Abstract
Heart disease is a leading cause of death in patients with Duchenne muscular dystrophy (DMD). Patients with DMD lack the protein dystrophin, which is widely expressed in striated muscle. In skeletal muscle, the loss of dystrophin results in dramatically decreased expression of the dystrophin associated glycoprotein complex (DGC). Interestingly, in the heart the DGC is normally expressed without dystrophin; this has been attributed to presence of the dystrophin homologue utrophin. We demonstrate here that neither utrophin nor dystrophin are required for the expression of the cardiac DGC. However, alpha-dystroglycan (α-DG), a major component of the DGC, is differentially glycosylated in dystrophin-(mdx) and dystrophin−/utrophin− (dko) deficient mouse hearts. In both models the altered α-DG retains laminin binding activity, but has an altered localization at the sarcolemma. In hearts lacking both dystrophin and utrophin, the alterations in α-DG glycosylation are even more dramatic with changes in gel migration equivalent to 24 ± 3 kDa. These data show that the absence of dystrophin and utrophin alters the processing of α-DG; however it is not clear if these alterations are a consequence of the loss of a direct interaction with dystrophin/utrophin, or results from an indirect response to the presence of severe pathology. Recently there have been great advances in our understanding of the glycosylation of α-DG regarding its role as a laminin receptor. Here we present data that alterations in glycosylation occurs in the hearts of animal models of DMD, but these changes do not affect laminin binding. The physiological consequences of these alterations remain to be determined, but may have significant implications for the development of therapies for DMD.
Keywords: Dystrophic cardiomyopathy, dystrophin, utrophin, alpha-dystroglycan, glycosylation
INTRODUCTION
DMD and Becker muscular dystrophy (BMD) are caused by mutations in the DMD gene, which encodes the cytoskeletal protein dystrophin (Hoffman et al. 1987). Dystrophin is a large protein that facilitates the connection between the extracellular matrix and the cytoskeleton (Ervasti and Campbell 1991, 1993b; Ohlendieck et al. 1991). Critical to this connection is dystrophin’s interaction with the glycosylated sarcolemmal proteins α- and β-dystroglycan (DG), the sarcoglycan complex (α-, β-, γ-, δ-SG), and sarcospan. Together, these proteins make up the DGC. It is important to note, however, that assembly of this complex does not require dystrophin to be present. For example, the DGC is fully retained in the dystrophin deficient myocardium (Bies et al. 1997; Matsumura et al. 1992; Townsend et al. 2007). It is hypothesized that the linkage between the extracellular matrix (ECM) and the cytoskeleton is critical for the maintaining the integrity of the sarcolemma and cellular calcium gradient (Ervasti and Campbell 1991; Hutter et al. 1991; Menke and Jockusch 1991; Ervasti and Campbell 1993a; Petrof et al. 1993). Reductions in dystrophin and associated proteins renders the myocyte susceptible to increased cell damage and necrosis, contributing to the muscular dystrophy phenotype (Ervasti and Campbell 1993b; Ohlendieck and Campbell 1991). Mice lacking dystrophin (mdx) show markedly reduced levels of all DGC proteins in skeletal muscle (Ohlendieck and Campbell 1991). Without dystrophin, skeletal muscle DGC proteins are degraded by the proteosome (Bonuccelli et al. 2003; Bonuccelli et al. 2007). Several components of the DGC also are down-regulated at the transcriptional level in mdx compared to wild type (WT) skeletal muscle (Turk et al. 2005). It therefore seems likely that the marked decrease in the DGC observed in mdx skeletal muscle results from a combination of increased degradation and decreased production. The role of these processes in the dystrophic heart is unknown.
Understanding how the functions of dystrophin differ between skeletal muscle and the heart provides unique insight into the function of dystrophin. Furthermore, understanding cardiac dystrophin has therapeutic relevance given the clinical importance of heart disease in the management of DMD (Bushby et al. 2010b, a). The mdx mouse, lacking dystrophin, is a genetic model of DMD that displays a subtle cardiac phenotype at baseline and becomes highly apparent with stress testing (Yasuda et al. 2005; Townsend et al. 2007; Bostick et al. 2008). In contrast to skeletal muscle, the components of the DGC are present in the dystrophin-deficient heart (Matsumura et al. 1992; Townsend et al. 2007; Yang et al. 1994), although their function in the absence of dystrophin is not clear. Increased expression of dystrophin’s autosomal homologue, utrophin, has been suggested to functionally replace dystrophin in both skeletal muscle and the heart (Matsumura et al. 1992; Tinsley et al. 1998). Furthermore, mice lacking both dystrophin and utrophin (dko) have a very severe disease (Grady et al. 1997; Connolly et al. 2001; Janssen et al. 2005; Deconinck et al. 1997). It is hypothesized that utrophin expression in the mdx heart is responsible for the continued expression of the DGC without dystrophin. Evaluating the status of the DGC in hearts with neither dystrophin nor utrophin is an important objective of the studies reported here.
The presence of the DGC in the dystrophin-deficient heart permits the unique ability to examine the post-translational processing and trafficking of the DGC without the presence of dystrophin. This is of particular interest for α-DG, which requires glycosylation to perform its primary function of binding laminin (Ibraghimov-Beskrovnaya et al. 1992; Ervasti and Campbell 1993b; Ibraghimov-Beskrovnaya et al. 1993). The core α-DG backbone alone has a predicted mass of 40 kDa, however additional post-translational glycosylation of α-DG in skeletal muscle creates a final protein that migrates equivalent to a 156 kDa protein (Ibraghimov-Beskrovnaya et al. 1992). This glycosylation occurs in a tissue-specific manner, with cardiac α-DG migrating slightly faster than the skeletal muscle form, while brain α-DG migrates equivalent to a 120 kDa protein (Gee et al. 1993; Ibraghimov-Beskrovnaya et al. 1992; Ervasti et al. 1997). Because glycosylation greatly alters the structure, localization, and binding characteristics of a protein, these tissue-specific differences in glycosylation likely reflect necessary modifications in α-DG function for each tissue and stage of development. α-DG is composed of three domains: two globular N-glycosylated domains at the N- and C-terminus and a central heavily O-glycosylated mucin domain (Brancaccio et al. 1995, 1997). Within this mucin domain are a large number of O-glycan structures, including both N-acetyl-galactosamine and mannose initiated O-glycans (Stalnaker et al. 2010). The O-mannose glycans are of particular importance to the laminin binding function of α-DG, as loss of the enzymes that synthesize the O-mannose result in the complete loss of laminin binding (Yoshida et al. 2001; Willer et al. 2002; Beltran-Valero de Bernabe et al. 2002). A subset of these O-mannose initiated structures are further modified by the addition of xylose-glucuronic acid moieties which have an important role in binding to laminin (Yoshida-Moriguchi et al. 2010; Inamori et al. 2012). Defects in the synthetic pathway of these O-mannose initiated glycans constitute an entire class of muscular dystrophies, characterized by muscle degeneration and a high prevalence of neurological deficits. The role of either dystrophin and/or utrophin in regulating the post-translational modification of α-DG is unknown and understanding this is an important focus of these studies.
EXPERIMENTAL PROCEDURES
Animal Care and Use
All animal experiments described here were reviewed and approved by the University of Minnesota’s Institutional Animal Care and Use Committee. C57BL/10 and mdx mice were obtained from locally maintained SPF colonies replenished every four generations with breeders from Jackson Labs (Bar Harbor, ME). Utrophin and dystrophin double knockout (dko) mice were provided by a colony maintained by the Muscular Dystrophy Center at the University of Minnesota (Landisch et al. 2008).
