Abstract
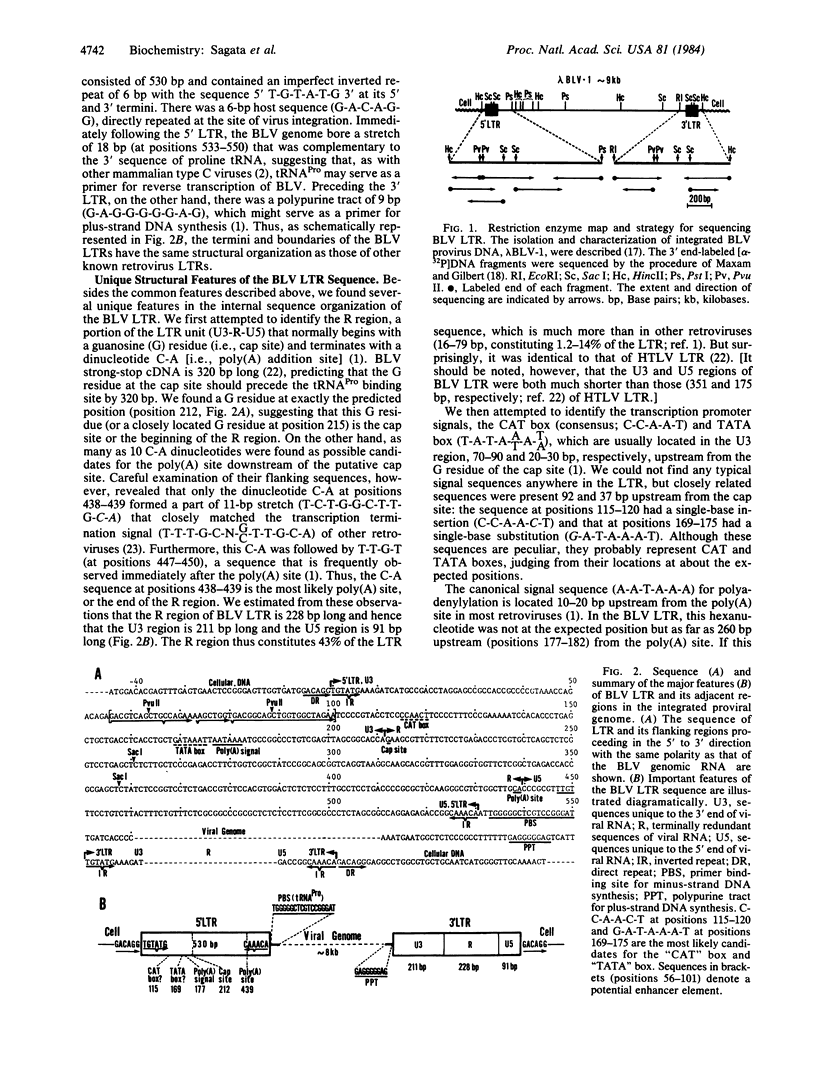
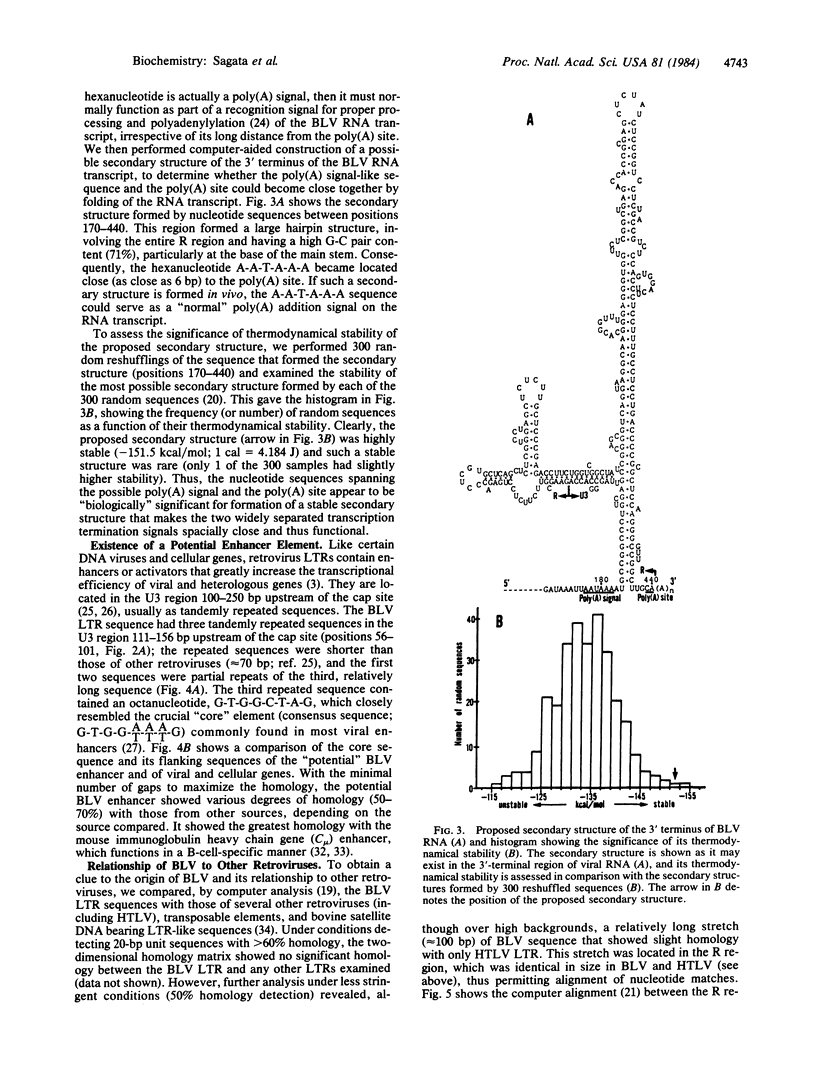
The nucleotide sequence of the long terminal repeat (LTR) of bovine leukemia virus, a unique oncogenic retrovirus of cattle, was determined. The LTR consisted of 530 base pairs (bp) with an inverted repeat of 6 bp at its 5' and 3' ends, flanked by a direct repeat of 6 bp of host cell origin. A tRNAPro binding site for minus-strand DNA synthesis followed the 5' LTR. The U3 region contained putative transcriptional promoters, "CAT" box and "TATA" box, but they had peculiar sequences (C-C-A-A-C-T and G-A-T-A-A-A-T). The U3 region also contained a potential enhancer element, whose sequence partially resembled those of other viral and cellular, especially of immunoglobulin, enhancers. The most striking structural feature of the LTR was an exceptionally long R region (228 bp), which separated a poly(A) addition signal (A-A-T-A-A-A) from a poly(A) site as far apart as 260 bp. The long R region was suggested to form a large stable hairpin structure on a nascent RNA chain, making the two transcription termination signals close together and thus ensuring normal termination of the chain. This structural feature of the bovine leukemia virus LTR was analogous to that of human T-cell leukemia virus LTR and, in fact, slight sequence homology (at most 50%) was observed between the R regions of these two retroviruses, indicating their evolutionary relationship. The unique structural feature of bovine leukemia virus and human T-cell leukemia virus LTRs may thus bear some relation to the biological features commonly shared by these retroviruses.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Bonner T. I., O'Connell C., Cohen M. Cloned endogenous retroviral sequences from human DNA. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4709–4713. doi: 10.1073/pnas.79.15.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couez D., Deschamps J., Kettmann R., Stephens R. M., Gilden R. V., Burny A. Nucleotide sequence analysis of the long terminal repeat of integrated bovine leukemia provirus DNA and of adjacent viral and host sequences. J Virol. 1984 Feb;49(2):615–620. doi: 10.1128/jvi.49.2.615-620.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps J., Kettmann R., Burny A. Experiments with cloned complete tumor-derived bovine leukemia virus information prove that the virus is totally exogenous to its target animal species. J Virol. 1981 Nov;40(2):605–609. doi: 10.1128/jvi.40.2.605-609.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Fujimura F. K., Deininger P. L., Friedmann T., Linney E. Mutation near the polyoma DNA replication origin permits productive infection of F9 embryonal carcinoma cells. Cell. 1981 Mar;23(3):809–814. doi: 10.1016/0092-8674(81)90445-1. [DOI] [PubMed] [Google Scholar]
