Abstract
In this study, we systematically identify Old Yellow Enzymes (OYEs) from a diverse range of economically important fungi representing different ecology and lifestyle. Using active site residues and sequence alignments, we present a classification for these proteins into three distinct classes including a novel class (Class III) and assign names to sequences. Our in-depth phylogenetic analysis suggests a complex history of lineage-specific expansion and contraction for the OYE gene family in fungi. Comparative analyses reveal remarkable diversity in the number and classes of OYE among fungi. Quantitative real-time PCR (qRT-PCR) of Ascochyta rabiei OYEs indicates differential expression of OYE genes during oxidative stress and plant infection. This study shows relationship of OYE with fungal ecology and lifestyle, and provides a foundation for future functional analysis and characterization of OYE gene family.
Old Yellow Enzymes (OYEs) are flavin-containing oxidoreductases initially isolated from the brewer's bottom yeast Saccharomyces pastorianus (formerly known as S. carlsbergensis) by Warburg and Christian in 19321,2. Enzymes in this family use a reduced flavin mononucleotide (FMN) cofactor and active-site residues to reduce the carbon-carbon double bond in α/β-unsaturated substrates3,4. Subsequently, FMN of these enzymes is restored to its reduced state by NAD(P)H5. Therefore, OYE is often referred to as NADPH dehydrogenase. OYEs catalyze various reactions useful for both biotechnological and pharmaceutical applications; however, natural substrates and true physiological roles for many of these enzymes are presently unknown. Several bacterial OYEs have been shown to reduce a range of xenobiotic explosive compounds such as pentaerythritol tetranitrate (PETN), nitroglycerine (NG), 2,4,6-trinitrotoluene (TNT) and Royal Demolition Explosive (hexahydro-1,3,5-trinitro-1,3,5-triazine; RDX)6,7,8,9,10,11. In addition, certain prokaryotic OYEs reduce morphine alkaloids and N-ethylmaleimide7,12. But these compounds are far from being considered as the true substrates for OYE as they are not synthesized in vivo. Initial indication towards the physiological function of OYE came through the isolation of homologs from plants Arabidopsis thaliana and Solanum lycopersicum13,14. These enzymes catalyze reduction of 12-oxophytodienoate (OPDA), an intermediate in the biosynthesis of jasmonic acid. Later on, homologs of OYE have been shown to reduce a variety of α/β-unsaturated carbonyl compounds viz. acrolein, 9,11-endoperoxide prostaglandin (PG)H2 and chanoclavine-I aldehyde15,16,17,18,19,20,21,22. These findings indicate towards different substrates and diverse physiological functions of OYE homologs in different organisms.
Combinations of site-directed mutagenesis (SDM) and crystal structure studies have characterized active site residues of OYE1 (OYE from S. pastorianus), which are well conserved across the OYE family23,24,25. Oligomerization of OYEs suggests that most of the members are dimeric, whereas few are monomeric in nature. All the OYEs are shown to be a single-domain protein with a (β/α)8 barrel (or TIM barrel) fold. The FMN is bound within the barrel near the carboxy-terminus of the β-sheet. Crystal structure of YqjM (OYE from Bacillus subtilis) for the first time revealed a new class of OYE on the basis of certain unique structural features26. The enzyme exists as a homotetramer and was shown to be a dimer of catalytically dependent dimers. It has an arginine finger (Arg336) that protrudes from one monomer into the active site of the adjacent monomer. Thus displaying shared active site architecture. Further confirmation towards the occurrence of two different classes of OYEs came from the prediction of a set of sequence motifs specific to OYE1-like and YqjM-like proteins42. Homologs of YqjM have been isolated from other bacteria27,28. However, homolog of YqjM has not been reported from any eukaryotic organism till date.
Fungi detect and respond to a variety of signals. As saprophytes or parasites, fungi need to monitor the nutrient status, suppress host defenses and avoid or respond to osmotic or oxidative stress. However, the mechanisms involved in these responses are not properly understood. By catalyzing the reduction of the α,β-olefinic unit in a broad range of substrates, OYE could serve in a defensive role against oxidative stress. Despite availability of fungal genome sequences29,30, reports of OYEs in fungi mostly come from the members of Saccharomycetes, Schizosaccharomycetes and Eurotiomycetes. No homolog has been reported from Sordariomycetes, Leotiomycetes, Dothideomycetes classes of ascomycetes and also from the phylum basidiomycetes. A comprehensive comparative and phylogenetic analysis of OYEs across the fungal kingdom has not been performed yet. In addition, it is also unclear whether the distribution of OYEs in fungi is related to their ecology and lifestyle.
In this study, we identified 424 OYE proteins from 60 fungi and divided them into three conserved classes. Phylogenetic analysis was accomplished to explore the evolutionary significance of OYEs among fungi. In addition, a widespread comparison upon the distribution and abundance of OYEs in different fungi was performed. Subsequently, qRT-PCR based gene expression profiling of Ascochyta rabiei OYEs (ArOYEs) was carried out under oxidative stress and in planta. This study presents the first information about fungal OYE proteins, their nomenclature and classification. In addition, we report a new class of OYE in the genomes of fungi.
Results
Identification of novel OYE proteins in fungi
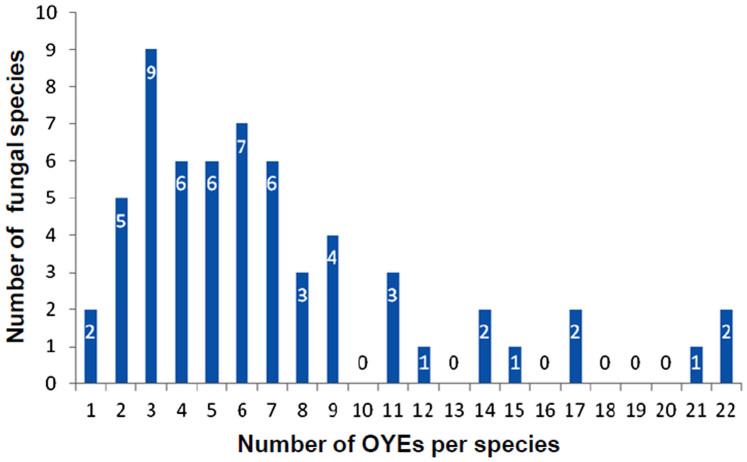
In order to find novel OYE proteins among fungi, genome-wide search was carried out in fungal species representing the dikarya or higher fungi (Supplementary Table S2). This analysis resulted in the identification of 424 OYEs proteins from 60 fungal genomes (Supplementary Table S4). The list includes previously isolated OYEs from Aspergillus fumigatus, Claviceps purpurea, Hansenula polymorpha, Kluyveromyces lactis and Pichia stipitis. All the identified 424 OYEs are well distributed among kingdom fungi and are single domain proteins having conserved TIM phosphate binding domain, which is backbone for OYE family. Within species, the number of OYE homologs per genome varies from one to as many as twenty two, suggesting several rounds of gene duplications and losses (Figure 1). Most species have three (9 species), four (6 species), five (6 species), six (7 species) or seven (6 species) OYE homologs in their genomes and over a half of the fungi analyzed contain more than five OYEs. Pair wise sequence comparisons indicates high sequence divergence among the homologs. However, few active site residues and the loop region containing the YGGS motif are well conserved in all the OYEs. The predicted protein localization of 424 OYEs varies from cytoplasm (259), mitochondria (127) to cytoskeleton (29). Interestingly, few OYEs are predicted to be localized in nucleus (3), peroxisomes (3), extracellular (2) and plasma membrane (1) (Supplementary Figure S1, Supplementary Table S4).
Figure 1. The distribution of OYE homologues in 60 fungi.
Blue bars showed the numbers of fungi that have the members of specific OYE family.
