Abstract
The ubiquitin conjugating enzyme Rad6B is overexpressed in breast cancer and induces β-catenin transcriptional activation and stabilization via K63-linked polyubiquitination. Here we identify β-catenin and Rad6B interacting regions, identify potential Rad6B ubiquitination sites in β-catenin, and characterize their breast cancer tissue expression. β-catenin and Rad6B colocalize in breast carcinoma and coimmunoprecipitate from MDA-MB-231 cells. Pull-down assays using GST-β-catenin and His-Rad6B deletion mutants identified amino acids 131–181 and 50–116, respectively, as necessary for their interaction. Ubiquitination assays using β-catenin deletion mutants mapped Rad6B-induced ubiquitination within β-catenin 181–422 encompassing Armadillo repeats 2–7. Lysine to arginine mutations within repeats 5–7 identified K394 as the major Rad6B ubiquitination site in vitro and in vivo, and confirmed by Rad6B ubiquitination of a β-catenin peptide encompassing K394. Ubiquitination of wild type- but not K394R-β-catenin was decreased by Rad6B silencing. Compared to wild type-, K312R-, K335R-, K345R-, or K354R-β-catenin, K394R mutation caused ~50% drop in TOP/Flash activity in Wnt-silent MCF-7 cells. Consistent with these data, expression of Rad6B, itself a β-catenin/TCF transcriptional target, was also reduced in K394R-β-catenin transfected cells. Steady-state K394R-β-catenin levels are decreased compared to wild type-β-catenin. The decreased expression is not due to proteasomal degradation as treatment with MG132 failed to rescue its levels. Lymph node-positive breast carcinomas express higher levels of Rad6 protein and Rad6 activity, and K63-linked ubiquitinated β-catenin than reduction mammoplasties. These data suggest that K394 is a novel site of β-catenin ubiquitination that may be important for the stability and activity of β-catenin in breast cancer.
Keywords: Breast cancer, Ubiquitination, Transcriptional activity, Wnt signaling
1. Introduction
The Rad6 gene, originally identified in S. cerevisiae, encodes a 17-kDa ubiquitin conjugating enzyme [1–3] that covalently adds ubiquitin to selected protein lysine residues. Ubiquitin conjugating activity is essential for Rad6 function as replacement of the conserved Cys88 with Ser or Ala produces a null phenotype [4,5]. The two human homologues of yeast Rad6, HHR6A (UBE2A) and HHR6B (UBE2B), referred as Rad6A and Rad6B, respectively, complement the DNA repair and UV mutagenesis defects of S. cerevisiae mutant rad6 [6,7]. We previously demonstrated that Rad6B is overexpressed in breast cancer. Normal human breast tissues weakly express Rad6B, but increased Rad6B expression is observed in hyperplastic breast tissues with overexpression in infiltrating breast carcinomas [8]. Constitutive Rad6B overexpression in nontransformed MCF10A human breast epithelial cells induces a transformed phenotype as indicated by anchorage independent growth in vitro and formation of hyperplastic lesions in vivo [8]. These data suggest an oncogenic role for Rad6B in early breast cancer development. Since constitutive Rad6B overexpression also induces β-catenin stabilization and transcriptional activation [9], it is conceivable that Rad6B oncogenic properties are in part coupled with increased β-catenin activation. The link between Rad6B and β-catenin is further strengthened by our finding that Rad6B is a β-catenin/T-cell factor transcriptional target. Rad6B promoter activation in MCF10A cells requires ectopic expression of β-catenin whereas it is constitutively active in metastatic MDA-MB-231 breast cancer cells with autocrine Wnt signaling [10].
The canonical Wnt signaling pathway regulates several processes including early neoplasia. Wnt pathway activation stabilizes β-catenin and leads to its nuclear translocation and stimulation of β-catenin responsive target gene expression [11–15]. Aberrant β-catenin activation is associated with carcinogenesis. Glycogen synthase kinase 3-β (GSK 3β), in complex with axin and adenomatous polyposis coli, regulates β-catenin via Ser33/Ser37/Thr41 phosphorylation [16,17], which promotes its ubiquitination by β-TrCP/SCF and subsequent proteasomal degradation [18]. Elevated β-catenin levels are detected in breast cancers, suggesting an important role for β-catenin in breast carcinogenesis [19–21]. Unlike many other cancers, however, mutations in β-catenin or its regulators that stabilize β-catenin are rare in breast cancer [22], suggesting alternative β-catenin stabilization mechanisms. Since Rad6B is overexpressed in breast cancer cells and stabilizes β-catenin via K63-linked β-catenin polyubiquitination that is insensitive to 26S proteasomal degradation, this suggests a novel β-catenin stabilization mechanism that is mutation independent [9].
Here we show that Rad6, Rad6 ubiquitin conjugating activity, and polyubiquitinated-β-catenin are co-upregulated in clinical breast carcinomas. Using coimmunoprecipitation and pull-down assays, we show that Rad6B and β-catenin proteins physically interact within the first armadillo repeat (ARM1) segment of β-catenin and amino acids 50–116 of Rad6B. By site-directed mutagenesis and in vivo and in vitro ubiquitination assays, we identified β-catenin K394 as a major site of Rad6B-induced β-catenin ubiquitination. β-catenin K394R exhibits decreased β-catenin-mediated TOP/Flash activity, and expression of Rad6B, itself a β-catenin/TCF transcriptional target, is reduced in K394R-β-catenin transfected MCF-7 cells. Steady-state K394R-β-catenin levels are decreased compared to wild type β-catenin and β-catenin containing K to R mutations at amino acids 312, 335, 345 or 354. Treatment with the proteasomal inhibitor MG132 failed to rescue its levels, indicating decreased expression is not due to proteasomal degradation. These data reveal K394 as a novel site of β-catenin ubiquitination and a potentially novel charge button that may be important for maintaining β-catenin structure and stability.
2. Materials and methods
2.1. Cell culture
MCF-7 breast cancer cells were maintained in Dulbecco's modified Eagle/F-12 medium (DMEM/F12) supplemented with 5% fetal calf serum and 10 μg/ml insulin [23]. MCF10A-Neo and MCF10A-Rad6B [8] cells were maintained in DMEM/F12 supplemented with 5% horse serum, 20 ng/ml Epidermal Growth Factor, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone and 0.1 μg/ml cholera toxin. MDA-MB-231, MDA-MB-231-nontarget shRNA, MDA-MB-231-Rad6BshRNA [9], MDA-MB-231-ΔLRP6 [24,25], WS-15, WS-15-nontarget shRNA, WS-15-Rad6BshRNA [9] and COS7 (ATCC) cells were maintained in DMEM/F12 supplemented with 5% fetal bovine serum.
2.2. Antibodies
β-catenin and β-actin monoclonal antibodies were from Sigma Chemical Company. Glutathione S-transferase (GST), His tag, c-Myc, HA tag, and ubiquitin (P4D1) monoclonal antibodies were from Santa Cruz Biotechnology. Antibody reactive to K63-linked ubiquitin conjugated chains was from Biomol International. Cyclin D1 and cytokeratin 18 monoclonal antibodies were from Dako Corporation. Rad6 antibody was generated and characterized in our laboratory [8]. MassSpec analysis of tryptic peptides of Rad6 proteins immunoprecipitated from MCF10A and MDA-MB-231 cells identified only parent ion masses identical to the predicted Rad6B, but not Rad6A, tryptic peptides, confirming identity of immunoreactive Rad6B bands (unpublished data). However, since the antibody is not Rad6B selective and sequences of Rad6 protein(s) detected by immunoblotting of every breast tissue used have not been verified, the endogenous Rad6 protein detected in immuno-blots is referred to as Rad6 rather than Rad6A or Rad6B. Myc tag antibody was a generous gift from Dr. Vitaly Balan (Wayne State University, Detroit, Michigan).