Immunohistochemistry
Excised heart tissue was embedded in Tissue-Tek O.C.T. Compound (Andwin Scientific, Woodland Hills, CA) and snap-frozen in liquid nitrogen-cooled isopentane. Frozen tissues were cut into 7µm sections and placed on glass slides and stored at −80°C. Slides stained with Cathepsin D required fixation (4% PFA for 10 minutes) and denaturing (1% SDS for 5 minutes) prior to the initial blocking step. At the time of staining, slides were removed from the freezer and allowed to warm to room temperature (RT); sections were washed with PBS and blocked for 30 minutes with 5% bovine serum albumin (BSA) and 0.5% Triton X-100 in PBS. Primary antibodies were diluted in PBS+0.5% Triton X-100+5% BSA and incubated for 1 hour at RT. Primary antibodies were diluted as follows: α-DG 1:100 (IIH6C4, EMD Millipore, Ballerica, MA), β-SG 1:200 (bSarc/5B1, Vector Laboratories, Burlingame, CA), Laminin 1:1000 (L9393, Sigma-Aldrich, St. Louis, MO), Cathepsin D 1:500 (ab75852, Abcam, Cambridge, MA). Slides were washed with 0.5% Triton X-100 in PBS. Goat-anti-mouse (GAM) and/or goat-anti-rabbit (GAR) secondary antibodies conjugated to either Alexa-488 or Alexa-594 were diluted to 1:500 in PBS+0.5% Triton X-100+5% BSA and incubated for 1 hour at RT. Slides were washed and mounted using ProLong Gold Antifade Reagent with DAPI (Invitrogen, Carlsbad, CA). The slide was allowed to cure 24 hours before imaging on the Zeiss Axio Imager M1 epifluorescent and/or Zeizz LSM 510 Meta confocal microscope (Carl Zeiss, Oberkochen, Germany). Laminin and α-DG membrane distributions were measured using a custom line scanning macro in ImageJ and Excel (NIH, Bethesda, MD and Microsoft, Redmond, WA). Briefly, line scans were taken across cellular borders, pixel intensities were normalized and the distance between 50% of peak height was determined, with linear approximations of inter-pixel intensities. Statistical analysis of data was performed by ANOVA with a Dunnett’s post-hoc test using Prism (GraphPad, La Jolla, CA); data from 419–596 individual line scans, from 21–30 images from 2–3 mice.
KCl Washed Membranes
KCl washed membranes were prepared as described previously (Ohlendieck and Campbell 1991). Briefly, excised tissues were snap-frozen and pulverized in liquid nitrogen. Tissues were homogenized and centrifuged at 20,000g for 20 minutes at 4°C. The supernatant was centrifuged again at 4°C at 100,000g for 35 minutes. The resulting pellet was incubated in buffer containing 0.6 M KCl for 1 hour at 4°C. KCl washed membranes were sedimented by a 150,000g spin at 4°C for 37 minutes. Samples were stored at −80°C prior to use. The protein content of the sample was determined using the BCA Protein Assay Kit (Thermo Scientific Pierce, Rockford, IL).
Gel Electrophoresis and Western Blotting
SDS-PAGE was performed on 4–20% gradient gels (Bio-Rad, Hercules, CA) using standard procedures (Towbin et al. 1979; Laemmli 1970). Nitrocellulose membranes were blocked with 5% nonfat milk in Tris-buffered saline (TBS, 50mM Tris, 150mM NaCl)+0.5% Tween-20 (TBS-T) at room temperature for 1 hour. Primary antibodies were diluted in PBS-T; α-DG 1:1000 (IIH6C4, Millipore), β-DG 1:100 (43DAG1/8D5, Vector Laboratories), γ-SG 1:100 (35DAG/21B5, Vector Laboratories), NaKATPase 1:10,000 (α6F Concentrate, Developmental Studies Hybridoma Bank, Iowa City, IA). Following a 1 hour incubation at RT, the membrane was washed with TBS-T and incubated with 1:5000 diluted GAM or GAR secondary antibody conjugated to a Alexa-688 (Invitrogen, Carlsbad, CA) or IR-800nm (Rockland Immunochemicals, Gilbertville, PA) for one hour at RT. After a final wash the membrane was visualized and quantified using the Odyssey Imaging System (Li-Cor, Lincoln, NE).
N-glycosidase F and Neuraminidase Treatments
Our protocols for N-glycosidase F and Neuraminidase treatments were performed as described previously (Ervasti and Campbell 1991). Briefly, 50 µg of protein were boiled in the presence of 1% SDS for five minutes. The sample was allowed to cool and 3 units of PNGase F (New England Biolabs, Ipswich, MA) or 0.0025 units of V. cholerae Neuraminidase (Sigma-Aldrich, St. Louis, MO) were added to the reaction. An equivalent amount of water was added for negative control reactions. The reactions were allowed to incubate at 37°C for 2 hours. 5X sucrose buffer that did not contain SDS was used to load each sample for SDS-PAGE. Migration measurements were calculated from the densest point in each band.
Lectin Binding Assays
Tissues were pulverized and homogenized as described for the KCl-washed membranes. An equal volume of 2X Solubilization Buffer (Tris-HCl 100mM, NaCl 1M, Digitonin 1mM, Pepstatin 0.6mM, Aprotinin 0.5mM, Leupeptin 0.5mM, PMSF 0.1mM, Benzamidine 0.75mM) was added to the tissue homogenate and allowed to incubate in a rotisserie at 4°C for 1 hour. Following this incubation, the homogenate was added to washed lectin beads (wheat germ agglutinin/WGA or Jacalin, Vector Labs, Burlingame, CA) and mixed with constant agitation for 2 hours at 4°C. The samples were spun at 225g for 1 minute and the supernatant was discarded. The samples were washed with Wash Buffer (Tris-HCl 50mM, NaCl 0.5M, Digitonin 0.1mM, and protease inhibitors) 3 times for 5 minutes at 4°C with constant agitation. Bound proteins were eluted by boiling 5 minutes in sample buffer. Beads were removed by centrifugation and eluted proteins were stored at −80°C until use.
Laminin Overlay Assays
Laminin overlay binding assays were preformed as previously described (Michele et al. 2002). Briefly, KCl washed microsome samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes. The blots were hydrated with water and rinsed well with Tris buffered saline+0.05% Triton X-100+Ca2+ (TBS-T+Ca2+, 20mM Tris pH 7.5, 100mM NaCl, 1mM CaCl2, 0.05% Tween-20). The blot was blocked for 1 hour at RT in laminin blocking buffer (laminin binding buffer (LBB; 10mM triethanolamine, 100mM NaCl, 1mM MgCl2, 1mM CaCl2, pH 7.6) + 5% nonfat dry milk, pH 7.4). The blot was washed 3 times for 10 minutes at RT with TBS-T+Ca2+. Engelbreth-Holm-Swarm (EHS) laminin (Invitrogen, Carlsbad, CA) was diluted 1µg/ml in laminin binding buffer and added to the blot which was incubated overnight at 4°C. Following 3 washes of 10 minutes each with TBS-T+Ca2+, anti-laminin primary antibody (L9393, Sigma) was diluted 1:1000 in LBB and added to cover the blot. The blot was incubated at RT for 1 hour, then washed with TBS-T+Ca2+ and incubated with the GAR secondary antibody (diluted 1:5000 in laminin blocking buffer) for 1 hour at RT. Following washes with TBS-T+Ca2+, blots were imaged on the Odyssey Imaging System (Licor, Lincoln, NE).