Classification, amino acid sequence comparison and nomenclature of OYEs
The high sequence divergence among members of the OYE family indicates for more than one Class of OYE present in fungal genomes. To investigate this, surveys of the active site residues of isolated sequences were carried out. Sequence analysis suggested that majority of sequences have the active site composition of OYE1 (OYE from S. pastorianus). Interestingly, many sequences showed the conserved active site residues of YqjM (OYE from B. subtilis). On the contrary, there was good number of sequences, which showed no conservation with the active site residues of either OYE1 or YqjM. These findings indicate towards three classes of OYE present in fungal genomes.
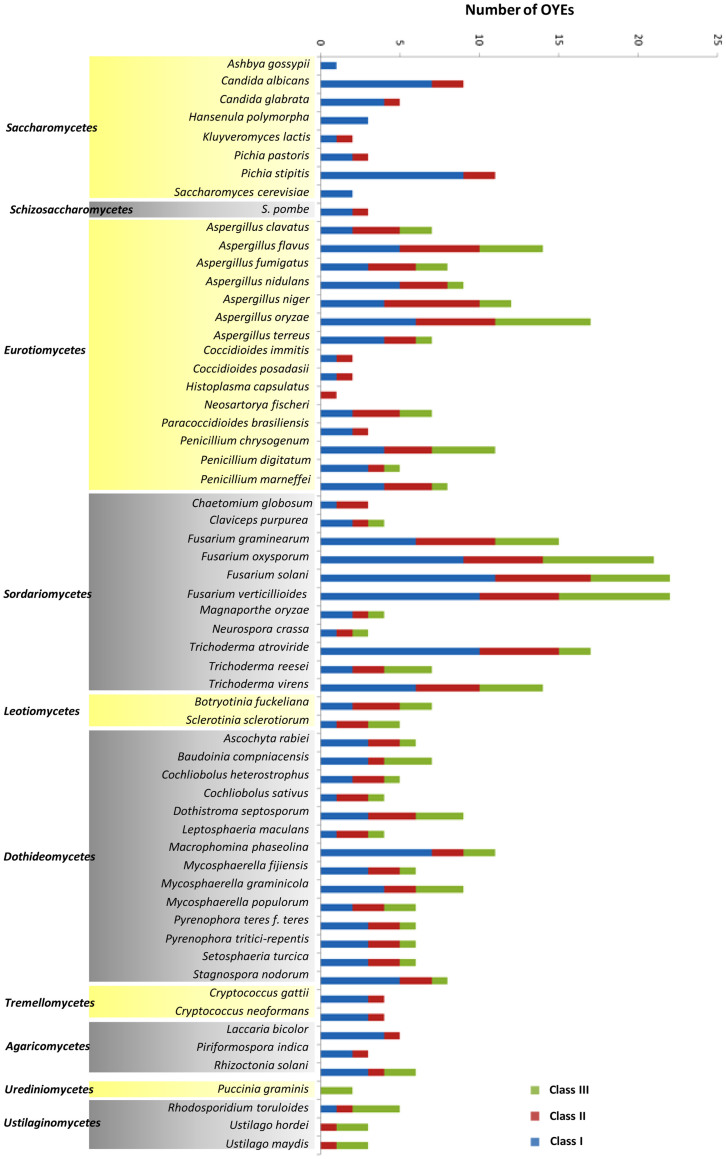
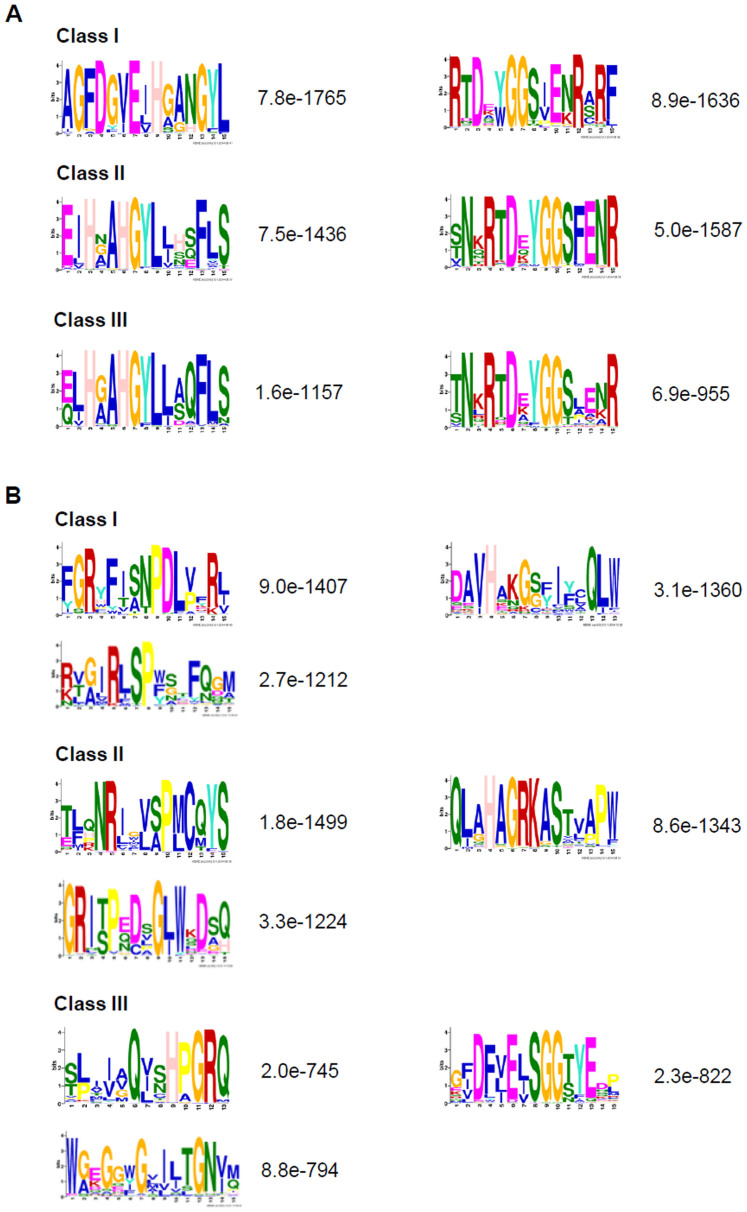
To gain further insight into different classes of OYEs, three HMM profiles representing three OYE classes were created. Thus, out of 424 sequences, 199, 127 and 98 sequences were grouped into Class I, II and III, respectively (Figure 2). In order to confirm this classification, comprehensive class wise sequence alignments were carried out. Comparing the sequences of Class I members clearly showed that the active site residues His191, Asn194, Tyr196, Phe250 and Tyr375 of OYE1 are either well conserved or substituted with similar amino acids in almost all proteins (Figure 3A, Supplementary Figure S2B). In a similar way, sequence comparison of Class II members was performed. The active site residues Cys26, Tyr28, Lys109, His164, His167, Tyr169 of YqjM are well conserved in OYE members with only few incidences of substitution with similar amino acids (Figure 3A, Supplementary Figure S2D, S2F). Sequence alignment of Class III members revealed strict conservation of YGGS motif and the core active site residues (His191, Asn194, and Tyr196 of OYE1 or His164, His167 and Tyr169 of YqjM) among the members (Figure 3A, Supplementary Figure S2G, S2H). In contrast, residues that make the active site of Class I or Class II proteins are not present in these OYEs. This suggests difference in active site residues of Class III OYEs. In addition to the conserved motifs present in all the three OYE classes we identified few motifs specific to certain OYE Class (Figure 3, Supplementary Figure S2). To analyze the structural conservation of these motifs, the conserved and Class I and II specific motifs were located in the structures of previously reported OYEs (OYE1 and YqjM) (Supplementary Figure S3). Taking together, these results indicate towards difference in active site organization and hence divergence in enzymatic and molecular functions of these OYEs.
Figure 2. Comparative analysis of fungal OYEs.
The numbers of OYEs in each species are represented as horizontal bars. Different colors of the bar represent different classes of OYE.
Figure 3. Conserved motifs in ArOYEs.