2.3. Vectors and recombinant protein expression
Human Rad6B 1–152 (wild type), rad6B 50–152 and rad6B 116–152 mutants were amplified by PCR from pCMV-wild type Rad6B[8] using forward primers 5′-caccatgtcgaccccggcccgg-3′ (for Rad6B 1–152), 5′-caccatggatggtacttttaaactagta-3′ (for rad6B 50–152), 5′-caccatgccgaatccaaacagtccagcc-3′ (for rad6B 116–152), and the reverse primer 5′-tgaatcattccagctttgttcaac-3′. Rad6B and rad6B mutants were subcloned into pET101 to encode Rad6B tagged to a C-terminal (His)6. Catalytically inert Rad6B C88A was generated by site-directed mutagenesis using wild type Rad6B pET vector. The following β-catenin constructs subcloned into pGEX-2T were used: full-length 1–781, β-catenin 131–422, β-catenin 181–422, K312R-, K335R-, K345R-, K354R-, and K394R-β-catenin. β-catenin 131–422 and β-catenin 181–422 plasmids were generous gifts from Dr. L.A. Pinna (University of Padova, Italy). Mutant β-catenin proteins in which lysine residues 312, 335, 345, 354, or 394 were substituted with arginine were generated by site-directed mutagenesis of GST-tagged wild type β-catenin or pCI-neo-myc-tagged β-catenin template. Myc-tagged β-catenin 1–781, 1–699, 131–699 and 555–781 subcloned into pCI-neo were generous gifts from Dr. Yutaka Hata (Tokyo Medical and Dental University, Japan). Rad6B and β-catenin sequences and β-catenin lysine to arginine substitutions were con-firmed by DNA sequencing.
2.4. Expression
His-tagged wild type and mutant Rad6B, and GST-tagged wild type and mutant β-catenin plasmids were expressed in Escherichia coli BL21 (DE3). His-tagged Rad6B proteins bound to Ni+2-nitrilotriacetic acid-agarose columns were eluted with 1 M imidazole following sequential washes with 5 and 60 mM imidazole. GST-β-catenin fusion proteins bound to glutathione-agarose beads were eluted with 20 mM glutathione. The purity of the isolated proteins was verified by silver staining.
2.5. Clinical samples
Nonidentifiable breast tissue samples (reduction mammoplasties, lymph node (LN)-negative, LN-positive, and metastatic lymph nodes) flash-frozen in liquid nitrogen were acquired by written consent of the Institutional Biorepository Core after review and approval by the Wayne State University Human Investigation Committee. Only surgical excisions that remained after diagnostic confirmation were used for experiments.
2.6. Immunoblotting and immunoprecipitation
Frozen samples ground to a fine powder were suspended in cold lysis buffer (25 mM HEPES [pH 7.5], 25 mM NaCl, 0.1% Nonidet P-40, 10% glycerol) containing protease inhibitor (Roche) and phosphatase inhibitor (1 mM sodium orthovanadate, 20 mM NaF, 10 mM sodium pyrophosphate and 10 mM β-glycerol phosphate) cocktails and homogenized. Homogenates were passed through a 19 ga needle until no longer viscous. Lysates were centrifuged at 18,000×g at 4 °C for 30 min, and the resulting cytosolic extracts containing 100 μg total protein were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by immunoblotting with Rad6 [8], β-catenin [9], β-actin and cytokeratin 18 [26] antibodies. For β-catenin ubiquitination status determination, precleared cytosols were incubated with β-catenin antibody (1 μg) and Protein A/G Sepharose. Bound proteins were resolved by SDS-PAGE and immunoblotted with ubiquitin or K63-linked ubiquitin chain antibody. Rad6 and ubiquitinated β-catenin levels were expressed relative to nascent β-catenin, and nascent β-catenin expressed relative to cytokeratin-18. For characterizing in vivo Rad6/β-catenin interactions, Protein A/G Sepharose precleared cytosols were incubated with Protein A/G Sepharose and anti-β-catenin (1 μg) or anti-Rad6 (1 μg) or the corresponding normal mouse or rabbit IgG. Co-precipitated β-catenin or Rad6 was then determined by immuno-blotting. Cytosols prepared from MCF10A-Neo, MCF10A-Rad6B, MDA-MB-231-nontarget shRNA, MDA-MB-231-Rad6BshRNA, MDA-MB-231-ΔLRP6, WS-15-nontarget shRNA and WS-15-Rad6BshRNA cells were analyzed by immunoblotting with Rad6 and β-catenin antibodies. Semi-quantitation of bands was performed by scanning densitometry and NIH ImageJ 1.45s software.
2.7. Rad6B ubiquitin conjugating activity assay
Histone H2A ubiquitination was assessed in extracts (100 μg) of reduction mammoplasties and tumor tissue as described above. Samples were incubated at room temperature for 1 h with histone H2A (2.5 μg; Roche Biotech), ubiquitin activating enzyme E1 (50 μg/ml, BioMol), ubiquitin (1.25 mg/ml; Roche), 2 mM MgCl2, 4 mM ATP and energy regeneration system (Boston Biochem) in reaction buffer (50 mM Tris– HCl [pH 7.5]) [9]. To verify the involvement of endogenous Rad6B in his-tone ubiquitination, some reactions were performed in the presence of catalytically inert rad6B (85 μg/mL; Boston Biochem), without histone H2A, or with Rad6B immunodepleted extracts. For immunodepletion, tissue extracts were incubated with anti-Rad6 or normal rabbit IgG (1.0 μg) and Protein A/G Sepharose overnight. After pelleting complexed proteins, depleted supernatants were used in ubiquitination assays. For a positive control, tissue extracts were substituted with wild type Rad6B (85 μg/ml; Boston Biochem). Reaction products were separated by SDS-PAGE and analyzed by silver staining or immunoblotting with ubiquitin and Rad6 antibodies.
2.8. Ubiquitination site mapping
Cytosolic extracts of COS7 cells expressing myc-tagged β-catenin 1–781, β-catenin 1–699, β-catenin 131–699 or β-catenin 551–781 were subjected to in vitro ubiquitination assays with or without recombinant Rad6B as described above. Reaction products were separated by SDS-PAGE and immunoblotted with myc tag antibody. Rad6B-induced in vitro ubiquitination of purified GST-tagged-β-catenin 131–422, β-catenin 181–422, or β-catenin containing mutations at 312, 335, 345, 354 or 394 was assessed as previously described [9]. Reactions were subjected to SDS-PAGE and immunoblotted with ubiquitin or GST antibody. The corresponding amounts of β-catenin proteins used in ubiquitination assays were determined by Coomassie staining. Ubiquitination of β-catenin carrying K312R, K345R, K354R, or K394R mutations was determined in MCF-7, or nontarget or Rad6BshRNA transfected MDA-MB-231 cells cotransfected with myc-tagged wild type or mutant β-catenin and pMT123 (HA-tagged ubiquitin expression vector [27]). Lysates were immunoprecipitated with myc tag antibody before HA tag antibody immunoblotting. To further verify the identified Rad6B induced β-catenin ubiquitination site, in vitro ubiquitination assays were performed with a synthetic 29-amino acid β-catenin peptide NLSDAATKQEGMEGLLGTLVQLLGSDDIN encompassing K394. Reactions were performed with methylated ubiquitin [9] and wild type or catalytically inert Rad6B. Methylated ubiquitin was used instead of ubiquitin as it limits addition of a single ubiquitin molecule and prevents ubiquitin dimerization resulting from ubiquitin self-conjugation, thus enabling cleaner visualization of the ubiquitinated peptide on silver stained gels.
2.9. In vitro pull-down assays
To map β-catenin regions interacting with Rad6B, purified His-tagged Rad6B 1–152 protein was incubated with control or GST-β-catenin (1–781, 131–422, or 181–422) proteins bound to glutathione (GSH)-Sepharose for 4 h at 4 °C in binding buffer (100 mM sodium phosphate [pH 7.2], 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol, and protease inhibitor cocktail). The beads were washed with binding buffer followed by washes in buffer containing 500 mM NaCl. The bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting with His tag or GST antibodies. To map Rad6B interaction sites on β-catenin, His-tagged Rad6B 1–152, rad6B 50–152, rad6B 116–152 or GST-β-catenin 1-781 proteins were expressed in Escherichia coli BL-21. Bacterial lysates containing equivalent amounts of Rad6B 1–152, rad6B 50–152 or rad6B 116–152, and GST-β-catenin 1–781 were loaded on Ni+ 2-nitrilotriacetic acid-agarose beads or GSHSepharose beads and processed as above. Interactions between His-tagged wild type or catalytically inactive C88A rad6B 1–152 and GST-β-catenin 1–781 and their elution profiles were determined by processing bacterial lysates containing Rad6B and β-catenin on Ni+ 2 nitrilotriacetic acid agarose or GSH-Sepharose columns. Ni+ 2 columns were washed with binding buffer, and sequentially with buffer containing 5 mM, 60 mM and 1 M imidazole. GSH-Sepharose columns were washed with binding buffer containing 500 mM NaCl and eluted with 20 mM glutathione. Column fractions were analyzed by immunoblotting with His tag or GST antibodies.