- Gilden R. V., Long C. W., Hanson M., Toni R., Charman H. P., Oroszlan S., Miller J. M., Van der Maaten M. J. Characteristics of the major internal protein and RNA-dependent DNA polymerase of bovine leukaemia virus. J Gen Virol. 1975 Dec;29(3):305–314. doi: 10.1099/0022-1317-29-3-305. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Hinuma Y., Gotoh Y., Sugamura K., Nagata K., Goto T., Nakai M., Kamada N., Matsumoto T., Kinoshita K. A retrovirus associated with human adult T-cell leukemia: in vitro activation. Gan. 1982 Apr;73(2):341–344. [PubMed] [Google Scholar]
- Hirose S., Takeuchi K., Suzuki Y. In vitro characterization of the fibroin gene promoter by the use of single-base substitution mutants. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7258–7262. doi: 10.1073/pnas.79.23.7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettmann R., Deschamps J., Cleuter Y., Couez D., Burny A., Marbaix G. Leukemogenesis by bovine leukemia virus: proviral DNA integration and lack of RNA expression of viral long terminal repeat and 3' proximate cellular sequences. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2465–2469. doi: 10.1073/pnas.79.8.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettmann R., Deschamps J., Couez D., Claustriaux J. J., Palm R., Burny A. Chromosome integration domain for bovine leukemia provirus in tumors. J Virol. 1983 Jul;47(1):146–150. doi: 10.1128/jvi.47.1.146-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Martin M. A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci U S A. 1983 May;80(9):2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Kugimiya W., Ikenaga H., Saigo K. Close relationship between the long terminal repeats of avian leukosis-sarcoma virus and copia-like movable genetic elements of Drosophila. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3193–3197. doi: 10.1073/pnas.80.11.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson B., Khoury G., Vande Woude G., Gruss P. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature. 1982 Feb 18;295(5850):568–572. doi: 10.1038/295568a0. [DOI] [PubMed] [Google Scholar]
- Lovinger G. G., Mark G., Todaro G. J., Schochetman G. 5'-terminal nucleotide noncoding sequences of retroviruses: relatedness of two old world primate type C viruses and avian spleen necrosis virus. J Virol. 1981 Jul;39(1):238–245. doi: 10.1128/jvi.39.1.238-245.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovinger G. G., Schochetman G. 5' terminal nucleotide sequences of type C retroviruses: features common to noncoding sequences of eucaryotic messenger RNAs. Cell. 1980 Jun;20(2):441–449. doi: 10.1016/0092-8674(80)90630-3. [DOI] [PubMed] [Google Scholar]