(A) Conserved core active site and YGGS motif arrangement among the three OYE Classes, and (B) OYE Class specific conserved motifs. The result shows the top five hits that were obtained through MEME analysis of each OYE Class. Numbers on the right shows the E-value of each motif. Some of these motifs correlate with the motifs identified by Oberdorfer et al.42.
Our study reveals that there are many fungal species which have more than one member of OYE in a certain Class. Since there wasn't any proper nomenclature system available for isolated OYEs therefore, it was decided to develop a proper naming method for OYE proteins. To accomplish this, two ways naming system was devised to name OYEs in a fungal species. Firstly, numbering was specified starting from Class I members followed by Class II and then Class III. Secondly, numbering within a class was assigned on the basis of similarity of members with the representative protein of that class. For example, to name Class I OYEs of a fungal species, a consecutive numbering based on the order of their similarity with yeast OYE1 was assigned. In the same way, similarity with YqjM was the basis for numbering Class II OYEs present among fungal species. ArOYE6, the first identified member of Class III, was taken as representative protein. In this way, all the 424 OYEs from 60 fungal species were properly named for the convenience of scientific community (Supplementary Table S4).
Comparing abundance of OYEs among fungi
Comparison of ascomycetes and basidiomycetes suggested more representation of OYEs in ascomycetes (389 homologs in 51 species) than basidiomycetes (35 homologs in 9 species) (Supplementary Figure S4A). In general, among ascomycetes Sordariomycetes (132 homologs in 11 species) and Eurotiomycetes (113 homologs in 15 species) contain more OYEs while Saccharomycetes (36 homologs in 8 species) and Leotiomycetes (12 homologs in 2 species) have fewer (Figure 2, Supplementary Figure S4B). A substantial increase in gene copy number was observed in species of Eurotiomycetes including Aspergillus flavus (14 genes), Aspergillus niger (12 genes) and Aspergillus oryzae (17 genes), as well as in the Sordariomycetes Fusarium oxysporum (21 genes), Fusarium solani (22 genes) and Fusarium verticillioides (22 genes).
Comparison of different OYE classes among fungi revealed interesting results. Class III OYEs are completely absent in all the Saccharomycetes species studied in this analysis (Figure 2, Supplementary Figure S4B). In addition, three out of eight species studied were lacking Class II proteins. Class I and II members are well represented in ascomycetes fungi except Histoplasma capsulatus which lacks Class I OYE. Among basidiomycetes, Class I is absent in 3 while Class II is absent in 4 out of 8 species analyzed. Species with all the three classes of OYE are Rhizoctonia solani and Rhodosporidium toruloides.
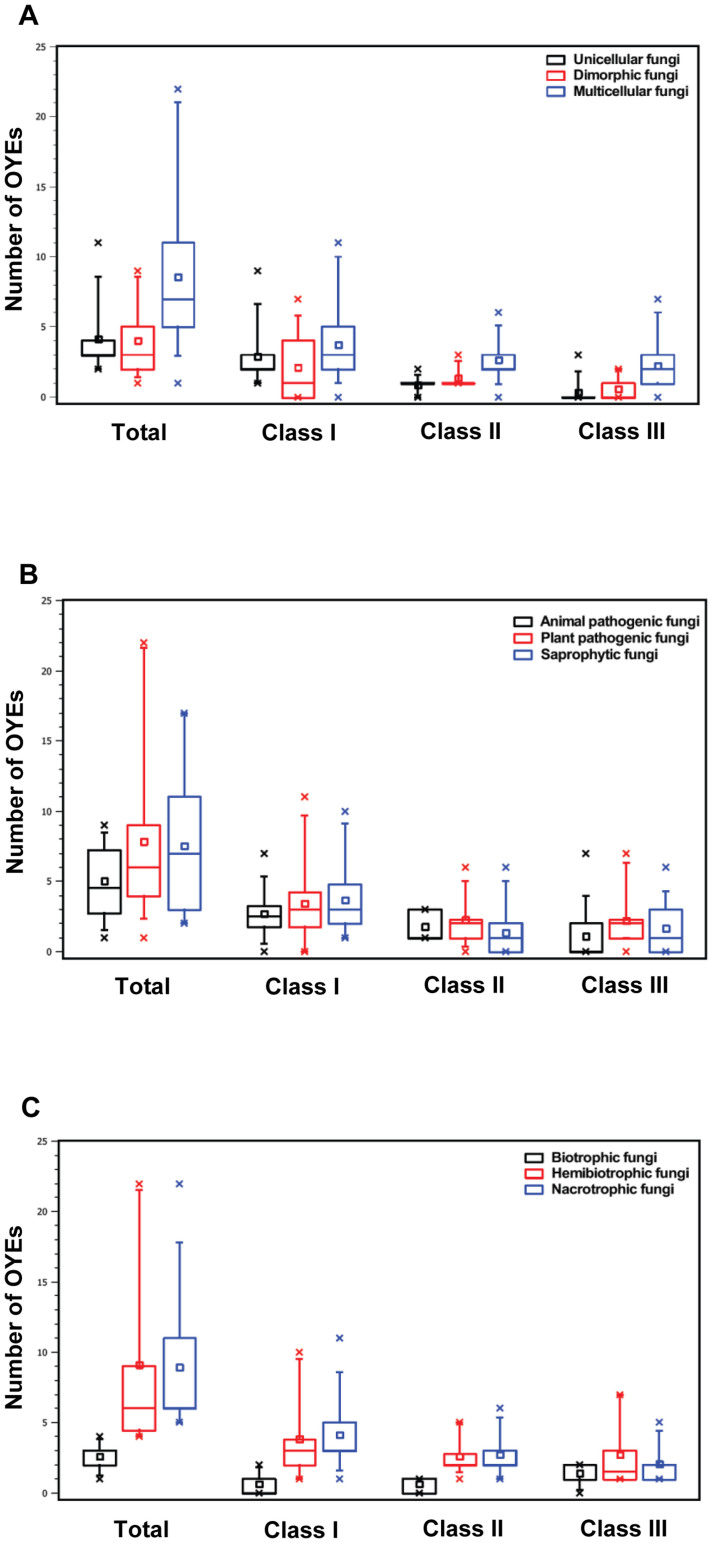
To analyse the distribution of OYE superfamily associated with fungal growth, fungi were classified as 1) unicellular fungi, 2) dimorphic fungi, and 3) multicellular fungi. The comparison suggested that unicellular fungi (37 homologs in 9 species) and dimorphic fungi (36 homologs in 9 species) have less number of OYEs compared to multicellular fungi (343 homologs in 40 species) (Figure 4A). Class I, II and III OYE proteins were mostly associated with multicellular fungi (43%, 54%, 72%) in comparison to dimorphic (24%, 28%, 18%) and unicellular (33%, 18%, 10%) fungi.
Figure 4. Comparison of OYEs among fungi, (A) among unicellular fungi (black), dimorphic fungi (red) and multi-cellular fungi (blue), (B) among animal pathogenic fungi (black), plant pathogenic fungi (red) and saprophytic fungi (blue) and (C) among biotrophic (black), hemibiotrophic (red), and necrotrophic (blue) fungi.
To test the distribution of OYE superfamily associated with species ecology, fungi studied were classified as 1) animal pathogenic fungi, 2) plant pathogenic fungi, and 3) saprophytic fungi. The comparison suggested that plant pathogenic fungi (220 homologs in 28 species) and saprophytic fungi (136 homologs in 18 species) have more number of OYEs compared to animal pathogenic fungi (60 homologs in 12 species) (Figure 4B). Class III OYE proteins were mostly associated with plant pathogenic (49%) and saprophytic (38%) fungi in comparison to animal pathogens (13%). Similarly, Class I and II proteins were almost equally distributed between plant pathogens and saprophytes but less in animal pathogenic fungi.