Solid-Phase Laminin Binding Assay
Solid phase binding assays were performed as previously described (Michele et al. 2002). Briefly, cleared heart homogenates were allowed to adhere to 96 well plates for 16 hours at 4°C. After washing, laminin was added to the wells at varying concentrations for 2 hours at room temperature. Following washing, 1:10,000 anti-laminin antibody (L9393, Sigma) overnight at 4°C. Plates were washed and 1:1000 secondary antibody was added for 30 minutes at room temperature. Signal intensity quantified using the Odyssey Imaging System (Licor).
RESULTS
Maintenance of the DGC in the Absences of Both Dystrophin and Utrophin
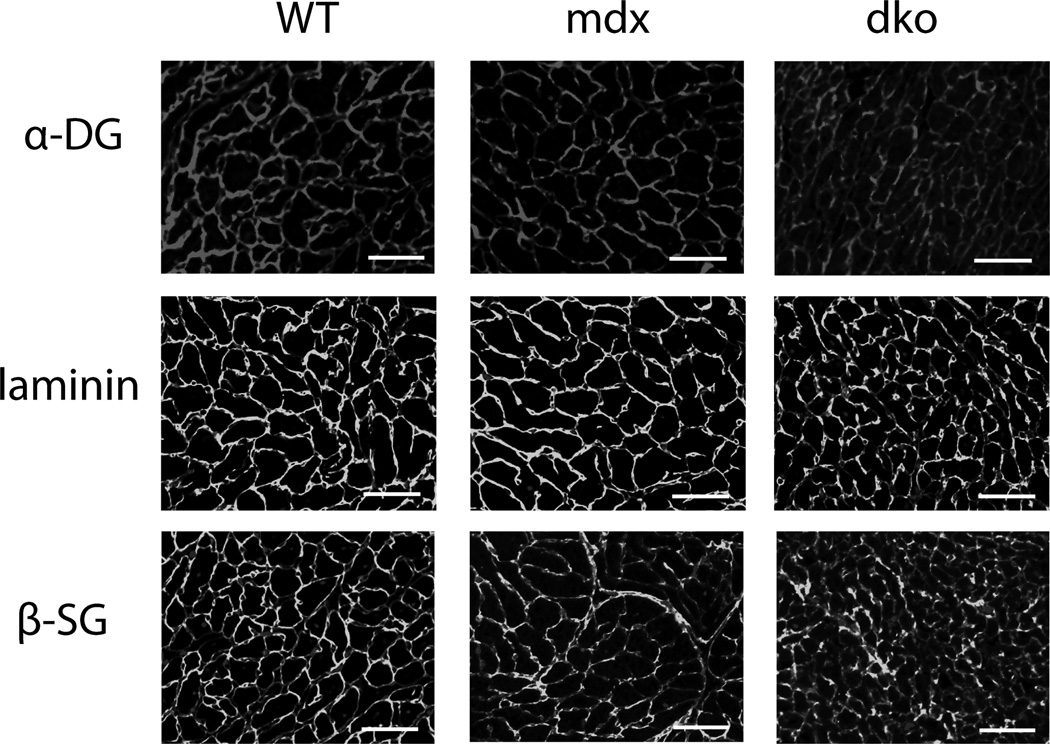
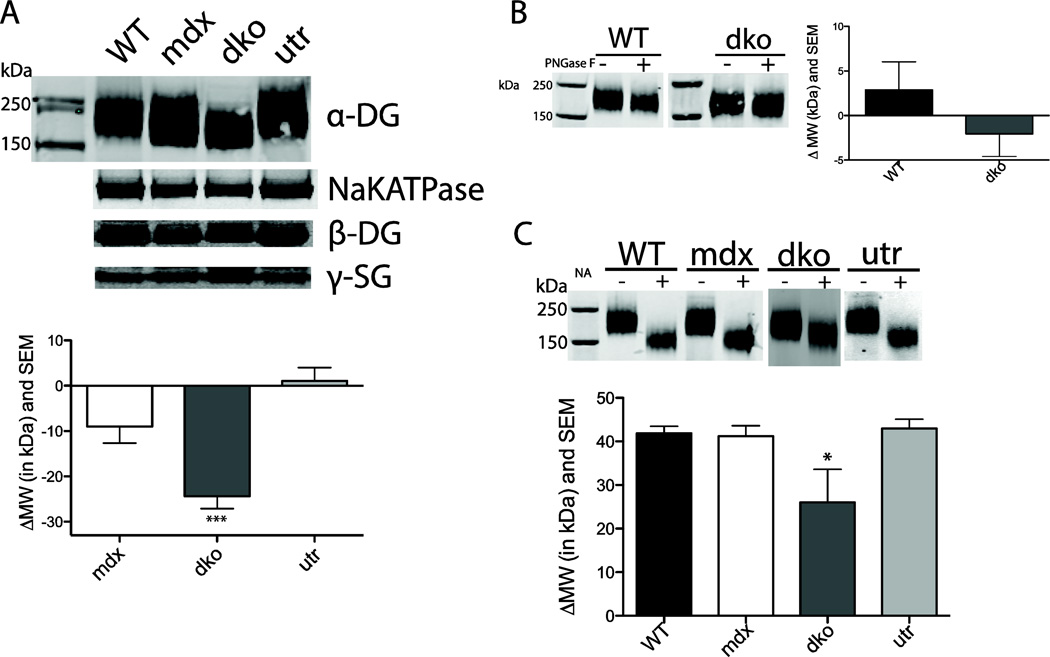
In contrast to skeletal muscle, α-DG and β-SG were expressed at the sarcolemma in the dystrophin-deficient heart (Fig. 1). Furthermore, these proteins in the DGC complex were also present at the sarcolemma in the absence of both dystrophin and utrophin. Quantitative western blotting of KCl-washed microsomal membranes isolated from the hearts of WT, mdx, dko, and utrophin-null (utr) mice demonstrated that α-DG, β-DG and γ-SG were detected at normal levels in the presence and absence of dystrophin and/or utrophin (Fig 2a). Interestingly, there were differences in the migration pattern of the α-DG from dko hearts, suggesting an alteration in the glycosylation of this protein. To begin assessing changes in α-DG glycosylation the differences in the overall migration of α-DG in WT, mdx, dko and utr mice were quantified. Migration of α-DG from mdx and utr hearts was not different from WT, however, dko α-DG migrated 24 ± 3 kDa faster (p<0.0001) than WT α-DG (Mean ± SEM) (Fig 2a). α-DG migration differences results are reported as a change in kDa relative to WT α-DG. However, it should be noted that these differences reflect changes in the overall size of the protein and/or its ability to migrate through a polyacrylamide gel.
Figure 1. Immunohistochemistry of mouse cardiac tissue.
Antibodies bound to α-DG, laminin and β-SG are observable in cardiac tissue of WT, mdx and dko mice. Line represents 100µm. n = 3
Figure 2. Western blots of mouse KCl washed cardiac microsomes and α-DG glycosylation.
a) Representative western blot results of KCl washed cardiac microsomes probed with α-DG, sodium-potassium ATPase (NaKATPase), β-DG and γ-SG antibodies for WT, mdx, dko and utr mice. Differences in α-DG migration are quantified based on the average WT molecular weight (MW) value. KCl migration WT n = 12, mdx n = 14, dko n = 14, utr n = 12. *** p < 0.0001. b) N-glycosidase-F (PNGase F), an enzyme that removes most types of N-linked glycosylation, does not significantly shift MW for WT or dko α-DG compared to untreated control samples (n = 4–5 sample). c) Treatment with neuraminidase (NA), which removes sialic acid and some glucosamines, shifts dko α-DG gel mobility significantly less than WT (n=3–4). * p < 0.05
Glycosidases Reveal Altered α-DG Glycosylation
Given that α-DG is heavily glycosylated, alterations in glycosylation processing seemed a potential mechanism for the increased mobility of α-DG observed in dko mice. To assess the level of N-linked glycosylation present on α-DG, we enzymatically removed N-linked carbohydrates via incubation with N-glycosidase F (PNGase F). No changes in α-DG mobility were observed between KCl washed microsome samples treated with or without PNGase F in WT and dko mice (Fig 2b). In contrast, incubation of samples with neuraminidase (NA), which removes sialic acid, resulted in a marked shift in α-DG mobility (Fig 2c). This shift was approximately equal for WT, mdx and utr α-DG (WT=42 ± 2), but was significantly lower for dko (26 ± 8) α-DG (p<0.05).