2.10. Luciferase assays and gene expression analysis
Transcriptional activities of K312R-, K335R-, K345R-, K354R-, or K394R-β-catenin mutants relative to wild type β-catenin were assayed by cotransfecting MCF-7 cells with myc-tagged β-catenin plasmid (1 μg), pTOP/FLASH or pFOP/FLASH (0.5 μg; Upstate Biotechnology), p300 (0.25 μg), and pRLTK (100 ng; Promega) vectors as previously described [9]. P300 was included in transfections since it enhances transcriptional activity of the expressed β-catenin [8]. In some cases, 1–4 μg of wild type- or K394R-β-catenin were transfected. Cyclin D1 promoter-driven luciferase or c-Myc promoter-driven luciferase reporters were substituted for pTOP/FLASH in some assays. Luciferase activity was measured ~48 h post-transfection with Promega Dual Luciferase assay system, and firefly luciferase activity was normalized against Renilla luciferase. The background luciferase activities from corresponding pFOP/FLASH transfections were subtracted. Duplicate dishes were assayed for each transfection, and three independent transfection assays were performed. Lysates from transfection assays were also analyzed by immunoblotting for Rad6, cyclin D1 [28], c-Myc [29], and myc-tagged β-catenin, and normalized against β-actin.
2.11. Semi-quantitative RT-PCR
Total RNA (1. 0 μg) isolated from MCF-7 cells transfected with myc-tagged wild type or mutant β-catenin (1.0 μg) and p300 (0.25 μg) was reverse-transcribed with Superscript II (Invitrogen, Carlsbad, CA). Rad6B amplification was performed with +17/+33 and +114/+97 [GenBank: NM_003337] forward and reverse primers, respectively. GAPDH expression was monitored by amplifications with forward (+186/+206) and reverse (+320/+302) [GenBank: XM_006959] primers. PCR reaction conditions that yielded linear product amplification were used: 5 min 95 °C, 1 min 55 °C, 2 min 65 °C for 21 cycles.
2.12. Immunohistochemical and immunofluorescence staining
Deparaffinized 5 μm sections from normal and infiltrating ductal breast carcinomas were permeabilized and incubated with Rad6 or β-catenin antibodies. After primary antibodies, slides were incubated with FITC- and Texas Red-conjugated secondary antibodies, or with appropriate biotinylated anti-rabbit or mouse IgG and HRP-conjugated streptavidin, and nuclei were counterstained with hematoxylin. Slides were stained in the absence of primary antibody or with isotype matched nonimmune IgG to assess nonspecific reactions. Images were collected on an Olympus BX60 microscope equipped with a Sony high resolution/sensitivity CCD video camera.
2.13. Statistical analysis
Kruskal–Wallis tests were used to compare relative levels of Rad6B, native β-catenin and high MW β-catenin from reduction mammoplasties or benign tissues (n=9), tumor samples from LN-negative (n=16) or LN-positive (n=11) breast cancer patients, and lymph node samples from women with metastatic disease (n=10). Significance of differences in transcriptional activities of β-catenin mutants was determined by Student's t test. Statistical calculations were conducted using GraphPad Prism 4 software.
3. Results
3.1. Rad6B and β-catenin physically interact
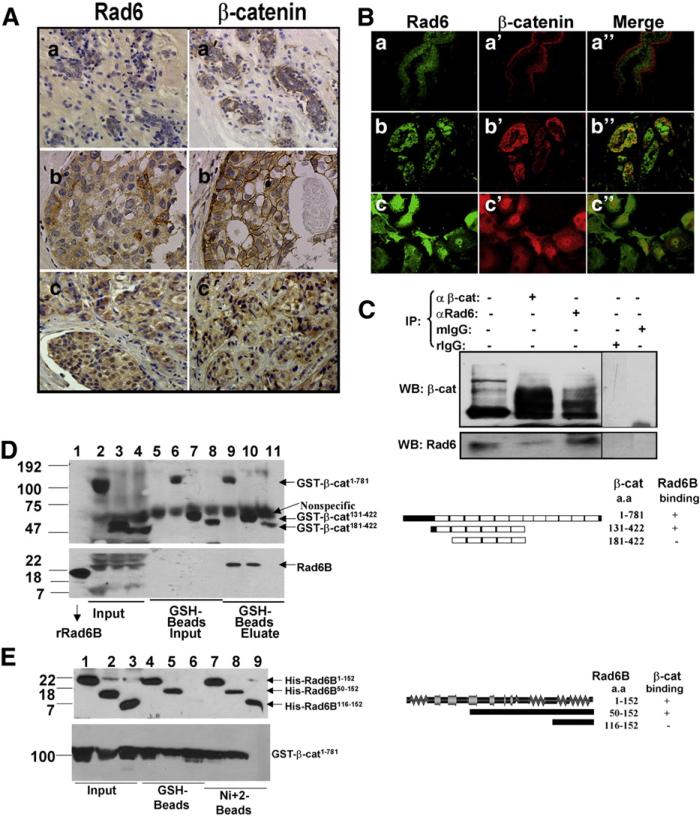
Cytoplasmic/nuclear β-catenin staining with or without loss of cell membrane associated β-catenin has been associated with poor prognosis of breast cancer [30]. A positive association between Rad6 and β-catenin immunostaining patterns is observed in ~72% of the breast carcinomas analyzed [10]. Immunohistochemical analysis of Rad6 and β-catenin in representative clinical samples show different cellular distributions (Fig. 1A). Whereas β-catenin is associated with cell membranes in normal breast tissues (reduction mammoplasties, Fig. 1Aa′), it is either present on cell membranes and cytoplasm/nuclei (Fig. 1Ab′) or in the cytoplasm/nuclei with loss of membrane staining (Fig. 1Ac′) in breast cancer. Rad6 also showed changes in cellular distribution. Rad6 staining is weak in reduction mammoplasties (Fig. 1Aa), but is detected in the cytoplasm (Fig. 1Ab) or in both cytoplasm and nuclei (Fig. 1Ac) of clinical breast carcinomas. Immunofluorescence staining showed Rad6 and β-catenin colocalization in infiltrating ductal carcinoma (Fig. 1Bb″) but not in reduction mammoplasty (Fig. 1Ba″). In WS-15 human breast cancer cells, Rad6 and β-catenin also colocalized (Fig. 1Bc–c″), corroborating clinical breast cancer immunostaining (Fig. 1A). Consistent with the immuno-fluorescence data, immunoprecipitation with β-catenin antibody (epitope 680–781) pulled down Rad6 along with nascent and high MW β-catenin from MDA-MB-231 cytosols. Likewise, Rad6 antibody ef-ficiently pulled down Rad6 along with nascent and high MW β-catenin, whereas normal IgGs failed to immunoprecipitate Rad6 or β-catenin (Fig. 1C).
Fig. 1.
Rad6B and β-catenin exhibit direct interactions. (A) Immunohistochemical analysis of Rad6 (a, b, c) and β-catenin (a′, b′, c′) in reduction mammoplasty (a, a′) and clinical breast carcinomas (b, b′, c, c′). (B) Immunofluorescence localization of Rad6 (a, b and c) and β-catenin (a′, b′ and c′) in reduction mammoplasty (a–a″), infiltrating ductal carcinoma (b–b″), and WS-15 breast cancer cells (c–c″). Magnification ×20 (a–a″ and b–b″) and ×100 (c–c″). (C) Co-immunoprecipitation analysis. Lane 1, 1/5th of MDA-MB-231 cytosol inputs. (D) Mapping β-catenin interacting regions. Pull-down assays from GSH-beads with His-tagged wild type Rad6B 1–152 (lanes 9–11) and GST-tagged β-catenin 1–781 (lane 9), β-catenin 131–422 (lane 10) or β-catenin 181–422 (lane 11) proteins. Lanes 1–4, inputs of His-Rad6B, GST-β-catenin 1–781, GST-β-catenin 131–422, and GST-β-catenin 181–422, respectively. Lanes 5–8, control GST, GST-β-catenin 1–781, GST-β-catenin 131–422, and GST-β-catenin 181–422, respectively. Blots were probed with GST (upper) and His (bottom) antibodies. (E) Mapping Rad6B interaction sites. Pull-down assays were performed with His-tagged Rad6B 1–152 (lanes 4 and 7), Rad6B 50–152 (lanes 5 and 8) or Rad6B 116–152 (lanes 6 and 9) and GST-β-catenin 1–781 (lanes 4–9) on GSH-beads (lanes 4–6) or Ni+ 2-beads (lanes 7–9) beads. Lanes 1–3, inputs of Rad6B 1–152, Rad6B 50–152, Rad6B 116–152, and GST-β-catenin 1–781. Blots were probed with His (upper) and GST (bottom) tag antibodies. Rad6B/β-catenin interaction regions are shown schematically in the right panels of D and E.