In order to analyze the distribution of OYE superfamily associated with plant pathogenic fungi lifestyles, they were grouped as 1) biotrophic fungi, 2) hemibiotrophic fungi, and 3) necrotrophic fungi. The analysis suggested that necrotrophic fungi (116 homologs in 13 species) and hemibiotrophic fungi (91 homologs in 10 species) have more number of OYEs compared to biotrophic fungi (13 homologs in 5 species) (Figure 4C). Class I, II and III proteins were mostly associated with necrotrophic (49, 46, 33%) and hemibiotrophic (44, 44, 44%) fungi in comparison to biotrophic (7, 10, 23%) fungi.
Phylogenetic and evolutionary analysis of OYE proteins
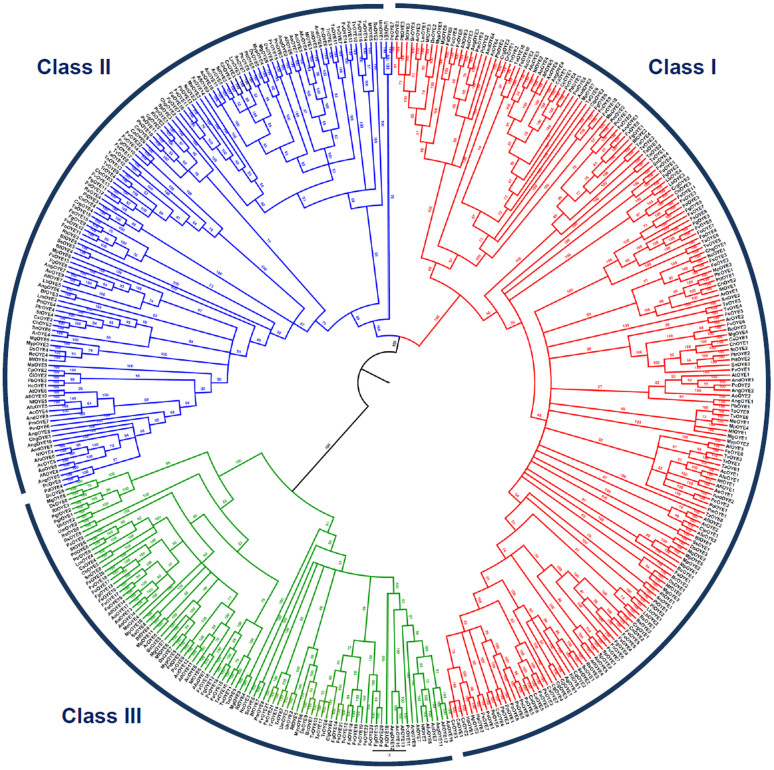
In order to gain better insights into the evolutionary relationships of OYEs among fungi, a comprehensive phylogenetic analysis was performed. Due to high sequence divergence among OYEs, the multiple sequence and structure-based alignment was carried out using PROMALS3D to generate phylogenetic trees using Bayesian inference (BI) and Maximum Likelihood (ML) approaches. Both the methods had nearly identical tree topologies; however, BI provided relatively higher posterior probabilities. Considering topology and robust clade support values, the tree classified the 424 OYEs proteins into three distinct groups on the basis of their class viz. Class I, Class II and Class III (Figure 5). It reassures the presence of three discrete classes of OYEs. The number of sub-groups in Class I are relatively more in comparison to Class II and III indicating that Class I is more diversified.
Figure 5. Evolutionary relationship of 424 deduced OYE proteins from 60 fungal species based on Bayesian inference analysis of the structure-based amino acid sequence alignment.
The multiple sequence alignment of full length OYE proteins was carried out using multiple sequence and structure alignment program PROMALS3D. The numbers at the nodes indicates the Bayesian posterior probabilities. The OYE proteins were classified into three distinct classes, designated as Class I, II, and III. Different colour was assigned to each class.
The phylogenetic tree showed absence of any species specific clades, except for the class Saccharomycetes, pointing towards high diversification of OYEs during evolution. Interestingly, all the OYE proteins from the species of Saccharomycetes clustered together in a single clade both in Class I as well as in Class II (Figure 5). One could thus infer that OYEs of Saccharomycetes deviated from that of other fungal classes in the beginning during the course of evolution and evolved distinctly. In contrast, OYE proteins of other fungal classes are clustered into several small groups supporting the orthology of these sequences across the entire phylogeny. Nevertheless, there are several indications of gene duplication as evident by the substantial number of putative paralogs. The phylum ascomycetes witnessed higher rates of gene duplication as compared to basidiomycetes. The gene duplication events were apparent at fungal class level also. In all the fungal classes, within species, OYEs of numerous fungi descended from the same node suggesting them as paralogs. Subsequent groupings can be seen accommodating the latter paralogs. These paralogs were derived from the nodes that had high branch support in both BI and ML analysis, pointing towards a common ancestor. In ascomycetes, large-scale expansion of OYEs was observed in species of Sordariomycetes due to multiple independent rounds of gene duplications. It is then followed by Eurotiomycetes and Dothideomycetes with a notable exception of Saccharomycetes and Leotiomycetes where fewer incidences of gene duplication were observed. However, among Saccharomycetes, C. albicans and P. stipitis displayed certain events of gene duplications (Figure 5).
Further investigation provided a unique observation. In Class I, OYE of F. solani (FsOYE11) belonging to ascomycetes, grouped together with OYEs of basidiomycetes L. bicolor, C. gattii and C. neoformans (LbOYE3, LbOYE4, CrgOYE3 and CnOYE3) in both BI and ML trees with high branch support values (Figure 5). Such evolutionary close relationship between protein sequences of taxonomically distant organisms over two phyla is unusual. When BLASTP analysis of FsOYE11 was performed, surprisingly, it failed to show any match in the fungal genome except for the above mentioned basidiomycetes OYEs. Similarly, in Class III, OYE of ascomycetes M. graminicola (MgOYE9) grouped together with OYE of basidiomycetes R. toruloides (RtOYE4). On performing their BLASTP analysis against nr database, no homolog could be found in any of the fungal genome. However, these sequences showed high sequence identity to many bacterial OYEs. Taking together these findings indicate that certain fungal OYEs diversified from the rest and evolved discretely.
Lineage specific expansion and contraction of OYE gene family
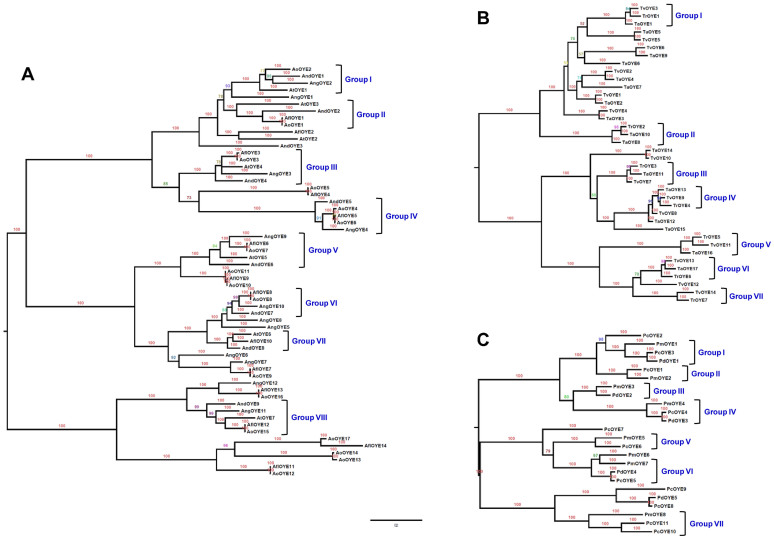
In order to gain better insight of the mode of evolution of OYE family and its lineage specific expansion and contraction, we examined OYE proteins from closely related species having a substantial number of OYE paralogs. For this, five species of Aspergillus (A. nidulans: 9, A. flavus: 14, A. oryzae: 17, A. terreus: 7 and A. niger: 12), and three species each of Trichoderma (T. reesei: 7, T. atroviride: 17, and T. virens: 14) and Penicillium (P. marneffei: 8, P. chrysogenum: 11 and P. digitatum: 5) were analysed (Figure 6). BI analysis of Aspergillus sp. identified eight distinct groups of orthologs (Group I–VIII; Figure 6A), which were supported by strong posterior probability values. Similarly, seven ortholog groups were present both in Trichoderma sp. and Penicillium sp. (Group I–VII; Figure 6B, 6C). In Aspergillus sp., Group III, VII and VIII contained orthologs from all the five species whereas Group IV and V consisted of orthologs from all except A. terreus, suggesting the loss of these orthologs in this species. Similarly, A. niger also seem to have lost the respective orthologs from Group II and VI. Interestingly, ortholog of AndOYE3 was missing from rest of the Aspergillus sp. indicating that this particular OYE has been lost during the course of evolution. There are several duplication events associated with A. oryzae, A. flavus and A. niger leading to the expansion of OYEs in these species. In Penicillium sp., Group I, IV and VI represented orthologs from all three species while Group II, V and VII orthologs were absent in P. digitatum pointing towards the contraction of OYEs in this speices. On the other hand, numerous duplication events indicate the expansion of OYEs in P. chrysogenum. In Trichoderma sp., out of seven, five groups (Group I, III, IV, V and VI) contained orthologs from all the three species. However, Group II and VII ortholog was absent in T. virens and T. atroviride, respectively. Many events of duplication were observed both in T. virens and T. atroviride signifying the lineage specific expansion of OYEs in these two species from the ancestral species, T. reesei. These findings indicate that OYE gene family has evolved due to multiple independent episodes of expansions and contractions across different fungal lineages.