Altered α-DG Lectin Binding in mdx and dko Hearts
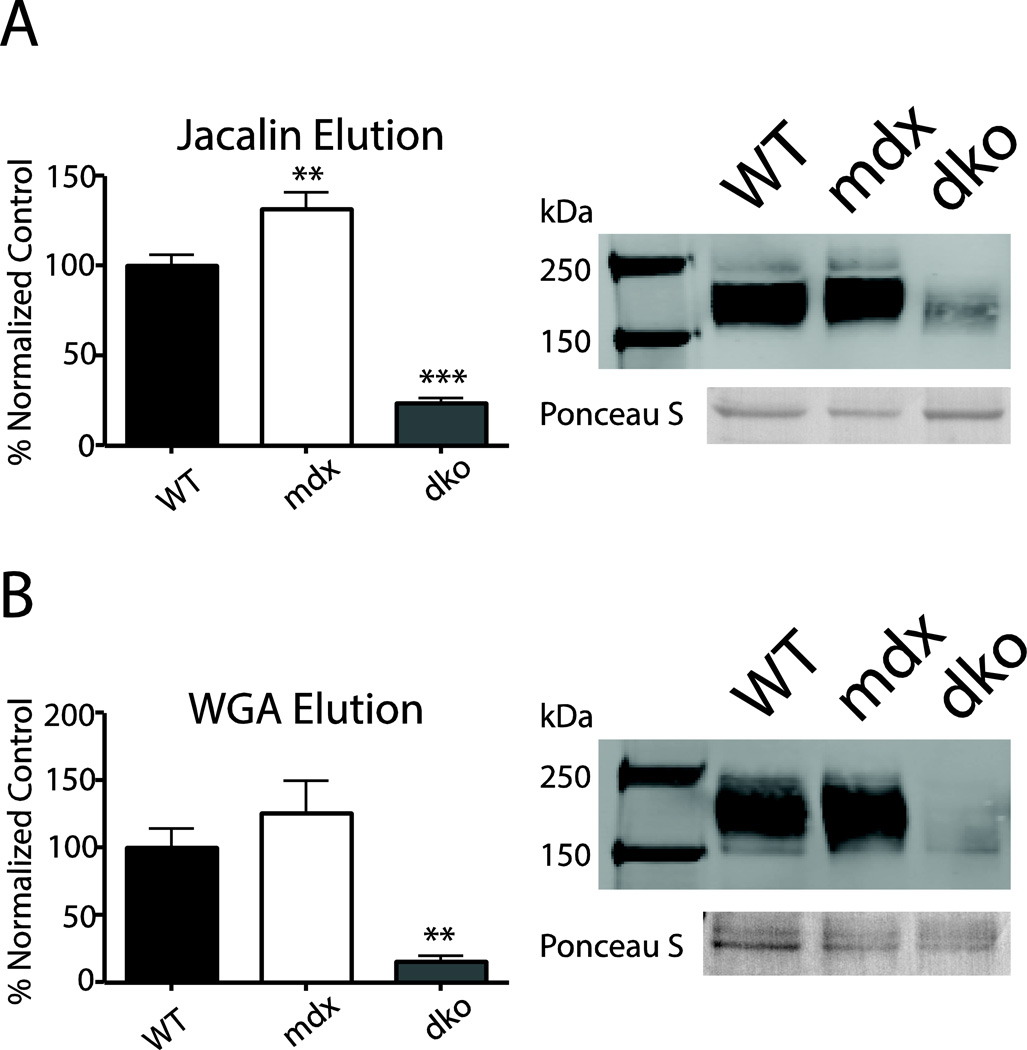
The binding of lectins are often used to characterize the properties of glycosylated proteins. To further assess the glycosylation properties of cardiac α-DG from WT, mdx, and dko mice, α-DG binding to wheat germ agglutinin-(WGA) and jacalin-conjugated agarose beads was determined in pull-down assays. These lectins preferentially bind to distinct carbohydrate moieties; jacalin binds to N-acetyl galactosamine in which the C-6 is free (Tachibana et al. 2006) and WGA has affinity for both N-acetyl-D-glucosamine and sialic acid moieties (Gallagher et al. 1985). In hearts without dystrophin or utrophin, significantly less α-DG bound to either WGA or jacalin compared to that in WT samples (p<0.0001, Fig 3). Interestingly, α-DG from mdx mice showed significantly more binding to jacalin compared to WT (p<0.001, Fig 3) indicating that the loss of dystrophin alone alters α-DG glycosylation in the heart.
Figure 3. Lectin binding assays.
Mouse cardiac homogenate proteins bound to jacalin- (a) or WGA- (b) conjugated agarose beads. Proteins were eluted, subjected to SDS-PAGE, and probed for α-DG on a western blot. α-DG expression is reported as a normalized percentage of WT α-DG signal +/− SEM. Significantly less dko α-DG was eluted from both WGA and jacalin lectins compared to WT, whereas mdx α-DG eluted more α-DG from the jacalin lectin compared to WT α-DG. Ponceau S stained band (~70 kDa) provided as a loading control. (n = 4–9) ** p < 0.001, *** p < 0.0001
Laminin Binding Unaffected by Altered Glycosylation of α-DG
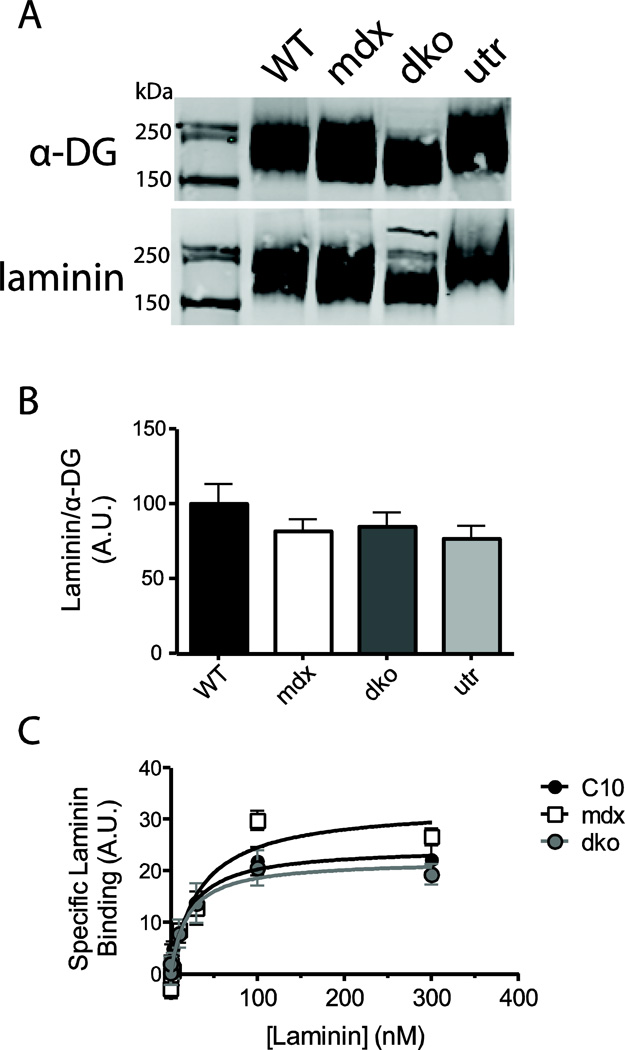
Glycosylation is critically important for α-DG ligand binding, thus alterations in glycosylation may effect the affinity of its laminin binding and perturb the connection to the extracellular matrix (Michele et al. 2002; Yoshida-Moriguchi et al. 2010; Inamori et al. 2012). We investigated this possibility by performing a laminin overlay binding assay in WT, mdx, dko and utr mice. Despite the significant differences in the gel migration and lectin binding α-DG in dko hearts, the affinity and levels of laminin binding to α-DG was not significantly different in any of the mouse genotypes examined here (Fig 4). Specific laminin binding affinities of 21.2 ± 7.6 nM, 33.0 ± 12.5 nM, and 18.8 ± 8.2 nM were observed in WT, mdx, and dko homogenates respectively. These data indicate that the changes in glycosylation observed in dko α-DG do not affect laminin binding.