3.2. Rad6B interacts with the first armadillo repeat (ARM1)-containing segment of β-catenin
To map Rad6B/β-catenin interactions, pull-down assays were performed. As shown in Fig. 1D (bottom panel), wild type Rad6B (lane 1) bound to GST-β-catenin 1–781 (lane 9) and GST-β-catenin 131–422 (lane 10) but not to GST-β-catenin 181–422 (lane 11). Binding was β-catenin specific, as Rad6B did not bind to control GSH-Sepharose (lane 5, Fig. 1D, bottom panel). Rad6B bound similarly to GST-β-catenin 1–781 and GST-β-catenin 131–422 (compare lanes 9 and 10, bottom panel) but did not bind to GST-β-catenin 181–422 (compare lanes 9 and 10 with 11), suggesting that Rad6B/β-catenin interaction requires the β-catenin 131–181 segment encompassing ARM1. Incubation with Rad6B did not affect GST-β-catenin protein binding to GSH-Sepharose since similar amounts of β-catenin proteins bound to beads in the absence (Fig. 1D upper panel, lanes 6–8, input) or presence (Fig. 1D upper panel, lanes 9–11) of Rad6B.
Pull-down assays of the Rad6B/β-catenin mixture on Ni+ 2-agarose (Fig. 1E) confirmed the Rad6B/β-catenin interaction data from GSH-Sepharose binding assays in Fig. 1D. β-catenin associated with Rad6B 1–152 and rad6B 50–152, but not rad6B 116–152 (Fig. 1E upper panel, lanes 7–9). β-catenin GSH-Sepharose pull-down assays further confirmed the inability of rad6B 116–152 to bind β-catenin (Fig. 1E upper panel, lane 6) whereas Rad6B 1–152 and rad6B 50–152 associated similarly with β-catenin 1–781 (Fig. 1E upper panel, lanes 4 and 5). These data suggest that Rad6B amino acids 50–116 interact with β-catenin amino acids 131–181 (Fig. 1D and E, right panels). These findings imply that the Rad6B and β-catenin colocalization observed in Fig. 1B and the positive association between Rad6B and β-catenin staining in clinical breast cancer (Fig. 1A; [10]) potentially reflect endogenous Rad6B/β-catenin interactions.
3.3. Interaction with β-catenin is not affected by mutation in the Rad6B catalytic site
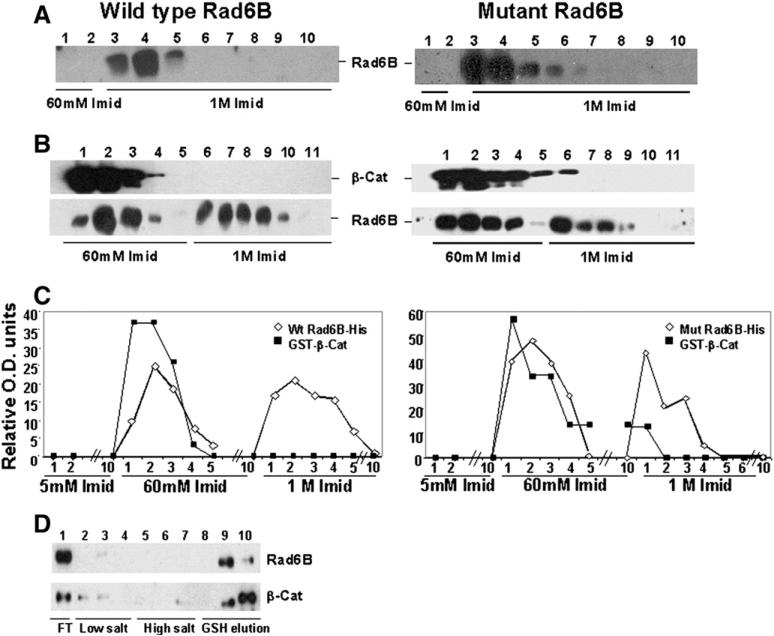
Since β-catenin is a Rad6B-mediated ubiquitination substrate, we tested whether this interaction requires the Rad6B catalytic site. Both wild type (Fig. 2A, left panel) and mutant (Fig. 2A, right panel) Rad6B bound very strongly to Ni+2 agarose as their elution required 1 M imidazole. However, when complexed with β-catenin, some Rad6B was eluted by 60 mM imidazole (Fig. 2B, left and right panels), whereas Rad6B uncomplexed to β-catenin was eluted by 1 M imidazole (Fig. 2B, left and right panels). These data suggest that complex formation with β-catenin altered His-tagged Rad6B affinity for Ni+2 agarose. The similar elution profiles for β-catenin complexed with wild type (Fig. 2B and C, left panels) and mutant (Fig. 2B and C, right panels) Rad6B suggest that β-catenin–Rad6B interaction is unaffected by the Rad6B catalytic site mutation. Rad6B and GST-β-catenin coelution from GSH beads by glutathione further corroborates Rad6B/β-catenin interaction data (Fig. 2D).
Fig. 2.
Physical interaction between Rad6B and β-catenin is not affected by mutation (Cys88 to Ala) in the ubiquitin conjugation catalytic site. (A, B) Elution profiles of His-tagged wild type Rad6B or catalytically mutant rad6B from Ni+ 2 beads in the absence (A) and presence (B) of GST-wild type β-catenin, and detected with His tag or GST antibodies. (C) Graphic representation of Rad6B and β-catenin elution profiles in A and B. (D) Elution profiles of wild type Rad6B and GST-wild type β-catenin from GSH-Sepharose.
3.4. Lysine 394 of β-catenin is a major site of Rad6B mediated ubiquitination
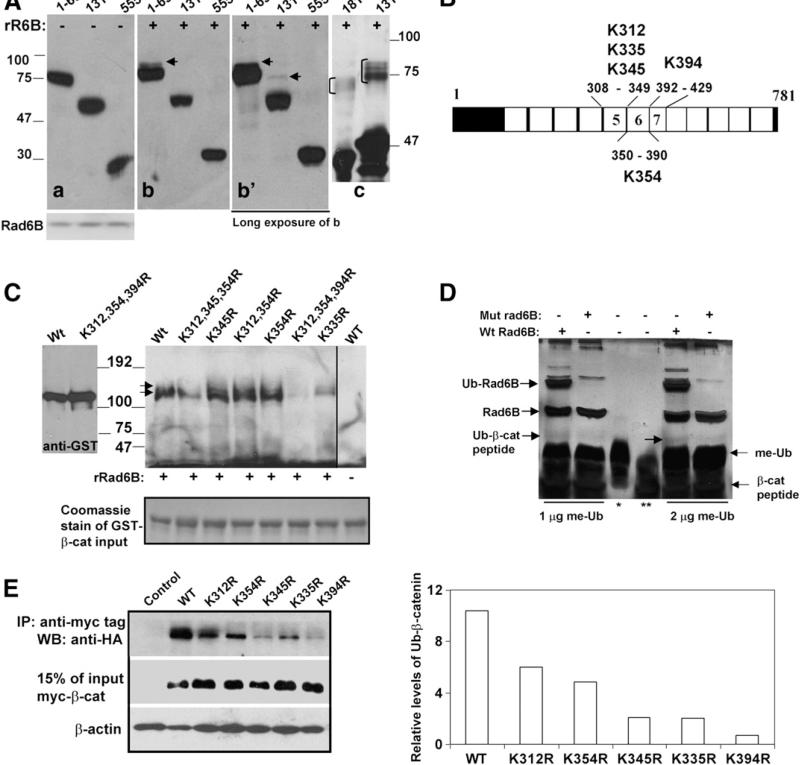
To map β-catenin lysine(s) that are ubiquitinated by Rad6B, in vitro ubiquitination assays were performed with recombinant Rad6B and extracts of COS7 cells transfected with various myc-tagged β-catenin deletion mutants. Rad6B ubiquitinated β-catenin 1–699 (Fig. 3A, arrow in b, b′) and β-catenin 131–699 (Fig. 3A, arrow in b′) but not β-catenin 555–781 (Fig. 3A, b and b′). The corresponding control reactions performed without recombinant Rad6B showed no β-catenin ubiquitination (Fig. 3A, panel a). GST-β-catenin 181–422 and GST-β-catenin 131–422 proteins were ubiquitinated by recombinant Rad6B in vitro (indicated by bracket in Fig. 3A, panel c). Taking together, these data suggest that lysine residues located between amino acids 181–422 of β-catenin are the major targets of Rad6B-induced ubiquitination.
Fig. 3.