- Luciw P. A., Bishop J. M., Varmus H. E., Capecchi M. R. Location and function of retroviral and SV40 sequences that enhance biochemical transformation after microinjection of DNA. Cell. 1983 Jul;33(3):705–716. doi: 10.1016/0092-8674(83)90013-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDonald H. C., Ferrer J. F. Detection, quantitation, and characterization of the major internal virion antigen of the bovine leukemia virus by radioimmunoassay. J Natl Cancer Inst. 1976 Oct;57(4):875–882. doi: 10.1093/jnci/57.4.875. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Onuma M., Sagata N., Okada K., Ogawa Y., Ikawa Y., Oshima K. Integration of bovine leukemia virus DNA in the genomes of bovine lymphosarcoma cells. Microbiol Immunol. 1982;26(9):813–820. doi: 10.1111/j.1348-0421.1982.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Oroszlan S., Sarngadharan M. G., Copeland T. D., Kalyanaraman V. S., Gilden R. V., Gallo R. C. Primary structure analysis of the major internal protein p24 of human type C T-cell leukemia virus. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1291–1294. doi: 10.1073/pnas.79.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagata N., Ogawa Y., Kawamura J., Onuma M., Izawa H., Ikawa Y. Molecular cloning of bovine leukemia virus DNA integrated into the bovine tumor cell genome. Gene. 1983 Dec;26(1):1–10. doi: 10.1016/0378-1119(83)90030-6. [DOI] [PubMed] [Google Scholar]
- Sasavage N. L., Smith M., Gillam S., Woychik R. P., Rottman F. M. Variation in the polyadenylylation site of bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1982 Jan;79(2):223–227. doi: 10.1073/pnas.79.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. The genome of human papovavirus BKV. Cell. 1979 Dec;18(4):963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- Seiki M., Hattori S., Hirayama Y., Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiki M., Hattori S., Yoshida M. Human adult T-cell leukemia virus: molecular cloning of the provirus DNA and the unique terminal structure. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6899–6902. doi: 10.1073/pnas.79.22.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck R. E. A multicopy insertion sequence in the bovine genome with structural homology to the long terminal repeats of retroviruses. Nature. 1982 Aug 19;298(5876):767–769. doi: 10.1038/298767a0. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981 Nov;27(1 Pt 2):1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Tsimanis A., Bichko V., Dreilina D., Meldrais J., Lozha V., Kukaine R., Gren E. The structure of cloned 3'-terminal RNA region of bovine leukemia virus (BLV). Nucleic Acids Res. 1983 Sep 10;11(17):6079–6087. doi: 10.1093/nar/11.17.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndall C., La Mantia G., Thacker C. M., Favaloro J., Kamen R. A region of the polyoma virus genome between the replication origin and late protein coding sequences is required in cis for both early gene expression and viral DNA replication. Nucleic Acids Res. 1981 Dec 11;9(23):6231–6250. doi: 10.1093/nar/9.23.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten M. J., Miller J. M., Boothe A. D. Replicating type-C virus particles in monolayer cell cultures of tissues from cattle with lymphosarcoma. J Natl Cancer Inst. 1974 Feb;52(2):491–497. doi: 10.1093/jnci/52.2.491. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