Figure 4. Laminin overlay binding assay.
The laminin binding capacity of α-DG was assessed using a far-western technique (a), and results were quantified by calculating the raw laminin signal over the raw α-DG signal (b). A solid-state binding assay was used to determine the affinity the laminin binding sites in cardiac homogenates (c). No significant difference in laminin binding capacity or affinity was observed between WT, mdx, dko and utr α-DG. (n=7–8)
Trafficking of α-DG in mdx and dko Hearts
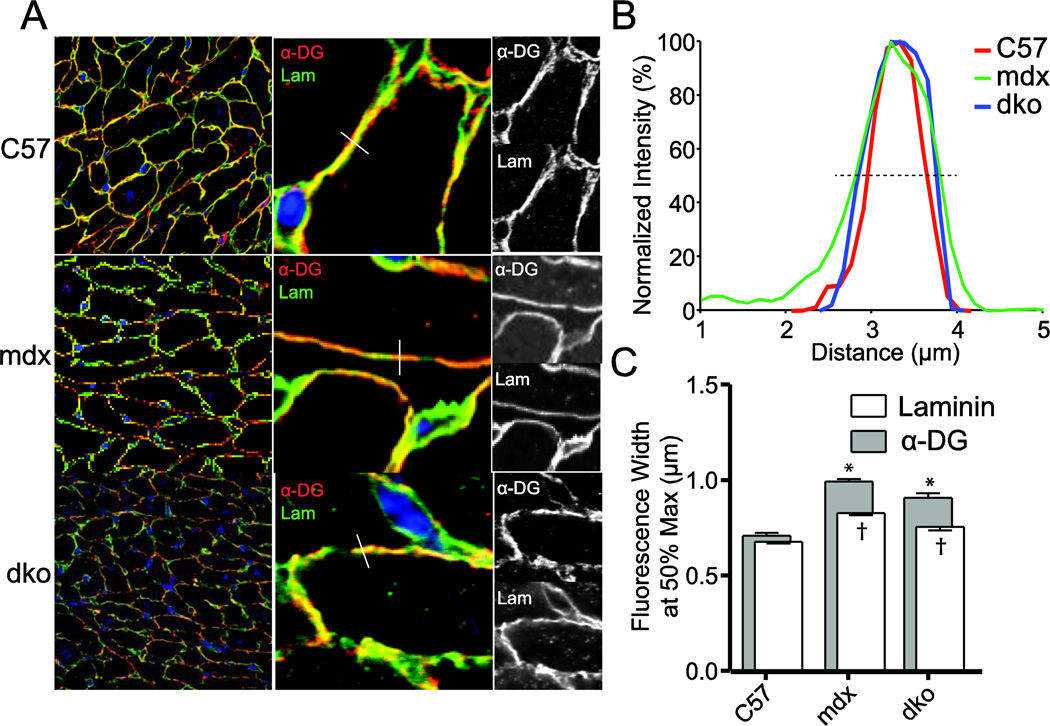
To test the possibility that the absence of dystrophin, with or without utrophin, may result in an unstable α-DG, the potential lysosomal degradation of α-DG was assessed via immunofluorescent label co-localization with Cathepsin D. These studies revealed that lysosomal content is increased in mdx hearts compared to WT hearts and is further increased in dko hearts; however, the majority of α-DG is not co-localized with Cathepsin D in the heart (Supplemental Fig 1). Using confocal microscopy, the distribution of laminin and α-DG was measured across cellular interface (Fig 5a). The width at 50% of the normalized height was determined for each line-scan for both α-DG and laminin for WT, mdx, and dko (Fig 5b and c). Interestingly, both mdx and dko hearts demonstrated significantly wider distributions of immunoreactivity for laminin and α-DG compared to WT values. Furthermore, the calculated difference between laminin and α-DG distributions is also significantly greater than that observed in WT hearts.
Figure 5. Line scanning of laminin and α-DG immunoreactivity.
Immunofluorescent colocalization of laminin and α-DG was measured by confocal microscopy. Line scans of membrane interfaces expressing both laminin and α-DG were collected and analyzed. (a) Representative images demonstrating example line scans. (b) Representative normalized line scans from WT, mdx, and dko sections. The dashed line represents 50%, used to calculate the distribution of immunofluorescence. (c) Summary of the distribution of immunoreactivity for laminin and α-DG. n= 419–596 individual line scans. * P<0.0001 versus WT α-DG distribution, † P< 0.0001 versus WT laminin distribution
DISCUSSION
We have shown normal localization of α-DG and β-SG at the cardiac sarcolemma in dystrophin-deficient mdx mice and demonstrate that α-DG, β-DG and γ-SG are expressed at normal levels in KCl-washed microsomes (Figs. 1 and 2a). These findings are in contrast to skeletal muscle, where the absence of dystrophin results in the nearly complete loss of the DGC (Ohlendieck and Campbell 1991; Ervasti et al. 1990). Interestingly, the expression of these proteins is reintroduced in skeletal muscle by over-expression of the dystrophin homologue utrophin (Matsumura et al. 1992). This observation has led to a potential therapeutic approach using utrophin over-expression to compensate for the loss of dystrophin (Rafael et al. 1998; Tinsley et al. 1996; Tinsley et al. 1998; Sonnemann et al. 2009; Matsumura et al. 1992). The up-regulation of utrophin in the mdx heart has been proposed as a mechanism by which the DGC is maintained in this tissue (Matsumura et al. 1992). The current study and several others (Matsumura et al. 1992; Yang et al. 1994; Townsend et al. 2007) provide evidence that the cardiac DGC is maintained in the absence of both dystrophin and utrophin, indicating that neither of these cytoskeletal proteins are required for sarcolemmal localization of DGC proteins in the heart. However, several studies have reported decreases in α-DG content from dystrophin deficient hearts (Bies et al. 1997; Lohan et al. 2005).