Lysine 394 of β-catenin is a major Rad6B ubiquitination site. (A) Cytosols from COS7 cells transfected with indicated myc-tagged β-catenin (a, b, b′) or purified GST-β catenin (181–422 and 131–422; panel c) were subjected to in vitro ubiquitination assays with recombinant Rad6B (panels b, b′, and c). Panel a, control reactions performed without Rad6B. Panel b′ is a longer exposure of b. Anti-myc and GST-tag reactive ubiquitinated β-catenin are indicated by arrows in b and b′, and bracket in c, respectively. (B) Schematic representation of lysines in ARMs 5–7. (C) Rad6B ubiquitination of purified GST-wild type (Wt) or K to R substituted β-catenin proteins. Last lane, wild type β-catenin reaction lacking Rad6B. Immunoblots were probed with ubiquitin antibody. Arrows indicate the positions of the ubiquitinated doublet bands of β-catenin. Left blot, GST-β-catenin proteins without reaction and detected by GST tag antibody. Bottom panel, Coomassie staining of GST-β-catenin proteins used in the reactions. (D) In vitro ubiquitination of K394 containing β-catenin peptide performed in the presence of wild type (Wt) or catalytically inert (mut) Rad6B and methylated ubiquitin (me-Ub). Lanes *, ** indicate me-Ub and β-catenin peptide, respectively. Reaction products were detected by silver staining. (E) In vivo ubiquitination of myc-tagged wild type or mutant β-catenin in MCF-7 cells. Right panel, graphic representation of wild type and mutant β-catenin ubiquitination.
The β-catenin 181–422 segment encompasses ARMs 2–7 and contains 10 lysine residues. ARMs 5–9 are implicated in interactions with TCF-4 and other cellular partners and regulate β-catenin transcriptional activity ([31] and references within). We have previously shown that ectopic Rad6B induces polyubiquitinated-β-catenin levels and β-catenin transcriptional activity in MCF10A cells. Conversely, Rad6B silencing in MDA-MB-231 and WS-15 breast cancer cells with autocrine Wnt signaling decreases polyubiquitinated-β-catenin levels and transcriptional activity [9]. Based on these data, we reasoned that the lysine(s) ubiquitinated by Rad6B and modulating β-catenin transcriptional activity may include the five lysines (312, 335, 345, 354, 394) located in ARMs 5–7 (Fig. 3B). GST-wild type-β-catenin and mutants GST-β-catenin K312R, K345R, and K354R were efficiently ubiquitinated by Rad6B in vitro as indicated by the presence of two anti-ubiquitin reactive bands (Fig. 3C, arrows). Relative to corresponding single mutants, K312/345/354R-β-catenin ubiquitination was reduced but not abolished, confirming these amino acids are not essential for Rad6B-induced ubiquitination (Fig. 3C). K335R-β-catenin was weakly ubiquitinated, while the K394R mutation abrogated ubiquitination (Fig. 3C). The reduced ubiquitination efficacy observed with K312/345/354R-β-catenin as compared to single mutants probably resulted from conformational changes that influenced accessibility of K394 to Rad6B. Wild type β-catenin ubiquitination did not occur in reactions lacking Rad6B, confirming reaction specificity (Fig. 3C). In vitro ubiquitination assays using a 29 amino acid β-catenin synthetic peptide containing K394 were performed to verify these results. As shown in Fig. 3D, β-catenin peptide ubiquitination occurs in the presence of wild type Rad6B but not catalytically inactive mutant rad6B. Rad6B ubiquitin conjugating activity was confirmed by detection of ubiquitin-conjugated Rad6B in reactions containing wild type but not catalytically inactive Rad6B (Fig. 3D).
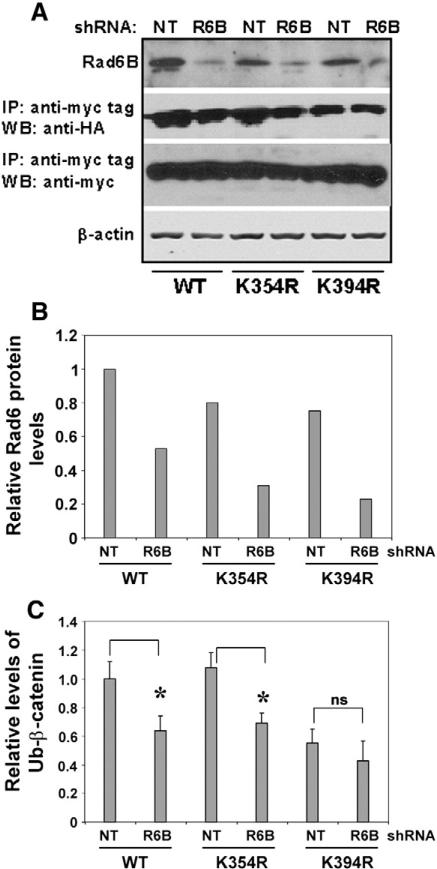
Next we characterized in vivo ubiquitination of the β-catenin K312R, K335R, K345R, K354R and K394R mutants in MCF-7 cells. Compared to wild type-β-catenin, mutants showed decreased ubiquitination, with β-catenin K394R showing the largest decrease (Fig. 3E and right panel). Variations in ubiquitination in vivo can result from variant expressions of transfected ubiquitin or mutant β-catenin, and/or from reduced efficiencies of ubiquitination of the mutant proteins. To verify whether in vivo ubiquitination of K394R-β-catenin is affected by Rad6B silencing, MDA-MB-231 cells were transfected with nontarget shRNA or Rad6BshRNA, and myc-tagged wild type, K354R-or K394R-β-catenin and ubiquitin expression plasmids. Wild type and K354R-β-catenin were efficiently ubiquitinated, and ubiquitination was inhibited by Rad6BshRNA (Fig. 4A and C). As in MCF-7 cells (Fig. 3E), K394R-β-catenin ubiquitination was compromised in MDAMB-231 cells and was minimally affected by Rad6B silencing (Fig. 4A, B and C). These data confirm that K394 is a physiologically relevant site of Rad6B-mediated β-catenin ubiquitination. However, since K394R-β-catenin ubiquitination, albeit low compared to wild type β-catenin, is observed, this suggests additional Rad6B-independent ubiquitinating events.
Fig. 4.
K394R-β-catenin ubiquitination is unaffected by Rad6B silencing. MDA-MB-231 cells were transfected with nontarget (NT) or Rad6B (R6B) shRNA and myc-tagged wild type (WT), K354R- or K394R-β-catenin and HA-ubiquitin. (A) Western blot analysis. (B and C) Graphic representation of Rad6 and ubiquitinated β-catenin, respectively. * and ns indicate significant (P<0.001) and no significant difference, respectively.
3.5. Lysine 394 mutation decreases β-catenin transcriptional activity
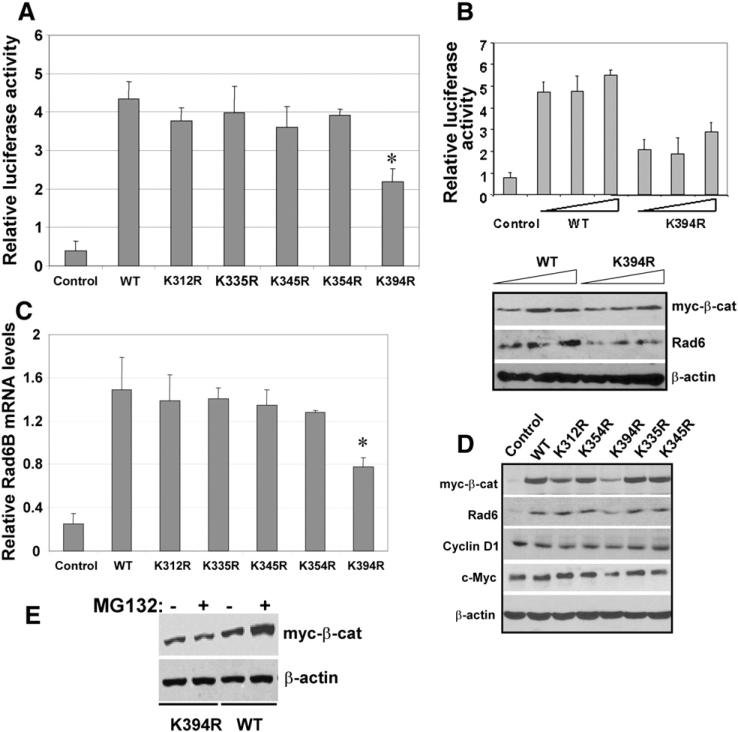
To characterize the role of lysines 312, 335, 345, 354 or 394 in β-catenin transcriptional activity, the myc-tagged mutants were tested for their ability to support reporter transactivation and expression of transcriptional targets Rad6B, cyclin D1 and c-Myc. Wild type β-catenin induced an ~11-fold increase in reporter activity as compared to empty vector-transfected cells (Fig. 5A). Compared to wild type, the increase in TOP/Flash activity was reduced by 10–23% in cells transfected with K312R-, K335R-, K354R- or K345R-β-catenin, but this difference was not significant. TOP activation by K394R-β-catenin, however, was reduced by ~50% (P<0.01) compared to wild type β-catenin (Fig. 5A). To verify the decreased transcriptional activity of K394R-β-catenin, TOP/Flash assays were performed with various amounts of wild type and K394R-β-catenin. Whereas 1, 2 and 4 μg of wild type β-catenin robustly supported TOP/Flash reporter transactivation, TOP/Flash activity was not significantly enhanced by increasing amounts of K394R-β-catenin (Fig. 5B). Simultaneous analysis of lysates for expression of myc-tagged wild type and K394R-β-catenin indicated that the drop in reporter activation by K394-β-catenin is not rescued commensurately by increasing levels of the mutant (Fig. 5B, lower panel). Expression of Rad6B, itself a transcriptional target of β-catenin/TCF, was also not enhanced by K394R-β-catenin, and resembled the TOP/Flash reporter data (Fig. 5B and lower panel).