Figure 6. Lineage specific expansion and contraction of OYE gene family.
Evolutionary relationships were inferred by the Bayesian Markov Chain Monte Carlo (MCMC) for (A) Aspergillus sp., (B) Trichoderma sp. and (C) Penicillium sp. For each node, Bayesian posterior probabilities supporting the phylogeny are indicated. The phylogeny shows clustering based mainly on orthology instead of paralogy, indicating that the expansion of OYEs occurred prior to speciation. In all the three species analysed, several groups display additional instances of gene duplications and losses supporting for birth-and-death model of evolution.
Chromosomal distribution - implications for evolution of OYE family
To explore the pattern of chromosomal distribution of OYE genes, the position of OYEs were mapped on chromosomes of eight fungal species viz. S. cerevisiae, A. fumigatus, C. neoformans, C. albicans, U. maydis, F. graminearum, M. oryzae and A. nidulans. In silico mapping suggested non-uniform distribution and density of the OYE genes on chromosomes (Supplementary Figure S5). All the Class I, II and III OYEs were arbitrarily distributed throughout the different chromosomes. In all the species, very few events of tandem duplication were observed and there were more segmental duplication events. Out of eight fungal species analysed, only C. albicans and F. graminearum showed few incidences of tandem duplication. However, segmental duplication was evident in all the eight fungal species. In C. albicans, two events of tandem duplication were observed. Four gene clusters belonging to Class I (CaOYE1, CaOYE2, CaOYE3 and CaOYE4) were located in tandem on chromosome 6 and similarly, CaOYE5 and CaOYE7 of same Class were tandemly duplicated on chromosome R. This suggests that multiple copies have arisen by consecutive duplication events. In F. graminearum also, FgOYE2, FgOYE3 and FgOYE5 of Class I were present in tandem duplication form on chromosome 2. Together it infers that successive segmental and tandem duplication events contributed to the expansion of OYE gene family.
Expression analysis of A. rabiei OYE genes
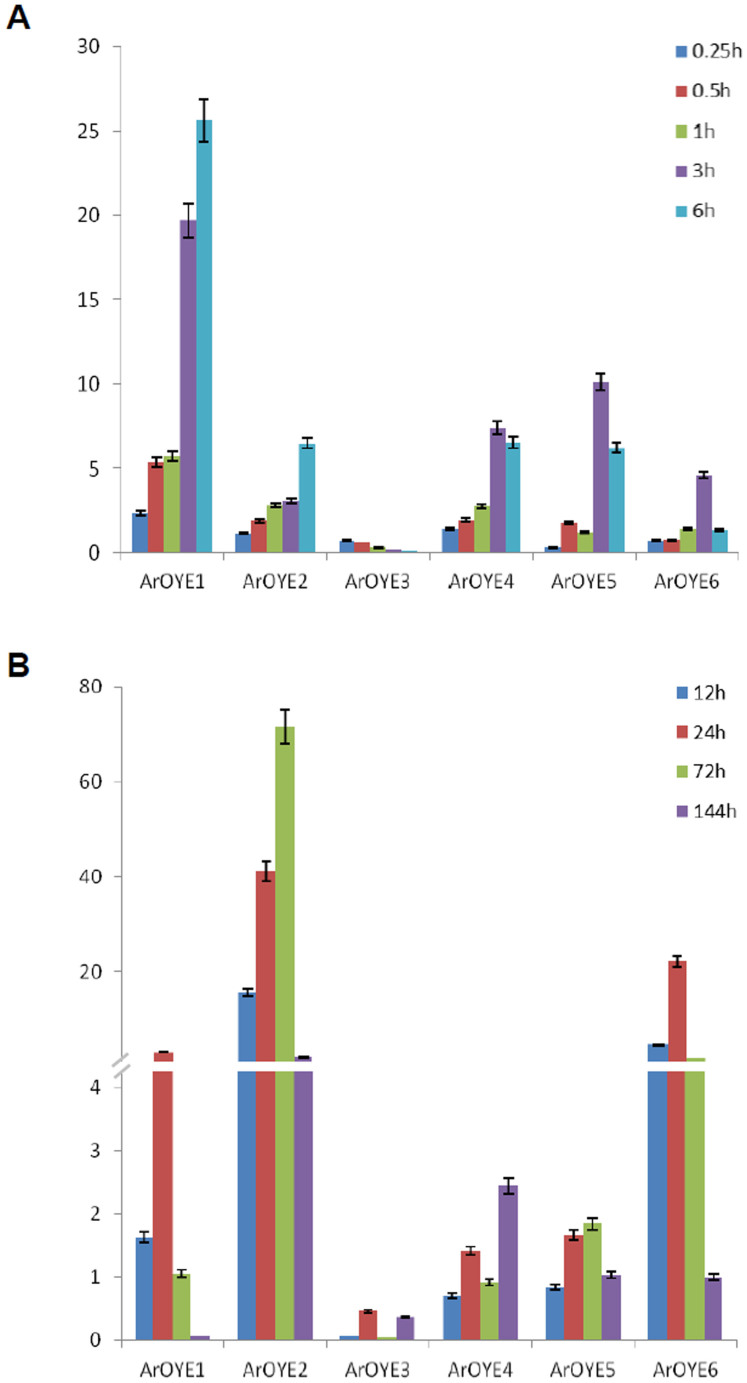
Previously in our lab, transcript profiling of A. rabiei under oxidative stress was performed to elucidate fungal survival during pathogenesis31. Expression analysis using qRT-PCR and northern blotting showed differential expression patterns of several genes under oxidative stress and plant infection (in planta). Therefore, to investigate the induction of ArOYEs during oxidative stress and in planta, expression levels of the OYE genes under menadione treatment and infected chickpea were assayed by qRT-PCR (Figure 7). All the genes were found differentially expressed under above mentioned conditions. In comparison with other OYE genes, the abundance of ArOYE3 transcripts was lowest in oxidative stress as well as in planta. The ArOYE1 gene had the highest expression level under oxidative stress but relatively lower expression in planta. In contrast, both ArOYE2 and ArOYE6 showed higher expression levels in planta (Figure 7A) but relatively lower expression under oxidative stress (Figure 7B), suggesting their important roles during plant infection. The ArOYE4 and ArOYE5 genes showed similar expression profiles with higher expression under oxidative stress and lower expression in planta, suggesting their more pronounced roles during oxidative stress.