It is possible that the α-DG may be present in a mislocalized form. Our data demonstrates that α-DG is not colocalized with the lysosomal marker Cathepsin D in WT, mdx or dko mouse cardiac cells, although cathepsin D content is increased in both mdx and dko hearts, similar to that seen in DMD patient (Whitaker et al. 1983) and mdx (Aoyagi et al. 1981) skeletal muscle. Confocal immunofluorescence studies demonstrate that the distribution of α-DG immunoreactivity is significantly greater in hearts lacking dystrophin; furthermore, the calculated difference between the laminin and α-DG distributions is significantly greater in hearts lacking dystrophin. These results are consistent with the hypothesis that the localization of α-DG is different without dystrophin, although the nature and functional consequences of this diffuse localization are unclear. It is important to recognize that the direct detection of α-DG in these studies uses the IIH6 monoclonal antibody, which has an epitope that is very tightly associated with the laminin binding function of α-DG (Ervasti and Campbell 1993b). While the overall levels of IIH6 immunoreactivity are the same as that seen in WT mice, it is possible that other, non-IIH6 binding forms of α-DG may be present in these dystrophic hearts.
The nature of the altered gel migration of α-DG observed in the dko hearts is of great interest. Changes in gel migration can result from changes in protein mass, structural stiffness, and/or net charge. The results presented here indicate that a combination of effects are likely responsible for the shift in migration observed in α-DG from dko hearts. Treatment with NA results in significant shifts in the migration of α-DG from all genotypes of mice examined here, but the change is markedly smaller in the dko. Interestingly, the final NA-treated α-DG has essentially equivalent migration in all genotypes. This observation indicates that the increased gel migration seen in the dko α-DG results from changes in the glycosylation, rather than proteolysis of the core peptide. Previous data has indicated that nearly ⅔ of the glycosylation sites of α-DG contain a sialic acid moiety (Stalnaker et al. 2010), the sugar removed by NA. This observation suggests that α-DG from dko hearts does not have the full complement of sialic acid moieties. The decreased change in gel migration following NA treatment indicates a reduced sialic acid content in the dko α-DG, which contributes to increased α-DG migration through reduction in size, charge, and protein stiffness. Modification of the sialic acids of α-DG has been shown to significantly modulate the severity of disease in the dystrophin-deficient mouse. Loss of a single hydroxyl group on a subset of α-DG carbohydrate chains significantly worsens the pathology of both skeletal and cardiac muscle (Chandrasekharan et al. 2010). In contrast, the addition of a N-acetyl-galactosamine, through the over-expression of Galgt2, provides significant protection to skeletal muscle of the mdx mouse (Martin et al. 2009).
The lectin WGA binds to sialic acid moieties (Combs and Ervasti 2005). The observation that WGA binds very poorly to α-DG from dko hearts is consistent with the hypothesis that sialic acid addition is disrupted without dystrophin and utrophin. However, WGA also binds to N-acetyl-glucosamine (Nagata and Burger 1974), allowing for the possibility that alterations in glycosylation extend beyond changes in sialic acid content. To explore the status of other forms of glycosylation, we examined the binding of α-DG to the lectin jacalin, which has an affinity for a subset of N-acetyl-galactosamine-containing carbohydrates (Tachibana et al. 2006). The marked reduction in jacalin binding to α-DG from dko hearts indicates that several forms of α-DG glycosylation are altered without dystrophin and utrophin. Interestingly, there is significantly greater binding of jacalin to α-DG from mdx hearts, suggesting that excess N-acetyl-galactosamine is present on the α-DG in the absence of dystrophin. Together these data strongly suggest that the glycosylation of the α-DG mucin domain is altered in these dystrophic hearts.
The main function of α-DG is to bind to the extracellular matrix proteins, such as laminin, an activity that is dependent on glycosylation (Combs and Ervasti 2005; Michele et al. 2002). Despite the changes in glycosylation, no significant differences in EHS mouse laminin binding were observed between WT, mdx, dko and utr α-DG in the laminin overlay assay. Similarly, no differences were observed between in the solid-state laminin binding assays, although the laminin binding affinities of the homogenates used in the current study were much lower than that observed with WGA-purified α-DG (Michele et al. 2009). These observations suggest that neither dystrophin nor utrophin are critically important for the addition of the carbohydrate structures that are essential for the binding of laminin (Inamori et al. 2012). This raises fundamental questions regarding the function of the α-DG glycosylation not participating in laminin binding. The high concentrations of serine, threonine, and proline in the central domain of α-DG are similar to the O-glycosylation domains of the mucin proteins (Ibraghimov-Beskrovnaya et al. 1992). In mucin proteins O-glycosylation has been proposed to function as a structural modifier. The extensive glycosylation of the central domain of α-DG would be expected to both stiffen and extend this domain, creating a rod like structure protruding ≈25 nm from the surface of the cell (Jentoft 1990; Hilkens et al. 1992). LARGE-modified glycosylation has been proposed to attach to the α-DG core protein at the end of this rod-like mucin domain (Yoshida-Moriguchi et al. 2010; Hara et al. 2011). The consequences of changes in the extent or nature of α-DG glycosylation is unknown, but changes to the mechanical properties of the protein would be expected to alter its function and may contribute to the severity of the dystrophic phenotype in the dko mouse.
The underlying mechanism by which the loss of dystrophin or dystrophin and utrophin result in altered α-DG glycosylation is not clear. One possibility is that the dystrophic disease process fundamentally alters protein processing in the cardiomyocyte. In heart failure there is evidence of disruptions of protein glycosylation and trafficking (Kiarash et al. 2004), although cardiac α-DG migrates normally in our studies of failing human hearts (data not shown). In skeletal muscle, the loss of dystrophin results in significant disruption of both the Golgi complex (Percival et al. 2010) and the microtubule network (Prins et al. 2009). If similar disruptions occur in the heart, they could contribute to the perturbations in α-DG processing observed in the current study. Alterations in glycosylation and trafficking of the DGC may be explained by a necessity for a direct interaction between the DGC and dystrophin or utrophin before insertion into the sarcolemma, however the failure to observe dystrophin association with any internal membranes argues against this hypothesis (Watkins et al. 1988; Byers et al. 1991). Although given the long half-life of dystrophin (Lu et al. 2005), the probability of observing such a transient complex would be low making it difficult to exclude a direct interaction of the DGC with dystrophin or utrophin in the internal membranes. Another possibility is that a portion of the post-translational processing of α-DG is completed following insertion in the surface membrane. This is supported by the observation that many glycosyltransferases can be found expressed on the surface membrane (Berger 2002), including enzymes implicated in α-DG processing (Beedle et al. 2007). A shift in gel migration of α-DG similar to that observed in the utrophin/dystrophin-null mice is observed in a subset of patients with mutations in the fukutin-related protein (FKRP) gene (Brockington et al. 2001). In the intact skeletal muscle, this protein is localized to the same surface membrane domain as the rest of the DGC (Beedle et al. 2007). Which, with the data presented here, supports a model by which the final maturation of α-DG occurs at the cell surface and this process is partially coordinated by dystrophin or utrophin. In summary, we demonstrate novel findings regarding the processing of dystrophin-associated proteins in the hearts of several relevant models of dystrophic cardiomyopathy. Specifically, the alterations in glycosylation of α-DG in the heart of the dko mouse may significantly contribute to the severity of the disease in this model of muscular dystrophy.