Fig. 5.
K394R mutation decreases β-catenin levels and transcriptional activity. (A) TOP/Flash activities (mean±S.E.M). *Indicates significant difference as compared to control (P<0.01). (B) TOP/Flash activity assays with increasing amounts (1–4 μg) of wild type or K394R-β-catenin. Lower panel, Western blot analysis of indicated proteins. (C) Rad6B mRNA expression relative to GAPDH (mean±S.E.M) in MCF-7 cells transfected with myc-tagged wild type or mutant β-catenin vectors. *Indicates significant difference as compared to control (P<0.01). (D). Steady-state levels of the indicated proteins in nontransfected (first lane) or transfected with the indicated β-catenin. (E) MG132 does not affect the steady state levels of K394R-β-catenin. Myc tag antibody immunoblots of K394R- or wild type (WT) β-catenin in MCF-7 cells with or without MG132 treatment.
To verify the reporter transactivation data in Fig. 5A, expression of myc-tagged β-catenin and β-catenin transcriptional targets Rad6B, cyclin D1, and c-Myc were analyzed in MCF-7 cells. Rad6 protein was induced by wild type, K312R-, K335R-, K345R-, or K354R-β-catenin (Fig. 5D). As in Fig. 5B (lower panel), induction of Rad6 was lower in K394R-β-catenin transfected cells (Fig. 5D). RT-PCR analysis confirmed that the decrease in Rad6B protein in K394R-β-catenin transfected cells is due to reduced Rad6B transcription (Fig. 5C). Analysis of β-catenin transcriptional target c-Myc regulation as indicated by c-Myc promoter-driven luciferase expression showed an ~25% decrease in K394R-β-catenin transfected cells compared to wild type β-catenin transfected cells (data not shown). c-Myc protein levels were also decreased, although not as dramatically as Rad6B, in K394R-β-catenin transfected cells (Fig. 5D). Cyclin D1 promoter activity (measured by cyclin D1 promoter-luciferase assays) was unaffected by the K394R mutation (data not shown), and cyclin D1 protein levels were minimally affected (Fig. 5D). The steady state levels of K394R-β-catenin were reduced compared to other mutants or wild type β-catenin (Fig. 5D), and addition of the 26S proteasome inhibitor MG132 did not rescue K394R-β-catenin levels (Fig. 5E). These data suggest that K394 may play an important role in maintaining the steady-state levels of β-catenin and its transcriptional activity.
3.6. Rad6 and ubiquitin-modified β-catenin levels are co-upregulated in breast carcinomas
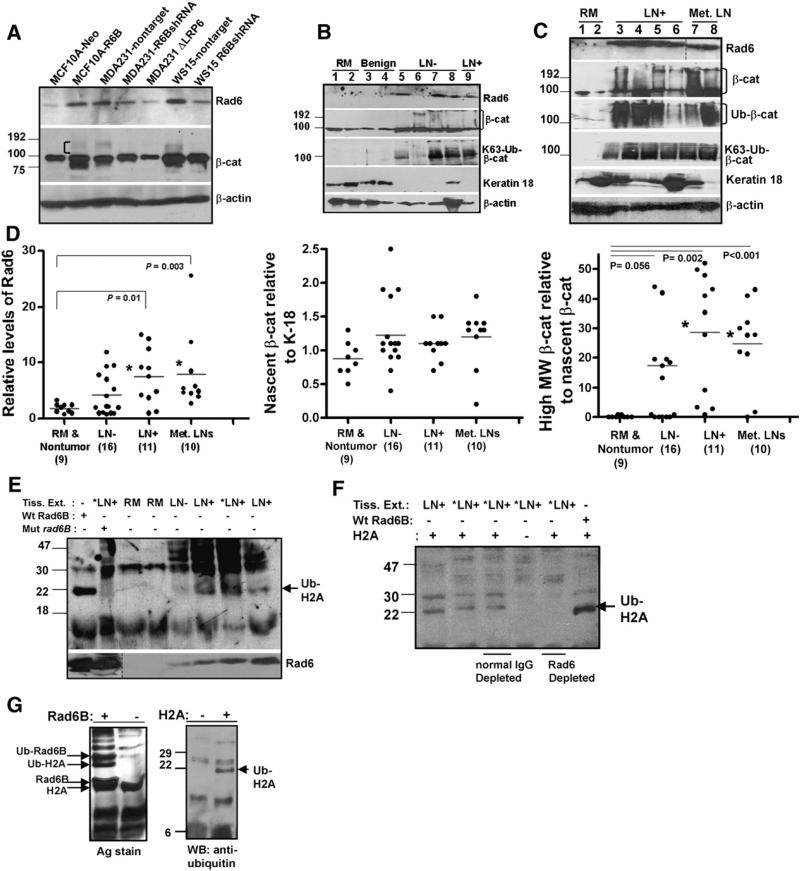
The relationship between the presence of high MW β-catenin and Rad6B seen in human breast cancer cells overexpressing Rad6B (MCF10A-Rad6B, MDA-MB-231, WS-15) or silenced for Rad6B gene expression (MDA-MB-231-Rad6BshRNA, WS-15-Rad6BshRNA) is shown in Fig. 6A. To determine whether a positive relationship between Rad6 levels and β-catenin ubiquitination also exists in clinical breast cancer, Rad6 and β-catenin protein levels were analyzed in cytosols from reduction mammoplasties (RM) or nontumor tissues, lymph node (LN)-negative, LN-positive, and metastatic lymph nodes. Representative blots and graphic summarization of Rad6 and β-catenin profiles are shown in Fig. 6B, C and D. Rad6 and high MW β-catenin (>90 kDa) levels were normalized against nascent β-catenin since it was present in all samples regardless of tumor grade. Steady-state levels of nascent β-catenin were normalized against cytokeratin-18, an epithelial-cell specific marker, rather than β-actin, since β-actin is not epithelial cell- specific and tissue extracts contain variable amounts of epithelial and stromal cells. Expression of cytokeratin-18 itself is variable in clinical samples, and loss of cytokeratin-18 has been correlated with breast cancer progression [32]. Significantly higher Rad6 levels were observed in LN-positive breast carcinomas (P=0.01) and metastatic lymph nodes (P=0.003) compared to RMs or benign breast tissues (Fig. 6B, C, and D). Nascent β-catenin levels were not significantly different in non-tumor and tumor tissues. β-catenin high MW forms were readily detected with β-catenin antibody and in Westerns of β-catenin antibody immunoprecipitates with ubiquitin or K63-linked ubiquitin chain antibody. β-catenin high MW forms were significantly higher in LN-positive breast cancer (P=0.002) and metastatic lymph nodes (P<0.001) as compared to RMs (Fig. 6C and D).
Fig. 6.