Figure 7. Expression profiles of six ArOYE genes during oxidative stress and in planta, assayed by qRT-PCR.
Bar diagrams representing the expression pattern of 6 genes are shown as the fold-change compared to the control. (A) Expression was analyzed at 0.25, 0.5, 1, 3 and 6 h after treatment with menadione. (B) Expression pattern of 6 genes from A. rabiei-infected chickpea samples are shown as fold-change compared to control. Expression was analyzed at 12, 24, 72 and 144 hours post inoculation (hpi). Error bars represent ±SE for three biological replicates. ArEF1α gene was used as an internal reference gene.
Discussion
Early studies of Old Yellow Enzyme demonstrated several important biochemical concepts, giving this enzyme a significant place in the history of enzymology. Since the identification of gene encoding OYE, several members of the OYE family have been isolated from many organisms including bacteria, yeasts, plants and filamentous fungi6,7,8,9,10,11,12,13,14,15,16. Structural and biochemical characterization of these enzymes have shown considerable differences between the enzymatic activities towards a diverse range of substrates. Given the broad substrate specificity of OYE family members in vitro, it appears unlikely that the enzyme has a single specific physiological substrate in vivo. In addition, more than one homolog of this protein is present in many organisms including yeasts. This indicates two possibilities, either the true physiological substrate has not been found yet or the metabolic function varies between organisms. Therefore, to gain some idea regarding the physiological function of OYE and to gain insight about distribution of these proteins in fungi, systematic identification of 424 OYEs was performed in the predicted proteomes of 60 fungal species. Earlier studies proposed differential function of OYEs within a species19. Our sub-cellular localization prediction suggested different locations of OYEs within a fungal species. The predicted localization of OYE homologs in a species varied from cytoplasm, mitochondria, to cytoskeleton and in few instances to nucleus, peroxisome, extracellular and plasma membrane. These results substantiates towards different physiological functions of the OYEs within a species. Based upon the phylogenetic analysis and conservation of active site residues, all identified OYE proteins were grouped into three classes. Most of the previously reported OYEs were found to be the members of Class I OYE. Till date, reports of Class II members are only from prokaryotes. Our analysis has revealed several members of this class in the genomes of fungi. However, there is no report of Class III member from any organism and our study suggests that this class is well represented in filamentous fungi. In this way, we report new classes of OYE in fungi. For the convenience of scientific community, all the OYEs were systematically named and provide a basis for the nomenclature of future homologs.
Genomic innovation is mainly driven by gene duplication which results in creating new genes. Functional diversification of such newly introduced genes works as a deciding factor for their retention in the genome. The evolutionary diversification of OYEs was explored to understand the biological roles of these proteins in fungi. Multiple paralogs present in several species indicates that OYE family diversified within the fungal kingdom. For instance, in several fungi of class Saccharomycetes, same nodes give rise to multiple OYEs descendants (Figure 5). This indicates their close relation to each other and/or same functional characteristics. Since OYE family is present both in ascomycetes and basidiomycetes, it is suggestive of its origin before the divergence of Dikarya. Our analysis and the presence of paralogs suggest several independent rounds of gene duplication events in OYE family during the course of evolution. Comprehensive phylogenetic analyses indicate lineage-specific variations in closely related species (Figure 6). For instance, the ancestral species, A. nidulans showed significant gene duplications, with each duplicated gene lineage, except AndOYE3, passed into a range of species (Figure 6A). Interestingly, all lineages are not represented in all the Aspergillus species. In A. terreus, there is loss of Group IV and V orthologs, suggestive of a birth-and-death model of gene evolution41. High rate of variations in the copy number is apparent even among closely related species. Moreover, chromosomal mapping suggests that OYE genes are clustered more on the basis of orthology instead of paralogy (Supplementary Figure S5). Taken together, these analyses indicate towards the diverse and significant expansion and contraction of the OYE gene family. Furthermore, the OYE paralogs exhibit high levels of sequence divergence and are thoroughly dispersed in the genome suggestive of rapid rate of evolution after duplication. The distribution pattern of OYEs on individual chromosomes revealed few physical regions that display comparatively higher accumulation of gene clusters. All these factors make OYE family highly diverse.
Comparing abundance of OYEs indicated highly variable distribution of OYE genes in the different fungal species. The driving force behind the patchy distribution of OYEs in different fungal species is unknown but likely relates to their uncharacterized physiological function. In addition, these analyses indicate that OYEs perform taxon-specific roles. They can provide conditional advantages in specific environmental niches. One possibility is that OYEs are gained or lost after speciation events according to niche-specific selection pressures. This might help in fine-tuning environmental and/or parasitic vigor. Such a dynamic response to the environmental stimuli involving high rates of gains and losses is frequently observed in stress-response genes. Particularly the genes involved in interactions with other organisms, adaptive immunity or patho-adaptation to new hosts have high rates of expansion and contraction. Our comparative analysis provides substantial evidences for this possibility. We found that animal pathogenic and plant pathogenic fungi differ significantly in the number of OYEs. Furthermore, lineage or species-specific gene duplication (gain) events also have occurred within many fungal taxa, particularly in plant pathogens. Since fungi are constantly exposed to host generated reactive oxygen species (ROS) and they are expected to grow and proliferate under such hostile environment, therefore, the correlation of fungal survival under oxidative stress and subsequent pathogenicity is quite important. Previous reports suggest that OYE is induced during oxidative stress19. Our qRT-PCR expression analysis of ArOYE1-6 suggests conditional induction of these genes during oxidative stress and in planta (Figure 7). This result strengthens the fact that various OYE homologs might have acquired different function during evolution when they were undergoing gene duplication or/and deletion. Our results thus indicate towards the functional relevance of ArOYE homologs with that of gene evolution.
In conclusion, this study provides the first report for identification of OYE proteins from diverse fungi and their classification into three different classes. Comparative analysis at different phylogenetic depths indicates a complex history of birth-and-death evolution of the OYE gene family. These analyses suggest the physiological roles of OYE family members in stress response and host-microbe interaction. Moreover, our results indicate different metabolic functions of these proteins among fungi. In addition, this study advocates the relationship of OYE gene number and Class with the ecology and lifestyle of fungi. Future work on molecular characterizations and functional studies that include gene deletion and/or repression may further improve our understanding of the diverse enzymatic and molecular functions of OYE proteins. Additionally, more comprehensive evaluations of the functions of each OYE under different stress conditions within a fungal species will provide valuable insights regarding the roles of these genes in stress. Furthermore, class-wise detailed analysis of OYEs should reveal the relationship of OYE classes required for host–fungal interactions.
Methods
Identification of OYE proteins from Ascochyta rabiei
In order to identify putative OYE proteins from the genome of Ascochyta rabiei, 21 OYE sequences previously reported from different species of bacteria, yeasts, and plants were retrieved from database (Supplementary Table S1). These sequences were used to perform a TBLASTN against the genome sequence of A. rabiei (unpublished data) using default parameters. This resulted in 6 hits as subject sequences. The full length nucleotide sequence of each hit was extracted from genome sequence. The nucleotide sequences were analyzed using BLASTX server to mark the start and stop codons as well as introns. After removal of the introns the protein sequences were deduced by translating them using EXPASY translate tool (http://web.expasy.org/tools/translate/). Individual BLASTP analysis of each putative OYE was carried out to check redundancy in the protein sequences.
