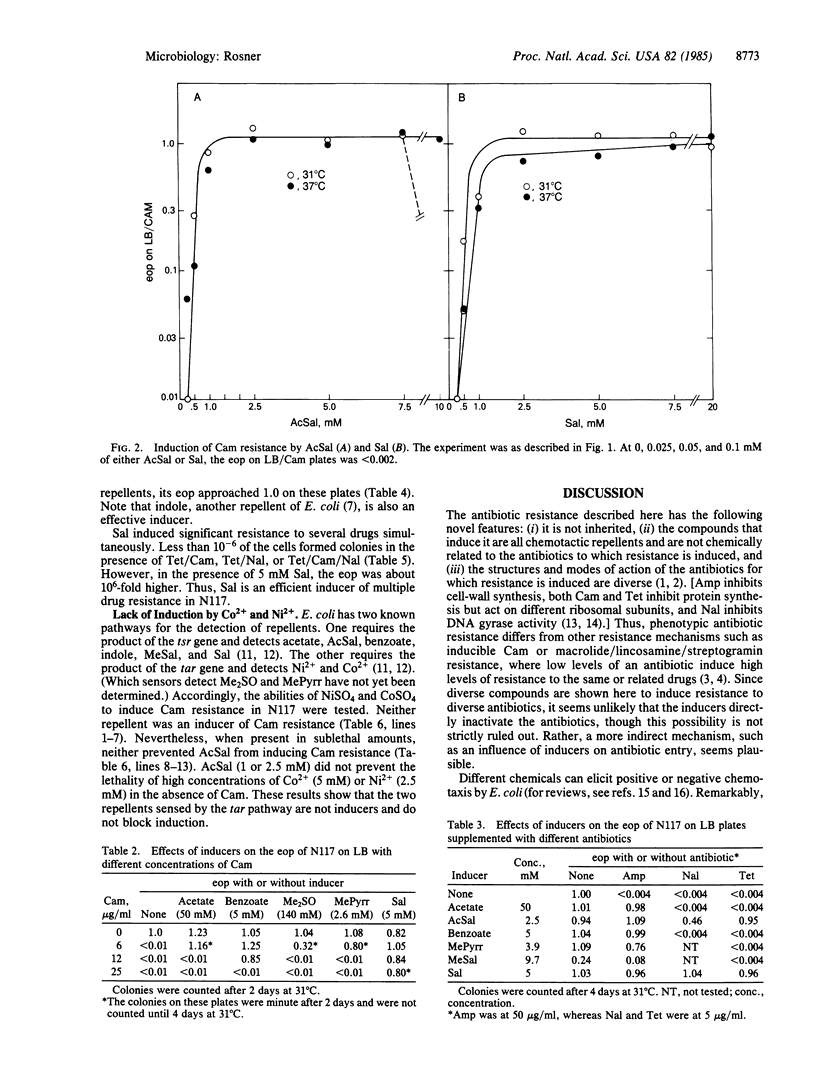
Phenotypic resistance to chloramphenicol and ampicillin was induced in sensitive Escherichia coli K-12 strains during incubation with the following substances: acetate, acetylsalicylate (aspirin), benzoate, dimethyl sulfoxide, 1-methyl-2-pyrrolidinone, and salicylate. In addition, acetyl-salicylate and salicylate induced resistance to nalidixic acid and tetracycline. The induction of resistance was highly efficient but varied somewhat with the strain and inducer used. In the presence of inducers, from 10% to 100% of the cells formed colonies on antibiotic media, an increase of 10- to 1000-fold over the controls without inducer. After growth in the absence of these inducers, the cells were normally sensitive to the antibiotics. Thus, the resistance was not due to a heritable change. These inducers also increased the level of chloramphenicol resistance of a strain carrying cat (whose gene product inactivates chloramphenicol by acetylation). All of the inducers are chemotactic repellents for E. coli, and they are detected by the tsr gene product (with the possible exceptions of dimethyl sulfoxide and methylpyrrolidinone, whose modes of detection are not known). Nickel sulfate and cobalt sulfate, repellents that are detected by the tar gene product, neither promoted resistance to chloramphenicol nor prevented the induction of resistance by acetylsalicylate. Since several of the inducers are present in common drugs or foods, it may be of medical importance to evaluate their effects on antibiotic therapies.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A. R., Randolph B. W., Rosner J. L. Detection of chemicals that stimulate Tn9 transposition in Escherichia coli K12. Mol Gen Genet. 1983;189(2):245–250. doi: 10.1007/BF00337812. [DOI] [PubMed] [Google Scholar]
- Eisenberg E. S., Mandel L. J., Kaback H. R., Miller M. H. Quantitative association between electrical potential across the cytoplasmic membrane and early gentamicin uptake and killing in Staphylococcus aureus. J Bacteriol. 1984 Mar;157(3):863–867. doi: 10.1128/jb.157.3.863-867.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968 Feb 14;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Macnab R. M. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981 Mar;145(3):1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Meyer J., Iida S. Amplification of chloramphenicol resistance transposons carried by phage P1Cm in Escherichia coli. Mol Gen Genet. 1979 Oct 3;176(2):209–219. doi: 10.1007/BF00273215. [DOI] [PubMed] [Google Scholar]
- Repaske D. R., Adler J. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol. 1981 Mar;145(3):1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso W. W., Adler J. Negative chemotaxis in Escherichia coli. J Bacteriol. 1974 May;118(2):560–576. doi: 10.1128/jb.118.2.560-576.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]