Supplementary Material
Bibliography
- Aoyagi T, Wada T, Kojima F, Nagai M, Umezawa H. Various enzyme activities in muscle and other organs of dystrophic mice. J Clin Invest. 1981;67(1):51–59. doi: 10.1172/JCI110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedle AM, Nienaber PM, Campbell KP. Fukutin-related protein associates with the sarcolemmal dystrophin-glycoprotein complex. J Biol Chem. 2007;282(23):16713–16717. doi: 10.1074/jbc.C700061200. [DOI] [PubMed] [Google Scholar]
- Beltran-Valero de Bernabe D, Currier S, Steinbrecher A, Celli J, van Beusekom E, van der Zwaag B, Kayserili H, Merlini L, Chitayat D, Dobyns WB, Cormand B, Lehesjoki AE, Cruces J, Voit T, Walsh CA, van Bokhoven H, Brunner HG. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. American journal of human genetics. 2002;71(5):1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger EG. Ectopic localizations of Golgi glycosyltransferases. Glycobiology. 2002;12(2):29R–36R. doi: 10.1093/glycob/12.2.29r. [DOI] [PubMed] [Google Scholar]
- Bies RD, Maeda M, Roberds SL, Holder E, Bohlmeyer T, Young JB, Campbell KP. A 5' dystrophin duplication mutation causes membrane deficiency of alpha-dystroglycan in a family with X-linked cardiomyopathy. J Mol Cell Cardiol. 1997;29(12):3175–3188. doi: 10.1006/jmcc.1997.0568. [DOI] [PubMed] [Google Scholar]
- Bonuccelli G, Sotgia F, Capozza F, Gazzerro E, Minetti C, Lisanti MP. Localized treatment with a novel FDA-approved proteasome inhibitor blocks the degradation of dystrophin and dystrophin-associated proteins in mdx mice. Cell Cycle. 2007;6(10):1242–1248. doi: 10.4161/cc.6.10.4182. [DOI] [PubMed] [Google Scholar]
- Bonuccelli G, Sotgia F, Schubert W, Park DS, Frank PG, Woodman SE, Insabato L, Cammer M, Minetti C, Lisanti MP. Proteasome inhibitor (MG-132) treatment of mdx mice rescues the expression and membrane localization of dystrophin and dystrophin-associated proteins. Am J Pathol. 2003;163(4):1663–1675. doi: 10.1016/S0002-9440(10)63523-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick B, Yue Y, Lai Y, Long C, Li D, Duan D. Adeno-associated virus serotype-9 microdystrophin gene therapy ameliorates electrocardiographic abnormalities in mdx mice. Hum Gene Ther. 2008;19(8):851–856. doi: 10.1089/hum.2008.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio A, Schulthess T, Gesemann M, Engel J. Electron microscopic evidence for a mucin-like region in chick muscle alpha-dystroglycan. FEBS Lett. 1995;368(1):139–142. doi: 10.1016/0014-5793(95)00628-m. [DOI] [PubMed] [Google Scholar]
- Brancaccio A, Schulthess T, Gesemann M, Engel J. The N-terminal region of alpha-dystroglycan is an autonomous globular domain. European journal of biochemistry/FEBS. 1997;246(1):166–172. doi: 10.1111/j.1432-1033.1997.00166.x. [DOI] [PubMed] [Google Scholar]
- Brockington M, Blake DJ, Prandini P, Brown SC, Torelli S, Benson MA, Ponting CP, Estournet B, Romero NB, Mercuri E, Voit T, Sewry CA, Guicheney P, Muntoni F. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. American journal of human genetics. 2001;69(6):1198–1209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, Kaul A, Kinnett K, McDonald C, Pandya S, Poysky J, Shapiro F, Tomezsko J, Constantin C. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010a;9(1):77–93. doi: 10.1016/S1474-4422(09)70271-6. 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, Kaul A, Kinnett K, McDonald C, Pandya S, Poysky J, Shapiro F, Tomezsko J, Constantin C. Diagnosis and management of Duchenne muscular dystrophy, part 2: implementation of multidisciplinary care. Lancet Neurol. 2010b;9(2):177–189. doi: 10.1016/S1474-4422(09)70272-8. 10.1016/S1474-4422(09)70272-8. [DOI] [PubMed] [Google Scholar]
- Byers TJ, Kunkel LM, Watkins SC. The subcellular distribution of dystrophin in mouse skeletal, cardiac, and smooth muscle. The Journal of cell biology. 1991;115(2):411–421. doi: 10.1083/jcb.115.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan K, Yoon JH, Xu Y, deVries S, Camboni M, Janssen PM, Varki A, Martin PT. A human-specific deletion in mouse Cmah increases disease severity in the mdx model of Duchenne muscular dystrophy. Sci Transl Med. 2010;2(42):42ra54. doi: 10.1126/scitranslmed.3000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs AC, Ervasti JM. Enhanced laminin binding by alpha-dystroglycan after enzymatic deglycosylation. Biochem J. 2005;390(Pt 1):303–309. doi: 10.1042/BJ20050375. 10.1042/BJ20050375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly AM, Keeling RM, Mehta S, Pestronk A, Sanes JR. Three mouse models of muscular dystrophy: the natural history of strength and fatigue in dystrophin-, dystrophin/utrophin-, and laminin alpha2-deficient mice. Neuromuscul Disord. 2001;11(8):703–712. doi: 10.1016/s0960-8966(01)00232-2. [DOI] [PubMed] [Google Scholar]
- Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, Watt DJ, Dickson JG, Tinsley JM, Davies KE. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90(4):717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Burwell AL, Geissler AL. Tissue-specific heterogeneity in alpha-dystroglycan sialoglycosylation. Skeletal muscle alpha-dystroglycan is a latent receptor for Vicia villosa agglutinin b4 masked by sialic acid modification. J Biol Chem. 1997;272(35):22315–22321. doi: 10.1074/jbc.272.35.22315. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66(6):1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. Dystrophin-associated glycoproteins: their possible roles in the pathogenesis of Duchenne muscular dystrophy. Molecular and cell biology of human diseases series. 1993a;3:139–166. doi: 10.1007/978-94-011-1528-5_6. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993b;122(4):809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345(6273):315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- Gallagher JT, Morris A, Dexter TM. Identification of two binding sites for wheat-germ agglutinin on polylactosamine-type oligosaccharides. Biochem J. 1985;231(1):115–122. doi: 10.1042/bj2310115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee SH, Blacher RW, Douville PJ, Provost PR, Yurchenco PD, Carbonetto S. Laminin-binding protein 120 from brain is closely related to the dystrophin-associated glycoprotein, dystroglycan, and binds with high affinity to the major heparin binding domain of laminin. J Biol Chem. 1993;268(20):14972–14980. [PubMed] [Google Scholar]
- Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90(4):729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- Hara Y, Kanagawa M, Kunz S, Yoshida-Moriguchi T, Satz JS, Kobayashi YM, Zhu Z, Burden SJ, Oldstone MB, Campbell KP. Like-acetylglucosaminyltransferase (LARGE)-dependent modification of dystroglycan at Thr-317/319 is required for laminin binding and arenavirus infection. Proc Natl Acad Sci U S A. 2011;108(42):17426–17431. doi: 10.1073/pnas.1114836108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilkens J, Ligtenberg MJ, Vos HL, Litvinov SV. Cell membrane-associated mucins and their adhesion-modulating property. Trends in Biochemical Sciences. 1992;17(9):359–363. doi: 10.1016/0968-0004(92)90315-z. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Hutter OF, Burton FL, Bovell DL. Mechanical properties of normal and mdx mouse sarcolemma: bearing on function of dystrophin. J Muscle Res Cell Motil. 1991;12(6):585–589. doi: 10.1007/BF01738447. [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355(6362):696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O, Milatovich A, Ozcelik T, Yang B, Koepnick K, Francke U, Campbell KP. Human dystroglycan: skeletal muscle cDNA, genomic structure, origin of tissue specific isoforms and chromosomal localization. Hum Mol Genet. 1993;2(10):1651–1657. doi: 10.1093/hmg/2.10.1651. [DOI] [PubMed] [Google Scholar]