Sequence and database search for fungal OYEs
In an attempt to obtain all the OYEs, the predicted proteomes of 58 fungi were downloaded mainly from Fungal Genome Initiative (FGI) site of Broad Institute (http://www.broadinstitute.org/science/projects/projects) and DOE Joint Genome Institute (JGI) site. For Claviceps purpurea, non-redundant protein database of NCBI was used. Supplementary Table S2 describes the list of fungi used in this study. The query consisted of 27 OYE sequences, consisting of 21 sequences used previously and ArOYE6 together with five best hits that we got during its BLASTP analysis from filamentous fungi including Mycosphaerella graminicola, Stagonospora nodorum, Leptosphaeria maculans, Pyrenophora teres f. teres and Pyrenophora tritici-repentis. The search was carried out in three steps. The first step consisted of performing PSI-BLAST32 to search against individual database for each fungus with a total of ~612,922 annotated protein sequences from 59 species using the 27 protein queries, with an E-value threshold of 10−5. This resulted in 447 hits as subject sequences. Six OYEs from A. rabiei were also included in this list making total of 453 hits. In the second step, HMM was built using hmmbuild based on the alignment of all query sequences and the sequences identified from the previous step were matched against these HMMs using HMMScan33 (HMMER version 3.1b1) with an E-value threshold of 10−2. This resulted in the reduction of the number of OYEs to 436. In the third step, self BLAST along with careful manual inspection of the sequences was carried out to remove the redundant and truncated sequences resulting in the identification of 424 OYE proteins from 60 fungal genomes. Subcellular localization prediction of each of the fungal OYEs was carried out using the WoLF PSORT server (http://www.genscript.com/psort/wolf_psort.html).
Classification of fungal OYEs
To gain further insight into different classes of OYEs, three HMM profiles representing three OYE classes were created. The first profile was built taking eight sequences together with OYE1 and its homologs from yeasts, A. fumigatus and C. purpurea. The second profile was created using 7 sequences including YqjM and its homologs. For the third profile, ArOYE6 along with its five best matches from the ascomycetes classes Eurotiomycetes, Sordariomycetes, Leotiomycetes and Dothideomycetes were used. All the three HMM profiles were individually matched against 424 OYE protein sequences using HMMScan. In this way, three scores were obtained for each protein sequence. On the basis of the best score, each protein was assigned to its respective class.
Sequence alignment, phylogenetic analyses and identification of conserved motifs
Full-length OYE protein sequences were aligned with multiple sequence and structure alignment program PROMALS3D (http://prodata.swmed.edu/promals3d)34 and trimmed using Gblocks (v0.91b) (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) with less stringent parameter settings35.The acquired alignment of 424 OYEs was used to carry out phylogenetic analyses using two different approaches. In the first approach, the phylogenetic tree was derived by Bayesian Inference (BI) implemented in MrBayes (v3.2.2)36. Under a mixed amino acid substitution model, the protein sequence alignments were run over 3,000,000 generations with two independent runs each containing four Markov Chain Monte Carlo (MCMC) chains and a sampling frequency of every 300 iterations with MCMC left at default settings to estimate the posterior probabilities for each nodes. The trace files generated by the Bayesian MCMC runs were used to inspect convergence of the runs using Tracer software (v1.5) (http://tree.bio.ed.ac.uk/software/tracer/). Finally, using the Sumt function of MrBayes consensus tree was generated and posterior probabilities were estimated by removing the burn-in generations for each run. In a second approach, Maximum Likelihood (ML) was performed using RAxML37 as executed in T-REX server (http://www.trex.uqam.ca/)38. ProtTest39 of OYE protein sequence alignment was performed to predict the best-fit evolutionary models for ML analysis. Best likelihood scores for all criteria (AIC and BIC) were predicted for LG + I + G model followed by WAG + I + G model. Thus, WAG + I + G model was used to construct ML tree by RAxML. Statistical reliability was determined by conducting bootstrap analysis for 100 replicates. For phylogenetic analyses of Aspergillus sp., Penicillium sp. and Trichoderma sp., the full-length OYE protein sequences aligned using PROMALS3D were directly used for Bayesian analyses with ngen = 1,000,000 and samplefreq = 100. All the phylogenetic trees were visualized using FigTree (v1.4.) (http://tree.bio.ed.ac.uk/software/figtree/). To detect potential conserved motifs, the program MEME (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) was used with following settings, distribution of motif was set to one per sequence, width of the motif was set to 12 for minimum and 15 for maximum, and the maximum number of motifs was set to 5.
Mapping OYE genes on chromosomes
The OYE genes of certain fungi were searched in their respective genome by using corresponding genome browser. The genome database of A. fumigatus, A. nidulans (http://www.aspergillusgenome.org/), C. albicans (http://www.candidagenome.org/), S. cerevisiae (http://www.yeastgenome.org/), C. neoformans (http://www.broadinstitute.org), M. oryzae and U. maydis (http://www.ncbi.nlm.nih.gov/projects/mapview/) were searched. Precise chromosomal positions of the genes encoding OYE proteins were determined by TBLASTN search of the sequences against the respective genome database. The genes were then located on the chromosomes as per their physical position in ascending order. Finally, the chromosomal locations of OYE genes were displayed using MapChart40.
Fungal isolates, stress treatment and plant infection
Cultures of Ascochyta rabiei (Delhi isolate, D-11) were grown on Potato Dextrose Agar (PDA; Difco Laboratories, USA) for 20 days or in Potato Dextrose Broth (PDB; Difco Laboratories, USA) with shaking at 120 rpm for five days at 22°C in the dark. For oxidative stress treatment, cultures of 100 ml PDB inoculated with 100 μl of a spore suspension (1 × 104 spores/ml) were incubated at 22°C for 5 days in an orbital shaker (120 rpm) and subjected to treatment with menadione (250 μM, Sigma-Aldrich, USA). The resulting mycelia were harvested at 0.25 h, 0.5 h, 1 h, 3 h and 6 h after treatment using miracloth. Ethanol treated samples were used as control. All mycelial samples were harvested from three biological repeats, immediately frozen using liquid nitrogen, and stored at −80°C until processed. Twenty one days old chickpea seedlings (Pusa-362) were spray inoculated with A. rabiei spore suspensions (2 × 105 spores/ml). Infected stems and leaves were collected at 12 h, 24 h, 72 h and 144 h post-inoculation (hpi) and stored as above.
RNA isolation and quantitative real-time PCR
Total RNA was isolated from A. rabiei using the TRIzol® reagent (Invitrogen, USA) and contaminating genomic DNA was removed by treatment with RNase-free RQ1 DNase (Promega, USA) according to manufacturer's protocol. First-strand cDNA was synthesized with 1 μg of total RNA primed with Oligo-dT using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) as per the instructions in the manual. Primer pairs were designed using the 3′-end exon and untranslated regions (UTRs) of the target genes using Primer Express® (version 3.0) software with the default parameters. Elongation factor1-alpha (ArEF1α) primers were used as an internal control. Specific amplification of only one desired band was observed using each primer combination for qRT-PCR analysis. Additionally, dissociation curve testing suggested only one melting temperature for each primer pair. The resulting cDNA preparations were diluted 10 times and 2 μl of each cDNA sample was used for qRT-PCR. The reactions were performed in MicroAmp® 96-well reaction plate (Applied Biosystems,USA) on a 7900HT Real-Time PCR System using Sequence Detection Systems Software version 2.3 (Applied Biosystems, USA). Each well contained 10 μl of 2× Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix (Agilent Technologies, USA), 2 μl of cDNA, and 18 pmol of each primer in a final volume of 20 μl. The thermal cycling conditions were 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. Transcripts of each ArOYEs and of reference gene ArEF1α were amplified using the primers described in Supplementary Table S3. The threshold cycles (CT) of each ArOYE were averaged from three replicate reactions. The obtained values for each ArOYE were then normalized according to the CT of the ArEF1α. The fold change value was calculated using ΔΔCT method.
Author Contributions
S.N. and P.K.V. designed and analyzed computational and experimental work. S.N., S.V. and R.K.G. performed computational work. S.N. and N.N.B. performed the experimental work. S.N., S.V. and P.K.V. wrote and reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We gratefully acknowledge a research grant from Department of Biotechnology, Government of India (File No: BT/PR10605/PBD/16/791/2008) and a core grant from National Institute of Plant Genome Research, New Delhi, India for funding this work. We are also thankful to the Director, National Institute of Plant Genome Research for providing facilities and help during this study. SN, SV and NNB acknowledge University Grants Commission, Council of Scientific and Industrial Research, and Department of Biotechnology, Government of India, respectively for their fellowships.