Breast carcinomas display elevated Rad6, Rad6 ubiquitin conjugating activity, and polyubiquitinated β-catenin. (A) Steady state levels of Rad6 and β-catenin. High MW β-catenin fraction is indicated by a bracket. (B and C) Steady-state levels of the indicated proteins in RM (reduction mammoplasty), LN– (lymph-node negative), LN+ (lymph-node positive) and Met LN (metastatic lymph nodes). (D) Graphic representation of Rad6, nascent β-catenin, and high MW β-catenin levels. *Indicates statistical significance. (E) In vitro ubiquitination of histone H2A with RM, LN–, and LN+ breast tissue extracts, and detected with ubiquitin antibody. *LN+ indicates the same LN+ breast cancer sample subjected to reactions with or without catalytically mutant (Mut) rad6B. Bottom, corresponding levels of Rad6 in the reactions. In the first lane, positive control reaction with histone H2A and recombinant wild type Rad6B. Arrow indicates the position of ubiquitinated histone H2A. (F) *LN+ indicates the same tissue extract sample that was subjected to ubiquitination reactions with or without H2A (lanes 2 and 4) or following immunodepletion with normal rabbit IgG or Rad6 antibody (lanes 3 and 5). The last lane shows a positive control reaction with H2A and recombinant Rad6B. Arrow indicates the position of ubiquitinated histone H2A. (G) Ubiquitination of histone H2A by Rad6B. (Left) Reactions with or without Rad6B and detected by silver stain. (Right) Reactions performed with or without H2A, and detected with ubiquitin antibody.
3.7. Rad6 ubiquitin conjugation activity is upregulated in invasive breast carcinomas
To determine whether elevated Rad6 expression levels in breast carcinomas [8] translate into commensurate increases in ubiquitinating activity, histone H2A ubiquitination assays were performed in extracts from normal (RM), LN-negative and LN-positive breast carcinomas. His-tones H2A and H2B are ubiquitinated by Rad6 [33,34]. Whereas monoubiquitinated histone H2A was undetectable in reactions involving RM (Fig. 6E), a conspicuous band with similar mobility to a band present in parallel recombinant wild type Rad6B reactions (Fig. 6E and F, arrow) was observed in LN-positive tumor extract reactions (Fig. 6E and F, LN+ lanes). Addition of catalytically inert rad6B in LN+ tumor reactions markedly inhibited histone H2A ubiquitination as compared to corresponding reactions not supplemented with mutant rad6B (Fig. 6E, compare *LN+ lanes with or without mutant rad6B). The ubiquitinated band was not detected in Rad6-immunodepleted tumor reactions (Fig. 6F, compare *LN+ reactions performed after Rad6 depletion vs. normal IgG control). Silver stains and ubiquitin antibody immunoblots of H2A ubiquitination reactions performed with or without recombinant Rad6B or histone H2A authenticate Rad6B ability to ubiquitinate H2A (Fig. 6G). These data confirm that the ubiquitinated band formed with LN+ tumor extracts indeed represents H2A monoubiquitination mediated by endogenous Rad6.
4. Discussion
The data presented here provide novel evidence for a Rad6B role in β-catenin stabilization and activation in breast cancer. Coimmunoprecipitation, immunolocalization, and pull-down assays showed that Rad6B and β-catenin physically interact, and the interacting regions reside between amino acid residues 50–116 of Rad6B and 131–181 of β-catenin encompassing ARM1. The amino acid 131–181 region is implicated in recognition and initiation of high-affinity β-catenin S45 phosphorylation by CK1, which is a prerequisite for subsequent phosphorylation by GSK3β at S33/S37/T41, ubiquitination, and proteasomal degradation [35–38]. Breast cancer cells with Rad6B overexpression show elevated levels of β-catenin unphosphorylated at S33/S37/T41. In vitro reactions examining the impact of recombinant Rad6B on CK1α-mediated GST-β-catenin S45 phosphorylation showed only modest inhibition. Besides S45 phosphorylation of β-catenin, N-terminal cleavage of GST-β-catenin occurred in reactions containing CK1α (data not shown). It is not clear at present if Rad6B inhibition of CK1α activity was compromised by CK1α cleavage of β-catenin.
Elevated levels of Rad6B ubiquitin conjugating activity and a significant positive association between Rad6B and polyubiquitinated β-catenin in invasive breast carcinomas support an important role for Rad6B in β-catenin regulation in clinical breast cancer. Rad6B can mediate β-catenin ubiquitination in vitro in the absence of an E3 ligase. It is likely, however, that the proficient β-catenin polyubiquitination observed in breast tumors involves E3 ligase(s). In this context it should be noted that Rad6B silencing in MDA-MB-231 and WS-15 breast cancer cells suppresses β-catenin polyubiquitination while sparing the nascent β-catenin protein, indicating a major requirement for the Rad6B E2 function in this process [9].
βTrCP ubiquitination of β-catenin K19 and K49 primes it for proteasomal degradation [39,40], and as expected, K19/K49R double mutant β-catenin levels are stabilized due to reduced βTrCP-dependent ubiquitination [40]. Overexpression of a dominant negative βTrCP1 potently inhibited mono- but not polyubiquitination of wild type, K19R-, K49R-, K19/K49R-, and S33Y-mutant β-catenin [40]. These data imply the presence of additional ubiquitinating mechanisms (possibly Rad6B) and ubiquitination sites besides K19 and K49. Among the lysine residues 312, 335, 345, 354, and 394 located in ARMs 5–7, we identified K394 in ARM7 as the major site of Rad6B-induced ubiquitination. Rad6B silencing reduced K354R- and wild type-β-catenin ubiquitination but not K394R-β-catenin, providing further support for a physiological role of Rad6B in K394-β-catenin ubiquitination. Interestingly, the UbPred program [41] predicted K394 as a potential ubiquitination site with similar confidence scores as K49 (score 0.67), an established ubiquitination site besides K19 (score 0.8).
Lysines 394 and 345 are surface-exposed residues [42], and like K345, K394 may be readily accessible for modification and/or serve as a contact site for β-catenin partners. Lysine 345 is a major site of acetylation, which increases β-catenin affinity for TCF4. The K345R mutation caused a 50% decrease in activation of TOP/Flash in HeLa cells [43]. However, in MCF-7 cells K345R mutation caused only a marginal drop in TOP/Flash activity, potentially reflecting cell type differences. K394R mutation caused an ~50% drop in TOP/Flash activity compared to wild type β-catenin, and consistent with the TOP reporter activation data, Rad6B expression, itself a β-catenin/TCF transcriptional target [10], was reduced in K394R-β-catenin transfected MCF-7 cells. Our data showed that compared to wild type and K312R, K335R, K345R and K354R β-catenin mutants, K394R-β-catenin steady-state protein levels were decreased in MCF-7 cells. The reduced K394R-β-catenin steady-state protein levels were not due to 26S proteasomal degradation, since treatment with proteasomal inhibitor MG132 failed to rescue it. The failure of MG132 treatment to enhance K394R-β-catenin levels suggests this additional ubiquitination does not occur at K19 and K49, as their ubiquitination is associated with proteasomal degradation. These data suggest that K394 is a novel ubiquitination site and potentially an important charge button for maintaining the overall structure and stability of β-catenin.
c-Myc promoter-driven luciferase expression (data not shown) and protein levels were downregulated, although not as dramatically as Rad6B, in K394R-β-catenin transfected cells. In contrast, cyclin D1 protein and cyclin D1 promoter-mediated reporter expression [44] were unaffected by β-catenin mutation. These data suggest that target gene cis-acting elements respond to and interact differently with modified β-catenin to regulate gene expression.
Rad6B mutations have not been found in breast cancer [8]. The frequent Rad6B overexpression and presence of activated β-catenin in clinical breast cancer suggests the possibility of in vivo β-catenin K394 ubiquitination. Rad6B, like β-catenin, is present in the cytoplasm and nucleus of Wnt active breast cancer cells [9], and like β-catenin, Rad6B shuttles in and out of the nucleus. This coexistence and ability to shuttle between the nucleus and cytoplasm could provide ample opportunity for β-catenin modification, stabilization and activation by Rad6B. Cytoplasmic or nuclear expression of β-catenin is reported in ~60% of human breast cancers and is shown to independently correlate with poor prognosis [45]. Considering the physiological link between Rad6B and β-catenin stabilization, targeting Rad6B may provide a novel strategy for disrupting activated canonical Wnt signaling in breast cancers [46–48].
5. Conclusions
The oncogenic activity of β-catenin is rendered through its function as a transcription factor. Breast cancer differs from most cancers in that elevated β-catenin mediated transcriptional activity is found despite the absence of β-catenin stabilizing mutations. The identification of Rad6B/β-catenin physical interaction, a Rad6B-induced ubiquitination site of β-catenin and its potential effect on β-catenin transcriptional activity provide a new model for β-catenin oncogenicity in breast cancer.
Acknowledgements
We thank Dr. L.A. Pinna for β-catenin 131–422 and β-catenin 181–422 constructs, Dr. Yutaka Hata for myc-tagged β-catenin 1–781, β-catenin 1–699, β-catenin 131–699 and β-catenin 555–781, Dr. Richard Pestell for cyclin D1 promoter-luciferase vector, Dr. Bert Vogelstein for c-Myc promoter-luciferase vector, and Dr. Vitaly Balan for the anti-myc tag antibody. We thank Dr. Gloria Heppner for helpful discussions and reading of the manuscript.