- Inamori K, Yoshida-Moriguchi T, Hara Y, Anderson ME, Yu L, Campbell KP. Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science. 2012;335(6064):93–96. doi: 10.1126/science.1214115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen PM, Hiranandani N, Mays TA, Rafael-Fortney JA. Utrophin deficiency worsens cardiac contractile dysfunction present in dystrophin-deficient mdx mice. Am J Physiol Heart Circ Physiol. 2005;289(6):H2373–H2378. doi: 10.1152/ajpheart.00448.2005. 10.1152/ajpheart.00448.2005. [DOI] [PubMed] [Google Scholar]
- Jentoft N. Why are proteins O-glycosylated? Trends in Biochemical Sciences. 1990;15(8):291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- Kiarash A, Kelly CE, Phinney BS, Valdivia HH, Abrams J, Cala SE. Defective glycosylation of calsequestrin in heart failure. Cardiovascular research. 2004;63(2):264–272. doi: 10.1016/j.cardiores.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landisch RM, Kosir AM, Nelson SA, Baltgalvis KA, Lowe DA. Adaptive and nonadaptive responses to voluntary wheel running by mdx mice. Muscle & nerve. 2008;38(4):1290–1303. doi: 10.1002/mus.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohan J, Culligan K, Ohlendieck K. Deficiency in Cardiac Dystrophin Affects the Abundance of the alpha-/beta-Dystroglycan Complex. J Biomed Biotechnol. 2005;2005(1):28–36. doi: 10.1155/S111072430440103X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J, Jadoon A, Bou-Gharios G, Partridge T. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci U S A. 2005;102(1):198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PT, Xu R, Rodino-Klapac LR, Oglesbay E, Camboni M, Montgomery CL, Shontz K, Chicoine LG, Clark KR, Sahenk Z, Mendell JR, Janssen PM. Overexpression of Galgt2 in skeletal muscle prevents injury resulting from eccentric contractions in both mdx and wild-type mice. American journal of physiology Cell physiology. 2009;296(3):C476–C488. doi: 10.1152/ajpcell.00456.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360(6404):588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- Menke A, Jockusch H. Decreased osmotic stability of dystrophin-less muscle cells from the mdx mouse. Nature. 1991;349(6304):69–71. doi: 10.1038/349069a0. [DOI] [PubMed] [Google Scholar]
- Michele DE, Barresi R, Kanagawa M, Saito F, Cohn RD, Satz JS, Dollar J, Nishino I, Kelley RI, Somer H, Straub V, Mathews KD, Moore SA, Campbell KP. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 2002;418(6896):417–422. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]
- Michele DE, Kabaeva Z, Davis SL, Weiss RM, Campbell KP. Dystroglycan matrix receptor function in cardiac myocytes is important for limiting activity-induced myocardial damage. Circ Res. 2009;105(10):984–993. doi: 10.1161/CIRCRESAHA.109.199489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y, Burger MM. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. The Journal of biological chemistry. 1974;249(10):3116–3122. [PubMed] [Google Scholar]
- Ohlendieck K, Campbell KP. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J Cell Biol. 1991;115(6):1685–1694. doi: 10.1083/jcb.115.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendieck K, Ervasti JM, Snook JB, Campbell KP. Dystrophin-glycoprotein complex is highly enriched in isolated skeletal muscle sarcolemma. J Cell Biol. 1991;112(1):135–148. doi: 10.1083/jcb.112.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival JM, Anderson KN, Huang P, Adams ME, Froehner SC. Golgi and sarcolemmal neuronal NOS differentially regulate contraction-induced fatigue and vasoconstriction in exercising mouse skeletal muscle. J Clin Invest. 2010;120(3):816–826. doi: 10.1172/JCI40736. 10.1172/JCI40736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci U S A. 1993;90(8):3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KW, Humston JL, Mehta A, Tate V, Ralston E, Ervasti JM. Dystrophin is a microtubule-associated protein. J Cell Biol. 2009;186(3):363–369. doi: 10.1083/jcb.200905048. 10.1083/jcb.200905048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafael JA, Tinsley JM, Potter AC, Deconinck AE, Davies KE. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nat Genet. 1998;19(1):79–82. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- Sonnemann KJ, Heun-Johnson H, Turner AJ, Baltgalvis KA, Lowe DA, Ervasti JM. Functional substitution by TAT-utrophin in dystrophin-deficient mice. PLoS Med. 2009;6(5):e1000083. doi: 10.1371/journal.pmed.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalnaker SH, Hashmi S, Lim JM, Aoki K, Porterfield M, Gutierrez-Sanchez G, Wheeler J, Ervasti JM, Bergmann C, Tiemeyer M, Wells L. Site mapping and characterization of O-glycan structures on alpha-dystroglycan isolated from rabbit skeletal muscle. J Biol Chem. 2010;285(32):24882–24891. doi: 10.1074/jbc.M110.126474. 10.1074/jbc.M110.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K, Nakamura S, Wang H, Iwasaki H, Maebara K, Cheng L, Hirabayashi J, Narimatsu H. Elucidation of binding specificity of Jacalin toward O-glycosylated peptides: quantitative analysis by frontal affinity chromatography. Glycobiology. 2006;16(1):46–53. doi: 10.1093/glycob/cwj038. [DOI] [PubMed] [Google Scholar]
- Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4(12):1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, Davies KE. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 1996;384(6607):349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend D, Blankinship MJ, Allen JM, Gregorevic P, Chamberlain JS, Metzger JM. Systemic administration of micro-dystrophin restores cardiac geometry and prevents dobutamine-induced cardiac pump failure. Mol Ther. 2007;15(6):1086–1092. doi: 10.1038/sj.mt.6300144. [DOI] [PubMed] [Google Scholar]
- Turk R, Sterrenburg E, de Meijer EJ, van Ommen GJ, den Dunnen JT, t Hoen PA. Muscle regeneration in dystrophin-deficient mdx mice studied by gene expression profiling. BMC Genomics. 2005;6:98. doi: 10.1186/1471-2164-6-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SC, Hoffman EP, Slayter HS, Kunkel LM. Immunoelectron microscopic localization of dystrophin in myofibres. Nature. 1988;333(6176):863–866. doi: 10.1038/333863a0. [DOI] [PubMed] [Google Scholar]
- Whitaker JN, Bertorini TE, Mendell JR. Immunocytochemical studies of cathepsin D in human skeletal muscle. Ann Neurol. 1983;13(2):133–142. doi: 10.1002/ana.410130205. [DOI] [PubMed] [Google Scholar]
- Willer T, Amselgruber W, Deutzmann R, Strahl S. Characterization of POMT2, a novel member of the PMT protein O-mannosyltransferase family specifically localized to the acrosome of mammalian spermatids. Glycobiology. 2002;12(11):771–783. doi: 10.1093/glycob/cwf086. [DOI] [PubMed] [Google Scholar]
- Yang B, Ibraghimov-Beskrovnaya O, Moomaw CR, Slaughter CA, Campbell KP. Heterogeneity of the 59-kDa dystrophin-associated protein revealed by cDNA cloning and expression. J Biol Chem. 1994;269(8):6040–6044. [PubMed] [Google Scholar]
- Yasuda S, Townsend D, Michele DE, Favre EG, Day SM, Metzger JM. Dystrophic heart failure blocked by membrane sealant poloxamer. Nature. 2005;436(7053):1025–1029. doi: 10.1038/nature03844. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Kobayashi K, Manya H, Taniguchi K, Kano H, Mizuno M, Inazu T, Mitsuhashi H, Takahashi S, Takeuchi M, Herrmann R, Straub V, Talim B, Voit T, Topaloglu H, Toda T, Endo T. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Developmental cell. 2001;1(5):717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, Oldstone MB, Schachter H, Wells L, Campbell KP. O-mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science. 2010;327(5961):88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.