Accessions codes: Nucleotide sequences of Ascochyta rabiei Old Yellow Enzymes (ArOYEs) are deposited in GenBank with accession numbers KF644454, KF644455, KF644456, KF644457, KF644458 and KF644459.
References
- Warburg O. & Christian W. Ein zweites sauerstoffubertragendes Ferment und sein Absorptionspektrum. Naturwissenschaften 20, 688 (1932). [Google Scholar]
- Warburg O. & Christian W. Ǘber das gelbe oxydationsferment. Biochem. Z. 263, 228–229 (1933). [Google Scholar]
- Saito K., Thiele D. J., Davio M., Lockridge O. & Massey V. The cloning and expression of a gene encoding old yellow enzyme from Saccharomyces carlsbergensis. J. Biol. Chem. 266, 20720–20724 (1991). [PubMed] [Google Scholar]
- Stott K., Saito K., Thiele D. J. & Massey V. Old Yellow Enzyme- the discovery of multiple isozymes and a family of related proteins. J. Biol. Chem. 268, 6097–6106 (1993). [PubMed] [Google Scholar]
- Massey V. & Schopfer L. M. Reactivity of old yellow enzyme with alpha-NADPH and other pyridine nucleotide derivatives. J. Biol. Chem. 261, 1215–22 (1986). [PubMed] [Google Scholar]
- French C. E. & Bruce N. C. Purification and characterization of morphinone reductase from Pseudomonas putida M10. Biochem. J. 301, 97–103 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C. E. & Bruce N. C. Bacterial morphinone reductase is related to Old-Yellow Enzyme. Biochem. J. 312, 671–678 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blehert D. S., Fox B. G. & Chambliss G. H. Cloning and sequence analysis of two Pseudomonas flavoprotein xenobiotic reductases. J. Bacteriol. 181, 6254–6263 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White G. F., Snape J. R. & Nicklin S. Biodegradation of glycerol trinitrate and pentaerythritol tetranitrate by Agrobacterium radiobacter. Appl. Environ. Microbiol. 62, 637–642 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C. E., Nicklin S. & Bruce N. C. Sequence and properties of pentaerythritol tetranitrate reductase from Enterobacter cloacae PB2. J. Bacteriol. 178, 6623–6627 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivinskas H. et al. Reduction of aliphatic nitroesters and N-nitramines by Enterobactercloacae PB2 pentaerythritol tetranitrate reductase. FEBS J. 275, 6192–6203 (2008). [DOI] [PubMed] [Google Scholar]
- Miura K. et al. Molecular cloning of the nemA gene encoding the N-ethylmaleimide reductase from Escherichia coli. Biol. Pharm. Bull. 20, 110–112 (1997). [DOI] [PubMed] [Google Scholar]
- Schaller F. & Weiler E. W. Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana. J. Biol. Chem. 272, 28066–28072 (1997). [DOI] [PubMed] [Google Scholar]
- Strassner J., Furholz A., Macheroux P., Amrhein N. & Schaller A. A homolog of Old Yellow Enzyme in tomato - spectral properties and substrate specificity of the recombinant protein. J. Biol. Chem. 274, 35067–35073 (1999). [DOI] [PubMed] [Google Scholar]
- Niino Y. S., Chakraborty S., Brown B. J. & Massey V. A new old yellow enzyme of Saccharomyces cerevisiae. J. Biol. Chem. 270, 1983–1991 (1995). [DOI] [PubMed] [Google Scholar]
- Miranda M. et al. Nucleotide sequence and chromosomal localization of the gene encoding the Old Yellow Enzyme from Kluyveromyces lactis. Yeast 11, 459–465 (1995). [DOI] [PubMed] [Google Scholar]
- Komduur J. A., Leao A. N., Monastyrska I., Veenhuis M. & Kiel J. A. Old yellow enzyme confers resistance of Hansenula polymorpha towards allyl alcohol. Curr. Genet. 41, 401–406 (2002). [DOI] [PubMed] [Google Scholar]
- Uchida K. et al. Acrolein is a product of lipid peroxidation reaction. J. Biol. Chem. 273, 16058–16066 (1998). [DOI] [PubMed] [Google Scholar]
- Trotter E. W., Collinson E. J., Dawes I. W. & Grant C. M. Old yellow enzymes protect against acrolein toxicity in the yeast Saccharomyces cerevisiae. App. l Environ. Microbiol. 72, 4885–4892 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubata B. K. et al. A key role for old yellow enzyme in the metabolism of drugs by Trypanosoma cruzi. J. Exp. Med. 196, 1241–1251 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. Z., Coyle C. M., Panaccione D. G. & O'Connor S. E. A role for old yellow enzyme in ergot alkaloid biosynthesis. J. Am. Chem. Soc. 132, 1776–1777 (2010). [DOI] [PubMed] [Google Scholar]
- Coyle C. M., Cheng J. Z., O'Connor S. E. & Panaccione D. G. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl Environ Microbiol. 76, 3898–3903 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. J., Deng Z., Karplus P. A. & Massey V. On the active site of Old Yellow Enzyme. Role of histidine 191 and asparagine 194. J. Biol. Chem. 273, 32753–32762 (1998). [DOI] [PubMed] [Google Scholar]
- Kohli R. M. & Massey V. The oxidative half-reaction of old yellow enzyme. The role of tyrosine 196. J. Biol. Chem. 273, 32763–32770 (1998). [DOI] [PubMed] [Google Scholar]
- Fox K. M. & Karplus P. A. Old yellow enzyme at 2 Å resolution: overall structure, ligand binding, and comparison with related flavoproteins. Structure 2, 1089–1105 (1994). [PubMed] [Google Scholar]
- Kitzing K. et al. The 1.3 Å crystal structure of the flavoprotein YqjM reveals a novel class of old yellow enzymes. J. Biol. Chem. 280, 27904–27913 (2005). [DOI] [PubMed] [Google Scholar]
- Griese J. J., Jakob R. P., Schwarzinger S. & Dobbek H. Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-hydroxycoumarin reduction. J. Mol. Biol. 361, 140–152 (2006). [DOI] [PubMed] [Google Scholar]
- Opperman D. J., Piater L. A. & Heerden E. V. A novel chromate reductase from Thermus scotoductus SA-01 related to old yellow enzyme. J. Bacteriol. 190, 3076–3082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A. et al. Life with 6000 genes. Science 274, 546–567 (1996). [DOI] [PubMed] [Google Scholar]
- Cuomo C. A. & Birren B. W. The fungal genome initiative and lessons learned from genome sequencing. Methods Enzymol. 470, 833–855 (2010). [DOI] [PubMed] [Google Scholar]
- Singh K., Nizam S., Sinha M. & Verma P. K. Comparative Transcriptome Analysis of the Necrotrophic Fungus Ascochyta rabiei during Oxidative Stress: Insight for Fungal Survival in the Host Plant. PLoS ONE 7, e33128. 10.1371/journal.pone.0033128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nuc. Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Kim B. H. & Grishin N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nuc. Acids Res. 36, 2295–2300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. and Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Alexandros S. RAxML-VI-HPC: Maximum Likelihood-based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 21, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- Boc A., Diallo Alpha B. & Makarenkov V. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nuc. Acids Res. 40, 573–W579 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abascal F., Zardoya R. & Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005). [DOI] [PubMed] [Google Scholar]
- Voorrips R. E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78 (2002). [DOI] [PubMed] [Google Scholar]
- Nei M. & Rooney A. P. Concerted and birth-and-death evolution in multigene families. Annu Rev Genet. 39, 121–152 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdorfer G., Steinkellner G., Stueckler C., Faber K. & Gruber K. Stereopreferences of old yellow enzymes: structure correlations and sequence patterns in enoate reductases. ChemCatChem. 3, 1562–1566 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information