Grant support: Department of Defense grants W81XWH07-1-0562 and W81XWH-09-1-0608 (MPV Shekhar), and funding from Karmanos Cancer Institute (strategic grant) and Wayne State University (bridge funding).
References
- 1.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence C. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays. 1994;16:253–258. doi: 10.1002/bies.950160408. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds P, Weber S, Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc. Natl. Acad. Sci. U. S. A. 1985;82:168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sung P, Prakash S, Prakash L. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2695–2699. doi: 10.1073/pnas.87.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung P, Prakash S, Prakash L. Stable ester conjugate between the Saccharomyces cerevisiae RAD6 protein and ubiquitin has no biological activity. J. Mol. Biol. 1991;221:745–749. doi: 10.1016/0022-2836(91)80169-u. [DOI] [PubMed] [Google Scholar]
- 6.Koken MH, Reynolds P, Jaspers-Dekker I, Prakash L, Prakash S, Bootsma D, Hoeijmakers JH. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8865–8869. doi: 10.1073/pnas.88.20.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koken MH, Smit EM, Jaspers-Dekker I, Oostra BA, Hagemeijer A, Bootsma D, Hoeijmakers JH. Localization of two human homologs, HHR6A and HHR6B, of the yeast DNA repair gene RAD6 to chromosomes Xq24-q25 and 5q23-q31. Genomics. 1992;12:447–453. doi: 10.1016/0888-7543(92)90433-s. [DOI] [PubMed] [Google Scholar]
- 8.Shekhar MP, Lyakhovich A, Visscher DW, Heng H, Kondrat N. Rad6 over-expression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res. 2002;62:2115–2124. [PubMed] [Google Scholar]
- 9.Shekhar MP, Gerard B, Pauley RJ, Williams BO, Tait L. Rad6B is a positive regulator of beta-catenin stabilization. Cancer Res. 2008;68:1741–1750. doi: 10.1158/0008-5472.CAN-07-2111. [DOI] [PubMed] [Google Scholar]
- 10.Shekhar MP, Tait L, Gerard B. Essential role of T-cell factor/beta-catenin in regulation of Rad6B: a potential mechanism for Rad6B overexpression in breast cancer cells. Mol. Cancer Res. 2006;4:729–745. doi: 10.1158/1541-7786.MCR-06-0136. [DOI] [PubMed] [Google Scholar]
- 11.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 12.Dierick H, Bejsovec A. Cellular mechanisms of wingless/Wnt signal transduction, Curr. Top. Dev. Biol. 1999;43:153–190. doi: 10.1016/s0070-2153(08)60381-6. [DOI] [PubMed] [Google Scholar]
- 13.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 14.Seidensticker MJ, Behrens J. Biochemical interactions in the wnt pathway. Biochim. Biophys. Acta. 2000;1495:168–182. doi: 10.1016/s0167-4889(99)00158-5. [DOI] [PubMed] [Google Scholar]
- 15.Wagenaar RA, Crawford HC, Matrisian LM. Stabilized beta-catenin immortalizes colonic epithelial cells. Cancer Res. 2001;61:2097–2104. [PubMed] [Google Scholar]
- 16.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 17.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 18.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. J. Mammary Gland Biol. Neoplasia. 2004;9:119–131. doi: 10.1023/B:JOMG.0000037157.94207.33. [DOI] [PubMed] [Google Scholar]
- 20.Brown AM. Wnt signaling in breast cancer: have we come full circle? Breast Cancer Res. 2001;3:351–355. doi: 10.1186/bcr321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe LR, Brown AM. Wnt signaling and breast cancer. Cancer Biol. Ther. 2004;3:36–41. doi: 10.4161/cbt.3.1.561. [DOI] [PubMed] [Google Scholar]
- 22.Candidus S, Bischoff P, Becker KF, Hofler H. No evidence for mutations in the alpha- and beta-catenin genes in human gastric and breast carcinomas. Cancer Res. 1996;56:49–52. [PubMed] [Google Scholar]
- 23.Shekhar MP, Santner S, Carolin KA, Tait L. Direct involvement of breast tumor fibroblasts in the modulation of tamoxifen sensitivity. Am. J. Pathol. 2007;170:1546–1560. doi: 10.2353/ajpath.2007.061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennan K, Gonzalez-Sancho JM, Castelo-Soccio LA, Howe LR, Brown AM. Truncated mutants of the putative Wnt receptor LRP6/Arrow can stabilize beta-catenin independently of Frizzled proteins. Oncogene. 2004;23:4873–4884. doi: 10.1038/sj.onc.1207642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerard B, Tait L, Nangia-Makker P, Shekhar MP. Rad6B acts downstream of Wnt signaling to stabilize beta-catenin: implications for a novel Wnt/beta-catenin target. J. Mol. Signal. 2011;6:6. doi: 10.1186/1750-2187-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor-Papadimitriou J, Stampfer M, Bartek J, Lewis A, Boshell M, Lane EB, Leigh IM. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: relation to in vivo phenotypes and influence of medium. J. Cell Sci. 1989;94(Pt 3):403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- 27.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 28.Genander M, Halford MM, Xu NJ, Eriksson M, Yu Z, Qiu Z, Martling A, Greicius G, Thakar S, Catchpole T, Chumley MJ, Zdunek S, Wang C, Holm T, Goff SP, Pettersson S, Pestell RG, Henkemeyer M, Frisen J. Dissociation of EphB2 signaling pathways mediating progenitor cell proliferation and tumor suppression. Cell. 2009;139:679–692. doi: 10.1016/j.cell.2009.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, Counter CM, Nevins JR, Means AR, Sears R. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 30.Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66:5487–5494. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- 31.Choi HJ, Huber AH, Weis WI. Thermodynamics of beta-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J. Biol. Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- 32.Woelfle U, Sauter G, Santjer S, Brakenhoff R, Pantel K. Down-regulated expression of cytokeratin 18 promotes progression of human breast cancer. Clin. Cancer Res. 2004;10:2670–2674. doi: 10.1158/1078-0432.ccr-03-0114. [DOI] [PubMed] [Google Scholar]
- 33.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 34.Sung P, Prakash S, Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988;2:1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- 35.Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, Ben-Neriah Y, Alkalay I. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bustos VH, Ferrarese A, Venerando A, Marin O, Allende JE, Pinna LA. The first armadillo repeat is involved in the recognition and regulation of beta-catenin phosphorylation by protein kinase CK1. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19725–19730. doi: 10.1073/pnas.0609424104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 38.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J. Biol. Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 39.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winer IS, Bommer GT, Gonik N, Fearon ER. Lysine residues Lys-19 and Lys-49 of beta-catenin regulate its levels and function in T cell factor transcriptional activation and neoplastic transformation. J. Biol. Chem. 2006;281:26181–26187. doi: 10.1074/jbc.M604217200. [DOI] [PubMed] [Google Scholar]
- 41.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Kries JP, Winbeck G, Asbrand C, Schwarz-Romond T, Sochnikova N, Dell'Oro A, Behrens J, Birchmeier W. Hot spots in beta-catenin for interactions with LEF-1, conductin and APC. Nat. Struct. Biol. 2000;7:800–807. doi: 10.1038/79039. [DOI] [PubMed] [Google Scholar]
- 43.Levy L, Wei Y, Labalette C, Wu Y, Renard CA, Buendia MA, Neuveut C. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol. Cell. Biol. 2004;24:3404–3414. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowlands TM, Pechenkina IV, Hatsell SJ, Pestell RG, Cowin P. Dissecting the roles of beta-catenin and cyclin D1 during mammary development and neoplasia. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11400–11405. doi: 10.1073/pnas.1534601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clapper ML, Coudry J, Chang WC. Beta-catenin-mediated signaling: a molecular target for early chemopreventive intervention. Mutat. Res. 2004;555:97–105. doi: 10.1016/j.mrfmmm.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 47.Dihlmann S, von Knebel Doeberitz M. Wnt/beta-catenin-pathway as a molecular target for future anti-cancer therapeutics. Int. J. Cancer. 2005;113:515–524. doi: 10.1002/ijc.20609. [DOI] [PubMed] [Google Scholar]
- 48.Dvory-Sobol H, Sagiv E, Kazanov D, Ben-Ze'ev A, Arber N. Targeting the active beta-catenin pathway to treat cancer cells. Mol. Cancer Ther. 2006;5:2861–2871. doi: 10.1158/1535-7163.MCT-06-0122. [DOI] [PubMed] [Google Scholar]